Abstract
The muscle-specific RING finger proteins MuRF1 and MuRF2 have been proposed to regulate protein degradation and gene expression in muscle tissues. We have tested the in vivo roles of MuRF1 and MuRF2 for muscle metabolism by using knockout (KO) mouse models. Single MuRF1 and MuRF2 KO mice are healthy and have normal muscles. Double knockout (dKO) mice obtained by the inactivation of all four MuRF1 and MuRF2 alleles developed extreme cardiac and milder skeletal muscle hypertrophy. Muscle hypertrophy in dKO mice was maintained throughout the murine life span and was associated with chronically activated muscle protein synthesis. During ageing (months 4–18), skeletal muscle mass remained stable, whereas body fat content did not increase in dKO mice as compared with wild-type controls. Other catabolic factors such as MAFbox/atrogin1 were expressed at normal levels and did not respond to or prevent muscle hypertrophy in dKO mice. Thus, combined inhibition of MuRF1/MuRF2 could provide a potent strategy to stimulate striated muscles anabolically and to protect muscles from sarcopenia during ageing.
Keywords: muscle hypertrophy and atrophy, striated muscle, ubiquitin ligases MuRF1 and MuRF2, Z-disks
Introduction
Striated muscle cells respond to changing functional requirements by a coordinated set of adaptations. For example, resistance training effectively remodels myofibrils, fiber type compositions, mitochondrial content, and muscle cell sizes (Seynnes et al, 2007), leading to body and skeletal muscle mass increases of 2–5 kg and strength gains of 5–20% within one week in athletes after optimal exercise and nutrition (for review, see Hartgens and Kuipers, 2004). Conversely, acute muscle unloading or starvation induces muscle protein catabolism (about 0.3% of calf muscle mass is lost per day in bed-rested patients (Rittweger et al, 2005), and up to 35% muscle loss in rats after 1 week of space flight (Fitts et al, 2001)). The trophic pathways promoting muscle growth and protein synthesis are modulated by a plethora of factors, including hormones, exercise, and myocellular stretch (for review, see Bassel-Duby and Olson, 2006), that enhance muscle tissue and myocytic cell growth. In vivo, these must be balanced by anti-anabolic signals that are less understood. Pathophysiologically, dominance of catabolism causes muscle wasting syndromes, for example, critical illness myopathies (Latronico et al, 2005), or blunted anabolism causes sarcopenia during ageing (Solomon and Bouloux, 2006). Because of their clinical importance, muscle catabolism-promoting factors are receiving increasing attention (for review, see Bodine, 2006), in particular, the ubiquitin ligases MAFbx/atrogin1 and MuRF1, because of their consistent upregulation during a variety of catabolic muscle states, including denervation, long-term immobilization, and microgravity (Lecker et al, 2004; Nikawa et al, 2004). Consistent with a critical role of MAFbx/atrogin1 and MURF1 for muscle catabolism, MAFbx/atrogin1 and MuRF1 knockout (KO) mice develop muscle atrophy more slowly after denervation (Bodine et al, 2001).
The shared binding of MuRF1 and MuRF2 to a set of seven structural muscle proteins (including titin; see Witt et al, 2005) prompted us to investigate the functional relationship between MuRF1 and MuRF2 by mouse genetics. For example, titin may regulate the E3-ligase activities of MuRF1 and MuRF2 in an activity-dependent fashion: titin is a giant intrasarcomeric protein that makes up a myofibrillar spanning system, confers elastic properties to the myofibrils, and associates with numerous signaling molecules whose expression is muscle-activity dependent (for review, see Miller et al, 2003; Granzier and Labeit, 2004; Lange et al, 2005; Peng et al, 2005). Thus we generated MuRF1 and MuRF2 KO mouse models for testing whether MuRF1 and MuRF2 functionally cooperate. Both MuRF1 and MuRF2 KO mice have normal fertility and life span (consistent with Bodine et al, 2001; Willis et al, 2007). In contrast, MuRF1/MuRF2 double knockout (dKO) mice develop an extreme and lifelong muscle hypertrophy. The molecular basis for the cooperative nature of MuRF1/2 signaling is likely their combined regulation of a large shared set of ligands, including proteins required for myofibrillar stretch sensing, translation, and transcription factors. Cooperative MuRF1/2 signaling is emerging as an important pathway that coordinates myofibrils, ribosomes, and nuclear gene expression in myocytes when being stressed metabolically or biomechanically.
Results
Increase of heart organ and cardiac myocyte size after deletion of MuRF1 and MuRF2
Our MuRF1 and MuRF2 KO mouse models correspond both to constitutive null models (for details on gene targeting, see Supplementary Figure 1B–D). The obtained MuRF1 and MuRF2 KO mice that are homozygous null for MuRF1 or MuRF2 have normal fertility and are viable (consistent with Bodine et al, 2001; Willis et al, 2007). Also, deletion of MuRF1 or MuRF2 has no effect on life span until month 24 (our oldest MuRF1 or MuRF2 KO mice so far). Fertility of single MuRF1 or MuRF2 KO mice allowed the generation of dKO strains by breeding them together. Newborn dKO mice have a severe phenotype: 74% of mice homozygous for both MuRF1 and MuRF2 KO alleles die within the first 7–16 days of life. Dissection of these young dKO mice revealed grossly enlarged hearts that filled the entire mediastinum (Figure 1A and B).
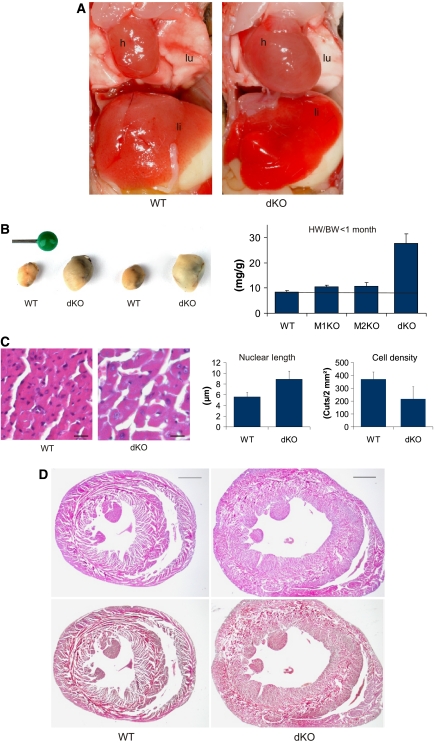
Figure 1.
Synergistic control of heart muscle mass and cardiac myocyte size by MuRF1 and MuRF2. (A) Dissection of 13-day-old MuRF1 and MuRF2-dKO mice revealed grossly enlarged hearts (h), causing caudally lung compression (lu); the liver appears hyperaemic (li). (B) Effect of MuRF1/MuRF2 genotypes on heart weights. Left: hearts isolated from two matched pairs of WT and dKO mice (13 days old). Right: dKO mouse hearts (ventricles only) had 231% (P=0.001) increased HW/BW ratios, whereas weights of MuRF1, MuRF2 KO, and WT hearts did not differ significantly (young mice between d8 and d24; MuRF1: 25.5% increase, P=0.1, MuRF2: 27%, P=0.2, heart ventricles weights, respectively; dKO n=18; MuRF1 KO n=6, MuRF2 KO n=7; WT n=7). (C) Left: hematoxylin/eosin (HE) sections indicated that cardiomyocytes from young dKO hearts were hypertrophic. Scale bar, 20 μm. Right: morphometric comparison of WT and dKO sections nuclei had 59% increased length and cardiomyocytes were also 58% larger/cell density was reduced by 58% (as indicated by the number of cells in 0.2 mm2 large sections). (D) HE (top) and Masson histology (bottom) of WT and dKO hearts indicated concentric-type physiological hypertrophy at month 12, with intact inner and outer circular fiber systems and absence of fibrosis. Scale bar, 1 mm.
Histology of mice that died spontaneously revealed microthrombi in the heart, edema in the lung, microbleedings, and compression of the neighboring organs (Supplementary Figure 2). Taken together, these pathological findings are consistent with death from chronic heart insufficiency and acute cardiac decompensation with heart failure.
Morphometric analysis of single cardiac myocytes demonstrated 59% enlarged myocytes and 58% enlarged nuclei, both indicating cellular hypertrophy and explaining muscle hypertrophy at least in part (see Figure 1C). The effects of deleting MuRF1 and MuRF2 alleles on heart to body weight (HW/BW) ratios were synergistic: young MuRF1 or MuRF2 KO mice had 10 and 8% HW/BW increases (P=0.1 and 0.2 respectively), whereas hearts of dKO mice had 231% increased HW/BW ratios (P=0.001, <1 month; Figure 1B right). The histological studies of older mice revealed for dKO myocardium a concentric-type hypertrophy. No fibrosis by Masson stainings or myofibrillar disarray was observed at light microscopic level (Figure 1D bottom). The inner and outer layers of the myocardium could still be distinguished (Figure 1D).
Taken together, the severe phenotype of dKO mice contrasts the absence of a noticeable phenotype in MuRF1 or MuRF2 KO mice and therefore demonstrates cooperativity of MuRF1 and MuRF2 on the genetic level.
dKO mice develop and maintain skeletal and cardiac muscle hypertrophy throughout their life span
Those dKO mice that survived the first two postnatal weeks became long-term survivors: All dKO mice alive at week 3 (n=27; 26% of total dKO offspring) were still alive at month 18 (unless being killed for our studies) and were able to have offspring. Adult dKO mice maintain cardiac hypertrophy (dKO: 84% increase, P=0.001; see Figure 2B). Consistent with this, electrocardiography indicated intact excitation conduction in dKO hearts from aged mice (see Supplementary Figure 3; contrasting, for example, the conduction blocks observed in Nkx2.5 KO mice; see Pashmforoush et al, 2004). Next, we examined the physiological cardiac performance by MRI imaging in more detail. This indicated massive persistent hypertrophy (see Figure 2A). Next, we estimated ejection fractions (EF) and stroke volumes by time-resolved MRI. This showed grossly reduced EFs (dKO: 0.2–0.3; wild type (WT): 0.7; see Figure 2A), whereas stroke volumes were 26% reduced under basal conditions.
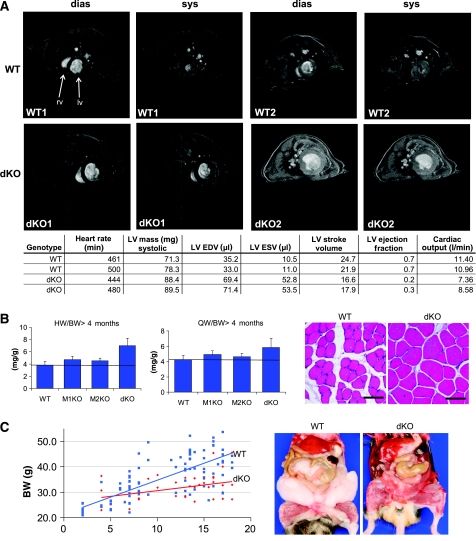
Figure 2.
Phenotype of dKO mice at 18 months of age. Perinatally, dKO mice have a high mortality of 74%. The 26% of dKO mice (n=27) that survived became long-term survivors and were all alive at month 18 (unless killed, see below), thus allowing phenotypic studies in aged dKO. (A) MRI scans detected hypertrophic hearts in adult dKO mice with reduced EFs and stroke volumes (for time-resolved MRI scans, see also Supplementary Videos 4, 5, 6 and 7). LV, left ventricle; RV, right ventricle; EDV, end-diastolic volume; ESV, end-systolic volume (total number of mice scanned: two mice of each genotype. (B) Left: effect of MuRF1/2 genotypes on heart (ventricles, left) and quadriceps skeletal muscle (right) to body weight ratios. dKO mice maintain cardiac hypertrophy during ageing (dKO, 84% increase, P=0.001; MuRF1, 24.5% increase; MuRF2, 19%). In addition, dKO mice have 38.1% increased QW/BW ratios (P=0.001), whereas MuRF1 KO and MuRF2 KO genotypes have moderate effects on skeletal muscle mass (MuRF1, 16% increase, P=0.05; MuRF2, 11% increase, P=0.08). Despite increasing body mass in WT during months 4–18, skeletal muscles remain more hypertrophic in dKO mice during ageing (4–18 months of age, dKO n=27; MuRF1 KO n=52; MuRF2 KO n=81; WT n=49). For absolute weights, please refer to Supplementary Figure 8. Right: hematoxylin/eosin sections indicated that skeletal myofibers from dKO muscle show hypertrophic fibers with slightly augmented cross-section areas alternating with normal-appearing fibers (mice aged 4 months). Scale bar, 100 μm. (C) Weight gain of WT and dKO mice during ageing. Left: between months 4–18, dKO mice gain less weight (red; P<0.001) than WT. Right: dissections revealed that aged dKO mice were leaner (mice aged 18 months).
Because dKO hearts support physiological circulation at least under non-challenged laboratory conditions for up to month 18, the effects of the absence of both MuRF1 and MuRF2 could also be studied in muscles of adult and aged animals. Similar to sustaining cardiac hypertrophy up to month 18 (our oldest dKO mice), MuRF1 and MuRF2 had also synergistic lifelong effects on skeletal muscle mass: after deletion of all four alleles, we found 38% increased quadriceps to body weight (QW/BW) ratios in dKO mice when compared with WT (P=0.001), whereas single MuRF1 or MuRF2 KOs had 17 and 11% increased QW/BW ratios (P=0.05 and 0.08, respectively; Figure 2B left). Similar as in heart muscle, skeletal muscle hypertrophy correlated with the hypertrophy of individual fibers (see histology of quadriceps muscle; Figure 2B right).
Intriguingly, during ageing, body weights of dKO and WT mice progressively diverged: whereas ageing WT mice substantially gained weight (64% increase when comparing months 5 and 18), weight gains were attenuated in dKO mice (17, 27, and 35% differences at 12, 15, and 18 months, respectively (P=0.001), Figure 2C left). Dissections of senescent WT and dKO mice indicated that a striking lack of body fat accumulation in senescent dKO mice accounted for their reduced weights (Figure 2C right). This was apparently not linked to a general cachexia, because skeletal muscle hypertrophy was maintained (Supplementary Figure 8 right).
Identification of binding partners shared by MuRF1 and MuRF2 and their convergent signaling on CARP, FHL2, and SQSTM1
Because muscle hypertrophy and the lean phenotype develop only after inactivation of all four MuRF1 and MuRF2 alleles, we searched for binding partners that are recognized by both MuRF1 and MuRF2 in an attempt to find molecular explanations for the phenotypic synergistic effects of MuRF1 and MuRF2 on muscle protein and lipid/energy metabolism.
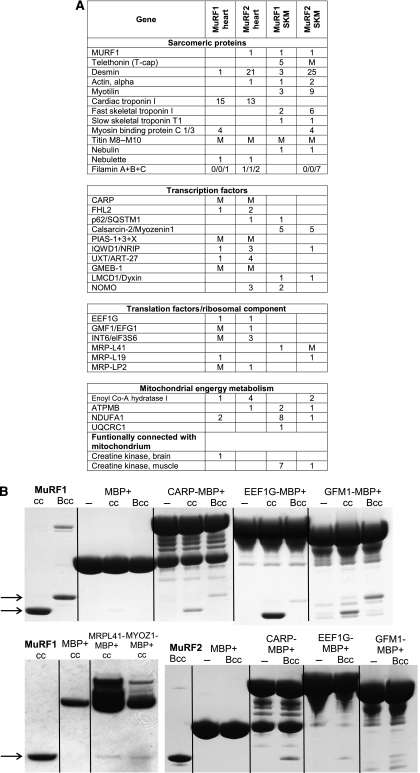
A total of 87 genes were identified as MuRF1 or MuRF2 interacting prey clones that coded for myofibrillar proteins (18, including 11 Z-disk proteins), transcriptional regulators (11), translation factors (4), and component of the mitochondrial proteome (including ATP-synthesis (9)). Of these 87 genes, a set of 35 genes was fished with both MuRF1 and MuRF2 baits and was further confirmed by mating studies. The group of ligands shared by MuRF1 and MuRF2 included a set of four myofibrillar Z-disk proteins and the transcriptional regulators CARP, myozenin1/calsarcin2, FHL2 (also associated with Z-disk region; for review, see Clark et al, 2002) (Figure 3A). To further test whether the Yeast Two-Hybrid (YTH) prey clones code indeed for MuRF1 and MuRF2 binding proteins, we performed in vitro pull-down studies using expressed CARP, myozenin1/calsarcin2 (two molecules selected as known transcriptional regulators of muscle gene expression), MRP-L41/pig3 (selected as a member of the mitochondrial ribosomal group, also being implicated in growth control; see Yoo et al, 2005), and EEF1G/EF-1γ (selected as a sophisticatedly regulated component of the translation machinery; see Belle et al, 1995) as well as its mitochondrial counterpart GFM1. Results indicated that a central MuRF1 fragment that comprises the MuRF1 residues 109–315 (‘MuRF1Bcc'; see Supplementary Figure 1A) is both sufficient and required for interaction with CARP, EEF1G, GFM1, myozenin1/calsarcin-2, and pig3/MRP-L41 (Figure 3B). Similarly, expressed MuRF2Bcc interacted in vitro with CARP, EEF1G, and GFM1 (Figure 3B). Finally, YTH mating suggested that MuRF3Bcc does not interact with CARP, myozenin-1/calsarcin-2, and pig3/MRP-L41 (data not shown).
Figure 3.
MuRF1 and MuRF2 interact with a shared set of myocellular proteins. (A) YTH screens with full-length MuRF1 and MuRF2 baits of both human cardiac (‘heart') and skeletal cDNA libraries (‘SKM') fished a total of 87 genes. The table summarizes those 35 prey clones identified independently in both MuRF1 and MuRF2 screens and thus predicted to interact with both MuRF1+2: 13 prey clone inserts code for sarcomeric proteins (4 of which are components of the Z-disk), 10 code for transcriptional regulators (2 of which are also associated with the Z-disk), 5 genes are involved in mitochondrial ATP production, and 6 genes participate in translation initiation and elongation. Numbers indicate independently identified prey clones in respective screens. M=interaction was found by mating. An SRF prey clone fished with the MuRF1 bait could not be confirmed by mating, as in our hands the 3′ UTR and not the coding sequence of SRF activated yeast growth during mating with MuRF1 and 2. (B) The interaction of selected proteins derived from the above-mentioned genes was studied in vitro by pull-downs using expressed MuRF1/MuRF2 Bcc (B-Box+coiled-coil domain) and MuRF1cc (coiled-coil domain) constructs (see also Supplementary Figure S1 and methods). MuRF1cc and MuRF1Bcc (arrows) co-eluted together with CARP, EEF1G, GFM1 MBP fusion proteins. Below: left—MuRF1cc co-eluted with myozenin-1/calsarcin-2, and MRP-L41/Pig3 MBP-fusion proteins; right—MuRF2Bcc co-eluted together with CARP, EEFG1, GFM1 MBP fusion proteins; controls—MBP plus MuRF1cc, Bcc, MuRF2Bcc, respectively, or fusion proteins only.
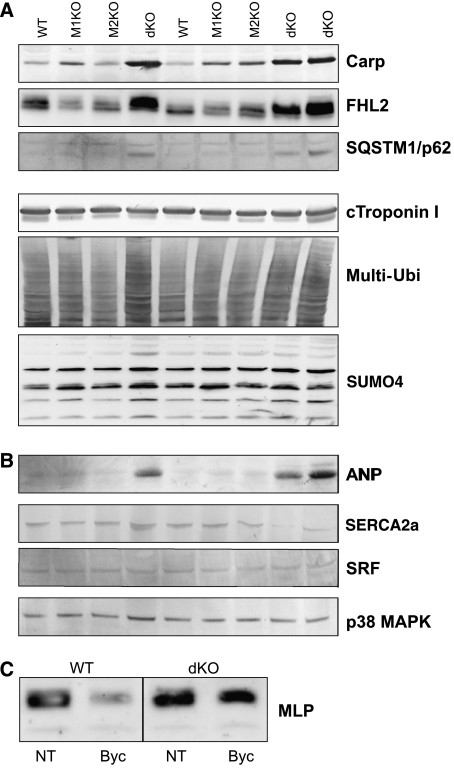
MuRF3 was recently shown to interact also with FHL2 and suggested to regulate its expression as an E3-ubiquitin ligase (Fielitz et al, 2007). Therefore, we tested next whether the expression of MuRF3 and FHL2 are affected in dKO mice. MuRF3 was expressed at normal levels in dKO mice (Array Express E-MEXP-1321), whereas the FHL2 protein was highly upregulated in dKO mice deficient for both MuRF1 and MuRF2 (Figure 5A). Thus MuRF1/2 signaling on FHL2 is cooperative and cannot be substituted by the related ubiquitin ligase MuRF3. Intriguingly, other catabolic factors, such as atrogin1, are expressed at normal levels in dKO myocardium (see Supplementary Table 9), suggesting that MuRF1/MuRF2 and atrogins are functioning in different pathways. In contrast, CARP and SQSTM1 (Sequestosome1/p62) became strongly upregulated only after inactivation of all four MuRF1 and MuRF2 alleles (Figure 5A). Gene expression profiling with Affymetrix system indicated that FHL2 and SQSTM1 mRNA levels are normal in dKO myocardium, and CARP is moderately upregulated (Supplementary Table 9). Therefore, upregulation of CARP, FHL2, and SQSTM1 in dKO hearts are primarily caused by post-transcriptional mechanisms.
Impaired mitochondrial ultrastructure and alteration of Z-disks after deletion of MuRF1 and MuRF2
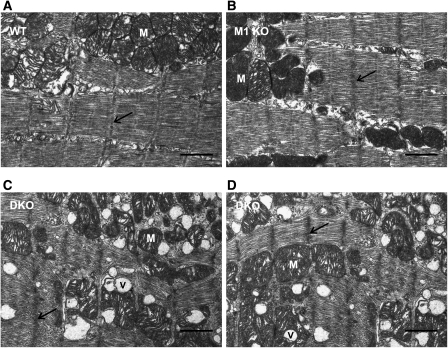
Because MuRF1 and MuRF2 interact with multiple components of the Z-disk and of the mitochondrium (Figure 3A), we studied the ultrastructural effects of the absence of MuRF1 and MuRF2 on Z-disks and mitochondria in myocardium by electron microscopy. We were unable to detect differences between WT, MuRF1, and MuRF2-KO myocardium (Figure 4A and B). In contrast, myofibrils in dKO myocardium were abnormal: myofibrils had more electron-dense Z-disks and were less regular (Figure 4C and D). Occasionally, myofibrils assembled in dKO hearts had free ends, somewhat reminiscent of growth tips found in proliferating skeletal myotubes (Ojima et al, 1999), and projected into regions rich in unassembled free filaments (not shown).
Figure 4.
Altered Z-disks and mitochondrial ultrastructure in dKO myocardium. (A, B) At 18 months, we noted no ultrastructural abnormalities in myocardium from WT (A), MuRF1-KO (B), and MuRF2 KO (not shown) mice. (C, D) In dKO myocardium, sarcomere lengths and myofiber alignments are less regular (maximal variation is 2.5-fold larger than in WT sarcomeres). Z-disks have a denser appearance (arrows). Vacuoles (V) are frequently found between or embedded within mitochondria (M). Mitochondria are less regular in shape including abnormally small mitochondria and are, unlike in WT, not tightly packed together. Scale bar, 1 μm.
Mitochondria were less regular in shape and less orderly packed together. dKO myocardium also contained vacuoles, often embedded into mitochondrial clusters (Figure 4C and D). Because of the mitochondrial defects, we tested the expression of PGC-1-α (as a master gene for mitochondrial biogenesis; see Rasbach and Schnellmann, 2007). PGC-1-α transcription was not dysregulated in dKO myocardium (see Supplementary Table S9). Future studies are required to determine the molecular basis of altered mitochondrial numbers and structures in dKO myocardium.
These ultrastructural changes were only present after inactivation of all four MuRF1 and MuRF2 alleles, again demonstrating that the MuRF1 and MuRF2 loci are genetically a complementation group.
Elevated ANP and MLP stretch signals in dKO muscles implicate MuRF1/MuRF2 in stretch signal inhibition
To gain insights into the mechanisms causing cardiac hypertrophy in dKO mice, we analyzed their transcriptomes by gene expression profiling. The transcriptional changes we found in dKO ventricles resembled those present in pressure induced aortic constriction (Zhao et al, 2004), including the upregulation of skeletal-type alpha actin 1, myosin light chains, atrial natriuretic peptide (ANP; isoforms A and B), and thrombospondin (see Supplementary Figure 9 and ArrayExpress accession E-MEXP-1321). Consistent with elevated stretch signaling, ANP was strikingly upregulated in dKO ventricles (Figure 5B). Normal to moderately elevated expression of other markers for cardiac hypertrophy suggested that the ANP induction was not a secondary consequence of heart failure and calcium overload (Figure 5A). SERCA2a (a marker for calcium overload during heart failure), serum response factor (SRF, previously suggested to transmit titin kinase/MuRF2-dependent stretch signals; Lange et al, 2005), and p38 MAPK (activated by the ERK/Map kinase pathway) were not affected by the absence of MuRF1 or MuRF2. In dKO myocardium, neither microarrays nor western blots showed an upregulation of SRF (Figure 5B and Supplementary Table 9).
Figure 5.
Characterization of altered signaling pathways in dKO myocardium by western blot studies. (A) Upregulation of the MuRF1/MuRF2 binding proteins CARP, FHL2, and SQSTM1 in dKO myocardium. Striking upregulation required the deletion of all four MuRF1/2 alleles, suggesting that both MuRF1 and MuRF2 synergistically control the transcriptional regulators CARP, FHL2, and SQSTM1. Below, cTnI and total multi-ubiquitinated protein species were not affected by the inactivation of MuRF1 and MuRF2 alleles. Among SUMO family members, we noticed for SUMO4 differential reactivity in the 8–30 kDa region. (B, C) Hyperactive stretch signaling in dKO as suggested by chronic upregulation of stretch-dependent signaling markers. (B) In myocardium, ANP is barely detectable in WT, MuRF1-KO, and MuRF2-KO hearts. ANP is strikingly upregulated in dKO myocardium (ventricles, 12 months old). Other markers for cardiomyopathy/hypertrophy remain normal or are moderately upregulated: SERCA2a (used as a marker for heart failure/calcium overload), SRF (previously implicated in stretch-dependent MuRF2 signaling), p38 Map kinase (a marker for ERK signaling and heart failure). (C) In dKO quadriceps muscles, hyperactive stretch signaling was suggested by the effect of 72 h immobilization on the stretch marker MLP: abnormally high levels of MLP/Crsp3 are maintained after 72 h immobilization in dKO quadriceps (NT=no treatment, byc=bycast immobilization).
Based on the elevated ANP levels in dKO ventricles, we hypothesized that stretch signaling is augmented in dKO muscles because of the failure to attenuate it. To test this hypothesis in skeletal muscle, we immobilized dKO skeletal muscles by a bycast. MLP/Csrp3 is required for stretch-regulated responses in myocardium and a binding partner of the Z-disk-associated protein TCap (Knoell et al, 2002) and is also expressed in skeletal muscle (in contrast to ANP). Therefore, we monitored MLP/Csrp3 as a marker protein for stretch signaling. MLP/Csrp3, already elevated at basal conditions, remained highly elevated after a 72 h bycast immobilization in dKO (see Figure 5C). Taken together, our data demonstrate that combined inactivation of MuRF1 and MuRF2 leads to chronic upregulation of stretch signals in both heart and skeletal muscle and failure to downregulate them.
Synergistic control of translational regulatory components by MuRF1/2
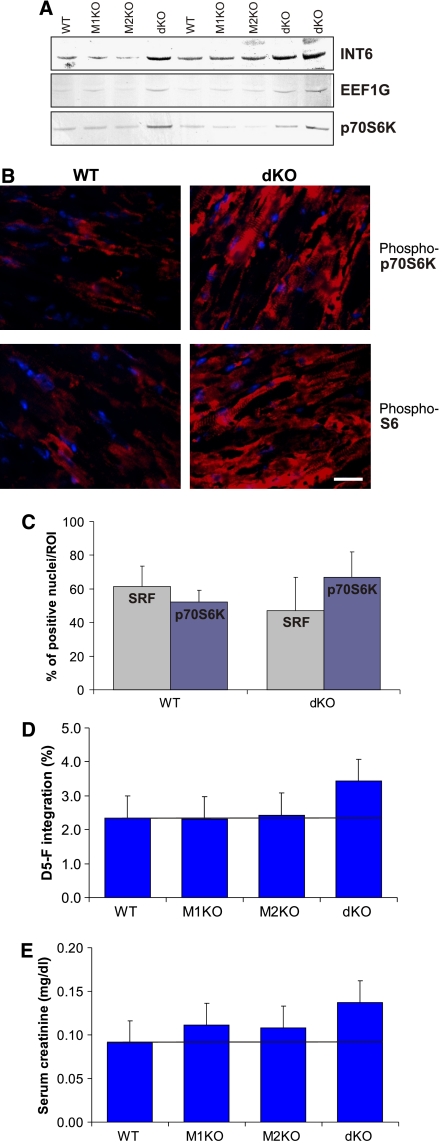
Possibly, the massive hypertrophic phenotype of dKO mice might be caused by reduced multi-ubiquitination and degradation of total muscle proteins (linked to inactivation of MuRF1/2 E3-ubiquitin-ligase activities). To test for reduced ubiquitination/degradation, we determined the levels of multi-ubiquitinated proteins on western blot panels displaying the different MuRF1/2 genotypes. Total levels of multi-ubiquitinated proteins did not change upon MuRF1/2 inactivation (Figure 5A). Among the family of ubiquitin-related modifiers, we only noted for SUMO4 an upregulation of SUMO4ylated species after MuRF1/2 inactivation (Figure 5A). Next, we speculated that, as an alternative mechanism, MuRF1 and MuRF2 might regulate the translational machinery directly (e.g., via interaction with INT6, EEF1G, GFM1; see Figure 3). When testing the myocardial expression of MuRF1/2-associated translation factors, we found that EEF1G and INT6 (subunits of EF-1 and elF3a, respectively; see Belle et al, 1995; Morris et al, 2007) were upregulated specifically after deletion of all four MuRF1/2 alleles (Figure 6A). Consistent with a general translational activation, we found upregulation of p70S6K (Figures 6A) and its activated phosphorylated form phospho-p70S6K, as well as its substrate phospho-S6 (markers for an activated Akt/mTor pathway; see Figure 6B). Phospho-p70S6K was recruited to the nucleus (Figure 6B), thus mimicking changes observed in exercise-induced translational activation (Koopman et al, 2006). For SRF, we noted no nuclear recruitment after MuRF1/2 deletions (Figure 6C).
Figure 6.
Elevated muscle protein synthesis after inactivation of MuRF1 and MuRF2. (A) The translation elongation factor EEF1G (interacting with MuRF1 and MuRF2; see Figure 3), its binding protein elF3a subunit INT6, and the translation-promoting p70S6 kinase are upregulated in dKO myocardium. (B) Immunohistochemistry with phospho-specific antibodies demonstrated the upregulation of the activated forms of p70S6K and its substrate S6 (phospho-p70S6K and phospho-S6, respectively) in dKO myocardium, suggesting activation of the AKT/mTOR pathway. Scale bar, 20 μm. (C) Nuclear entry of phospho-p70S6K was stimulated in dKO myocardium, whereas cellular distribution of SRF did not change significantly (data represent counts of 200 nuclei on 4HPF sections). (D) Fractional synthesis rate (% per 48 h) of total cardiac muscle proteins was determined by injecting D5-F i.p. into mice of each genotype (n=6). Total cardiac muscle protein synthesis is elevated in dKO myocardium (P=0.05 in dKO versus WT, mice aged between 6 and 16 months). (E) Serum level of creatinine was increased in dKO mice (n=6 in each group, P=0.01 in dKO versus WT mice, mice aged between 6 and 16 months).
To test more directly for translational activation, we injected deuterium-labeled phenylalanine (D5-F) into dKO and control mice, allowing the determination of fractional de novo muscle protein synthesis by comparing D5-F to phenylalanine contents in muscle protein lysates (see, e.g., Dardevet et al, 2002). Incorporation of D5-F into dKO myocardium after 2 days was 46.6% higher than in WT myocardium (Figure 6D), whereas incorporation of D5-F into WT, MuRF1-KO, and MuRF2-KO mice was statistically not different. Consistent with a chronically elevated rate of muscle metabolism, we found that serum creatinine levels (a degradation product of the muscle creatinine) were elevated by 55% in dKO compared to WT mice (Figure 6E).
Taken together, these data point at the synergistic activation of the translational machinery in the absence of both MuRF1 and MuRF2, causing marked upregulation of phospho-S6K (or S6), whose level is tightly correlated with total muscle protein mass in rodents (Baar et al, 2000).
Discussion
The fulminant cardiac phenotype of dKO mice documents the synergistic cooperation of the two homologous RING finger proteins MuRF1 and MuRF2: inactivation of the four MuRF1/2 alleles leads to 74% early postnatal lethality. Autopsy and histology point at acute heart failure as the most likely cause of early postnatal death (see Supplementary Figure 2). After birth, dKO mice develop three- to fourfold enlarged hearts, whereas mice possessing a single intact MuRF1 or MuRF2 allele are healthy (reminiscent of functional complementation, e.g., MyoD and Myf5, where only removal of both proteins leads to a severe phenotype (Rudnicki et al, 1993). Thus, this is to our knowledge the first report showing the convergent signaling of two ubiquitin ligases (i.e., MuRF1 and MuRF2) in control of the trophic muscle state, molecules that have previously been implicated in skeletal muscle wasting and atrophy (for MuRF1, see Bodine et al, 2001; Lecker et al, 2004; Nikawa et al, 2004) or in stretch-dependent control of muscle gene expression (for MuRF2, see Lange et al, 2005). Intriguingly, those dKO animals surviving the first 2 weeks have a normal life expectancy and are fertile, possibly because they ‘outgrow' the initial massive enlargement of their hearts after birth: chest cavities expand after birth and relative HW/BW ratios drop from about 231% at 2 weeks of age to 84% at month 18. Longevity of 26% of the dKO mice also allowed us to study the functional consequences of MuRF1/2 absence in ageing skeletal muscle: significant quadriceps skeletal muscle hypertrophy only developed after inactivation of both MuRF1 and MuRF2, and muscle hypertrophy was linked to fiber hypertrophy (Figure 2B).
A likely molecular explanation for MuRF1/2 cooperativity is their recognition of 35 or more shared targets: our YTH screens implicated both MuRF1 and MuRF2 in the recognition of specific components of the Z-disk, transcriptional regulators, the translational machinery, and the mitochondrial metabolism. Thus, MuRF1/2 recognize a broader range of targets than detected previously. Based upon a recent study on the interaction of cMuRF1 with titin A168–170 (Mrosek et al, 2007), we speculate that the coiled-coil domains of MuRF1/2 (MuRFcc) have an extended flexible shape enabling multiple protein/protein interactions, thereby allowing the MuRF1/2cc domains to function as a recognition domain for numerous physiological targets. Thereby, MuRF1/MuRF2 could possibly orchestrate Z-disk signaling, transcription, translation, and mitochondrial and cytoplasmic metabolism in a coordinated fashion.
Analysis of dKO muscle tissues provided insights into the molecular mechanisms causing MuRF-dependent muscle hypertrophy that previous studies might have missed, because MuRF2 might have complemented for MuRF1 and vice versa in single KO models (Bodine et al, 2001; Willis et al, 2007): Our studies identified two causes that are likely to promote muscle hypertrophy in dKO mice. First, translation is enhanced in dKO myocardium as demonstrated by the upregulation of the downstream control check point S6 and the increased de novo muscle protein synthesis. As a potential mechanism leading to downstream translational activation, our YTH studies identified the interaction of MuRF1/2 with multiple components of the translational machinery, including INT6 (recently identified as a regulator of translation initiation factor 3; see Morris et al, 2007), EEF1G (a component of the elongation factor complex EF-1 that is sophisticatedly regulated during development and the cell cycle; see Belle et al, 1995), and GFM1 (the mitochondrial elongation factor G1).
These interactions together with the translational activation in dKO myocardium suggest that enhanced muscle synthesis accounts for muscle hypertrophy, whereas reduced protein degradation by multi-ubiquitination is at least less important. Consistent with enhanced muscle protein turnover in addition to a globally reduced muscle protein catabolism, we found elevated creatinine levels in the sera of dKO mice (Figure 6E).
As a second mechanism explaining muscle hypertrophy in dKO mice, we found that both cardiac and skeletal dKO muscles expressed elevated levels of stretch markers, including ANP and MLP (which again are predicted to promote muscle protein synthesis). Both ANP and MLP are also components of the myofibrillar Z-disk. Interestingly, ANP, CARP (both also highly upregulated in young dKO mice; see Supplementary Figure 10), and MLP all target to the myofibrillar Z-disk region (Bang et al, 2001; Knoell et al, 2002; Hoshijima, 2006). Consistent with a higher content of Z-disk proteins in dKO muscles, our EM studies showed denser and widened Z-disks (Figure 4), a lattice structure that performs stretch-sensing functions in muscle tissues (Vigoreaux, 1994; Knoell et al, 2002). Finally, failure to downregulate MLP after immobilization suggests that dKO muscles chronically express high levels of MLP independently of muscle exercise (Figure 5C).
Hyperactive anabolic stretch-sensing pathways could also potentially explain the unexpected lean phenotype of dKO mice: our adult C57/BL6 laboratory mice with unlimited access to nutrition and limited physical activity gained about 4% weight per month by fattening. In contrast, our dKO mice (back-crossed onto the same genetic background) failed to accumulate fat (Figure 2C). This could be explained, at least in part, by the activation of muscle protein synthesis and metabolism, recruiting a larger fraction of dietary calories for protein anabolism. Alternatively, MuRF1 and MuRF2 may also exert direct effects on lipid metabolism, as enoyl-coenzyme hydratase I (interacting with both MuRF1 and MuRF2; see Figure 3A; for review, see Kim and Battaile, 2002) participates in the β-oxidation of lipids.
Overall, the phenotype of dKO mice together with previous data (Arya et al, 2004; Kedar et al, 2004; Willis et al, 2007) are consistent with a role of MuRF1/2 to serve as an inhibitory feedback loop to downregulate anabolic responses after stretch/strain and metabolic stress: inactivation of MuRF1 and MuRF2 results in chronically elevated stretch signals, both in the heart (for ANP, see Figure 5B and Supplementary Table 10; array data on ArrayExpress accession E-MEXP-1321) and in skeletal muscle (for MLP, see Figure 5C). Future molecular insights into the signaling pathways shared by MuRF1 and MuRF2 may allow the design of combined MuRF1/MuRF2 inhibitors to achieve the effects seen in dKO mice including elevated muscle protein synthesis, failure to accumulate body fat during ageing, and maintenance of elevated trophic stretch signals. Although such a strategy will require considering cardiac hypertrophy as an additional unwanted effect the survival of dKO mice until month 18 encourages strategies to target MuRF1/MuRF2 for stimulating muscle anabolically. Finally, the striking lipolysis in dKO mice warrants future studies on the potential role of MuRF1 and MuRF2 in lipid and energy metabolism.
Materials and methods
Inactivation of the murine MuRF1 and MuRF2 genes and mouse breeding
The murine MuRF1 and MuRF2 genes were targeted by homologous recombination into exons 2 and 5, respectively (Witt et al, 2001). Two independent ES cell clones for each targeting were obtained, homologous integration verified, and mouse generated essentially as previously described (Witt et al, 2001). Homologous recombination caused the complete loss of MuRF1 and MuRF2 transcripts, respectively, as detected by RT-PCR (see Supplementary Figure 1D). 129/C57/BL6 hybrid mice were back-crossed with C57/BL6 over generations to obtain a comparable genetic background. During breeding of 129/C57/BL6 mice, MuRF1 and MuRF2 null alleles segregated Mendelian. Because of the high lethality, dKO mice were obtained exclusively by breeding double heterozygous mice to avoid any selection of potential modifiers.
YTH interaction studies
For YTH screens and mating experiments, the human full-length MuRF1 and MuRF2 fragments were amplified from total human skeletal muscle cDNA with the following primer pairs (small letters denote cloning sites): MuRF1-S, tttccatg-GAGAACTTGGAGAAGCAGCTGATCTGC, 478S; MuRF1-R, ttttagatct-TCTGGGGGCCTCTCATTCATCCAGCTC, 1525R; MuRF2-S, tttagatct-GATAACTTAGAGAAGCAACTCATCTGTCCC, 184S; MuRF2-R, tttgtcgac-AGAGGGGCAGCAGTTGGAATGAATATC,1808R.
Sequencing confirmed identity to the data library entries and AJ291712 and AJ291713. For MuRF2, the isoform amplified from total human heart corresponded to the 50 kDa version (see McElhinny et al, 2004). The full-length MuRF1,2 fragments were inserted into pGBKT7 (Clontech) and the recombinant MuRF1,2 baits were transformed into Saccharomyces cerevisiae, strain AH109. For screening, AH109 cells were co-transformed with 40 μg of amplified human skeletal muscle cDNA and cardiac cDNA libraries, prepared in pACT2 prey vector (Matchmaker skeletal cDNA library no. 638818 and cardiac library no. 2020484, Clontech). The YTH screens were essentially performed as described by the manufacturer (Yeastmaker Yeast Transformation System 2, Clontech). Co-transformed cells were incubated for 5 days at 30°C on SD/Leu-/Trp-/His-plates. Subsequent determination of β-galactosidase activities, further mapping studies by mating of deletion constructs, rescue of plasmid DNA from interacting prey clones, and their sequence analysis with AD3 primer were as described previously (Witt et al, 2005).
Cine MRI
Cine MRI was performed on a high field 7.05 Tesla Biospec magnet (Bruker, Ettlingen, Germany) using an ECG and respiratory triggered fast gradient echo FLASH (Haase et al, 1986) sequence (for imaging parameters, see Ruff et al, 1998; Wiesmann et al, 2000; flip angle 25–40°; echo time 1.7 ms; repetition time 7–16 ms (depending on the heart rate and number of frames per heart cycle); field of view 30 mm2; image matrix 128 × 128; in-plane resolution 230 μm and slice thickness 1.0 mm. A total of 12–18 frames per heart cycle and 10–12 contiguous ventricular short-axis slices were acquired to cover the entire heart. A birdcage probehead with an inner diameter of 35 mm for transmission and reception of the MR signal and a microimaging system with rapid gradient performance (maximum gradient strength 870 mT m−1 and 280 μs rise time) were used. The size of myocardial and ventricular compartments was determined by semi-automated segmentation in the short-axis slice images covering the entire heart. Endocardial borders in all end-diastolic and end-systolic frames were delineated, clearly distinguishable due to the high MRI-derived non-saturated spin contrast between the blood-filled ventricular cavity and the myocardial tissue. The obtained compartment areas were multiplied with the slice thickness to determine the myocardial and ventricular slice volumes. Ventricular volumes were calculated as follows: stroke volume=SV, end-diastolic volume=EDV, end-systolic volume=ESV; EF=SV/EDV and cardiac output CO=SV × heart rate. Myocardial masses were calculated from the multiplication of the total volumes with the specific gravity of the myocardium (1.05 g cm−3). Volumes and masses were calculated with similar reproducibility and accuracy as known from human MR studies and to autopsy data (Ruff et al, 1998).
Protein expression, antibodies, western blot analysis and pull-downs
Full-length CARP, MRP-L41/pig3, and an N-terminal fragment of myozenin-1/calsarcin-2 was cloned into pETM-44 region, and maltose-binding protein (MBP) fusions were expressed in BL21-DE3 as described below. For cloning, the following primer pairs were used to amplify the desired fragments from total cardiac muscle cDNA: CARP-253S, tttagatct-ATGGTACTGAAAGTAGAGGAACTGGTCACT; CARP-1210R, tttgtcgac-TCAGAATGTAGCTATGCGAGAGGTCTTGTA; myozenin-1/calsarcin2 378S, tttctcgagc-GGAACCCCGGCCCCTAATAAGAAG; myozenin-1/calsarcin2 830R, tttacgcgtta-TCCTCCTCTGCCAGCCTGGCC; MRP-L41/pig3-106S, tttcc-ATGGGCGTCCTGGCCGCAGC; MRP-L41/pig3-519R, tttggatc-CTAGCGCAGGAAGTTCCTGGGGTAGAG; GFM1-1491S, tttccatg-GATCTGGAAAAATTTTCAAAAGGTATTGGC; GFM1-2384R, tttggatcc-GTCAACTCACAGTAAGCAAAGTTAGTTCTTGGCT; EEF1G-140S, ATCACCATGGCGGCTGGGACCCTGTACACG; EEF1G-1460R, tttggtacc-TCACTTGAAGATCTTGCCCTGATTGAAGGC.
For protein expression in Escherichia coli, constructs were transformed into BL21-DE3, grown in LB supplemented with kanamycin (20 μg/ml) to an OD of 0.6, and T7-driven expression of insert proteins was induced by the addition of IPTG to a final concentration of 200 μM. Harvesting of cells, their lysis, and purification of His-tagged protein complexes on Ni-NTA agarose was essentially as described by manufacturers for induction of protein expression (see Novagen, www.emdbiosciences.com/html/NVG/literature; for His-tag fusion protein purification, see Qiagen). After Ni-NTA purification, specifically bound proteins were eluted with 300 mM imidazole. Pull-down experiments were performed essentially as described by Mrosek et al (2007). In brief, MuRF1 fragments (see Supplementary Figure 1) or MuRF1/MuRF2 ligands were cloned into pETM-44 and expressed as MBP fusions. Complexes of MuRF1/MuRF2 with its ligands were purified on amylose agarose (New England Biolabs) essentially as described by the manufacturer.
Antibodies: CARP specific antibodies have been described previously (Miller et al, 2003; Witt et al, 2004). Other antibodies used: 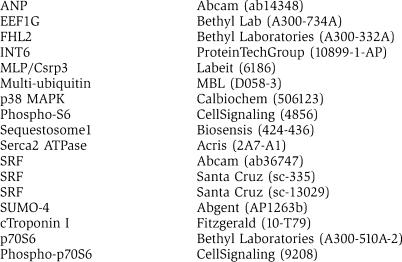
For western blot analysis, blots of mouse ventricle tissue lysates were probed with affinity purified primary antibodies. Muscle tissue lysates were prepared from snap-frozen mouse ventricles. Briefly, pulverized heart muscle was solubilized in RIPA buffer (10 ml NP-40, 150 mM NaCl, 50 mM Tris–HCl, 1 mM EDTA, 5 ml sodiumdeoxycholate, pH=7.2). Western blot analysis was performed essentially as described previously (Witt et al, 2006).
Expression profiling (Affymetrix gene chip analysis) was performed essentially as described previously (Witt et al, 2004). We used the Mouse Genome 430 2.0 array and compared two WT/dKO pairs from matched litters at 4 months of age. Data were analyzed with SAS. The raw data have been deposited on ArrayExpress (http://www.ebi.ac.uk/arrayexpress, accession E-MEXP-1321); selected genes are presented in Supplementary Table 9.
Immunofluorescence and electron microscopy
Immunofluorescence microscopy was performed on muscle strips dissected from left ventricular walls from 3-month-old WT, dKO, MuRF1, and MuRF2 KO mice. Cryo-sectioning, immunolabeling, and electron microscopy were essentially as described previously (Witt et al, 2006). Images were acquired with a Zeiss AxioPlan2 microscope. For morphometric analysis, the amount of cells per 2 mm2 was analyzed with Zeiss AxioVision LE 4.3 software.
Bycast immobilization of quadriceps muscles was performed essentially as described by Kemp et al (2000) using 5-month-old mice and for every genotype in duplicate. Right-sided quadriceps muscles of KO and matched WT mice were immobilized by a plaster bycast for 72 h. Mice were killed, quadriceps muscles were analyzed by Affymetrix arrays, and western blots were performed as described previously. Induction of a subset of genes including MAFbx/atrogin1 was monitored as a positive control for immobilization in by-casted skeletal muscles.
D5-phenylalanine incorporation
D5-phenylalanine (D5-F) was purchased from Sigma. A total of 50 μmol/100 g mouse weight (prepared in 0.9% NaCl) was injected i.p. into mice that were kept for 48 h and fed ad libitum. Incorporation of D5-F into mouse hearts was determined essentially as described (Dardevet et al, 2002). Briefly, mice were killed 48 h after the D5-F injection, TCA insoluble total muscle proteins were lysed in 6 M HCl, and total amino acids were purified by passing over cation exchange column (AG50 W-X8, Bio-Rad). Fractional total muscle protein synthesis was estimated by determining the Mw 166.1 (Phenylalanine) and 171.1 (D5-F) ratios in the respective samples.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Video 4
Supplementary Video 5
Supplementary Video 6
Supplementary Video 7
Supplementary Figure 8
Supplementary Table 9
Supplementary Figure 10
Supplementary Information
Acknowledgments
We thank Christopher Bleck for expert electron microscopy analysis and Thomas Franz (EMBL EM and proteomic core facilities), Alexander Gasch, and Alexander Schuster for protein expression analysis and technical advice, Ralf Erber for immunofluorescence support, Karl-Heinz Hiller and the Bavaria MRI Center for expert MRI studies, Christian Wolpert for mouse ECG scans, Carsten Sticht for microarray analysis, and Katja Müdder for expert mouse-breeding support. We are indebted to the DFG and the NAR/Landesstiftung Baden-Württemberg for generous financial support.
References
- Arya R, Kedar V, Hwang JR, McDonough H, Li HH, Taylor J, Patterson C (2004) Muscle ring finger protein-1 inhibits PKC{epsilon} activation and prevents cardiomyocyte hypertrophy. J Cell Biol 167: 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar K, Torgan CE, Kraus WE, Esser K (2000) Autocrine phosphorylation of p70(S6k) in response to acute stretch in myotubes. Mol Cell Biol Res Commun 4: 76–80 [DOI] [PubMed] [Google Scholar]
- Bang ML, Mudry RE, McElhinny AS, Trombitas K, Geach AJ, Yamasaki R, Sorimachi H, Granzier H, Gregorio CC, Labeit S (2001) Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J Cell Biol 153: 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN (2006) Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem 75: 19–37 [DOI] [PubMed] [Google Scholar]
- Belle R, Minella O, Cormier P, Morales J, Poulhe R, Mulner-Lorillon O (1995) Phosphorylation of elongation factor-1 (EF-1) by cdc2 kinase. Prog Cell Cycle Res 1: 265–270 [DOI] [PubMed] [Google Scholar]
- Bodine SC (2006) mTOR signaling and the molecular adaptation to resistance exercise. Med Sci Sports Exerc 38: 1950–1957 [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708 [DOI] [PubMed] [Google Scholar]
- Clark KA, McElhinny AS, Beckerle MC, Gregorio CC (2002) Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol 18: 637–706 [DOI] [PubMed] [Google Scholar]
- Dardevet D, Sornet C, Bayle G, Prugnaud J, Pouyet C, Grizard J (2002) Postprandial stimulation of muscle protein synthesis in old rats can be restored by a leucine-supplemented meal. J Nutr 132: 95–100 [DOI] [PubMed] [Google Scholar]
- Fielitz J, van Rooij E, Spencer JA, Shelton JM, Latif S, van der Nagel R, Bezprozvannaya S, de Windt L, Richardson JA, Bassel-Duby R, Olson EN (2007) Loss of muscle-specific RING-finger 3 predisposes the heart to cardiac rupture after myocardial infarction. Proc Natl Acad Sci USA 104: 4377–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitts RH, Riley DR, Widrick JJ (2001) Functional and structural adaptations of skeletal muscle to microgravity. J Exp Biol 204: 3201–3208 [DOI] [PubMed] [Google Scholar]
- Granzier HL, Labeit S (2004) The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ Res 94: 284–295 [DOI] [PubMed] [Google Scholar]
- Haase A, Matthaei D, Hanicke W, Frahm J (1986) Dynamic digital subtraction imaging using fast low-angle shot MR movie sequence. Radiology 160: 537–541 [DOI] [PubMed] [Google Scholar]
- Hartgens F, Kuipers H (2004) Effects of androgenic-anabolic steroids in athletes. Sports Med 34: 513–554 [DOI] [PubMed] [Google Scholar]
- Hoshijima M (2006) Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z-disk, titin, and associated structures. Am J Physiol Heart Circ Physiol 290: H1313–H1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C (2004) Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci USA 101: 18135–18140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp TJ, Sadusky TJ, Saltisi F, Carey N, Moss J, Yang SY, Sassoon DA, Goldspink G, Coulton GR (2000) Identification of Ankrd2, a novel skeletal muscle gene coding for a stretch-responsive ankyrin-repeat protein. Genomics 66: 229–241 [DOI] [PubMed] [Google Scholar]
- Kim JJ, Battaile KP (2002) Burning fat: the structural basis of fatty acid beta-oxidation. Curr Opin Struct Biol 12: 721–728 [DOI] [PubMed] [Google Scholar]
- Knoell R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W, Yokoyama M, Schork NJ, Omens JH, McCulloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP et al. (2002) The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111: 943–955 [DOI] [PubMed] [Google Scholar]
- Koopman R, Zorenc AH, Gransier RJ, Cameron-Smith D, van Loon LJ (2006) Increase in S6K1 phosphorylation in human skeletal muscle following resistance exercise occurs mainly in type II muscle fibers. Am J Physiol Endocrinol Metab 290: E1245–E1252 [DOI] [PubMed] [Google Scholar]
- Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edstrom L, Ehler E, Udd B, Gautel M (2005) The kinase domain of titin controls muscle gene expression and protein turnover. Science 308: 1599–1603 [DOI] [PubMed] [Google Scholar]
- Latronico N, Peli E, Botteri M (2005) Critical illness myopathy and neuropathy. Curr Opin Crit Care 11: 126–132 [DOI] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL (2004) Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 18: 39–51 [DOI] [PubMed] [Google Scholar]
- McElhinny AS, Perry CN, Witt CC, Labeit S, Gregorio CC (2004) Muscle-specific RING finger-2 (MURF-2) is important for microtubule, intermediate filament and sarcomeric M-line maintenance in striated muscle development. J Cell Sci 117: 3175–3188 [DOI] [PubMed] [Google Scholar]
- Miller MK, Bang ML, Witt CC, Labeit D, Trombitas C, Watanabe K, Granzier H, McElhinny AS, Gregorio CC, Labeit S (2003) The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol 333: 951–964 [DOI] [PubMed] [Google Scholar]
- Morris C, Wittmann J, Jack HM, Jalinot P (2007) Human INT6/eIF3e is required for nonsense-mediated mRNA decay. EMBO Rep 8: 9596–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrosek M, Labeit D, Witt S, Heerklotz H, von Castelmur E, Labeit S, Mayans O (2007) Molecular determinants for the recruitment of the ubiquitin-ligase MuRF-1 onto M-line titin. FASEB J 21: 1383–1392 [DOI] [PubMed] [Google Scholar]
- Nikawa T, Ishidoh K, Hirasaka K, Ishihara I, Ikemoto M, Kano M, Kominami E, Nonaka I, Ogawa T, Adams GR, Baldwin KM, Yasui N, Kishi K, Takeda S (2004) Skeletal muscle gene expression in space-flown rats. FASEB J 18: 522–524 [DOI] [PubMed] [Google Scholar]
- Ojima K, Lin ZX, Zhang ZQ, Hijikata T, Holtzer S, Labeit S, Sweeney HL, Holtzer H (1999) Initiation and maturation of I-Z-I bodies in the growth tips of transfected myotubes. J Cell Sci 112 (Part 22): 4101–4112 [DOI] [PubMed] [Google Scholar]
- Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, Evans SM, Clark B, Feramisco JR, Giles W, Ho SY, Benson DW, Silberbach M, Shou W, Chien KR (2004) Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117: 373–386 [DOI] [PubMed] [Google Scholar]
- Peng J, Raddatz K, Labeit S, Granzier H, Gotthardt M (2005) Muscle atrophy in Titin M-line deficient mice. J Muscle Res Cell Motil 26: 381–388 [DOI] [PubMed] [Google Scholar]
- Rasbach KA, Schnellmann RG (2007) PGC-1alpha over-expression promotes recovery from mitochondrial dysfunction and cell injury. Biochem Biophys Res Commun 355: 734–739 [DOI] [PubMed] [Google Scholar]
- Rittweger J, Frost HM, Schiessl H, Ohshima H, Alkner B, Tesch P, Felsenberg D (2005) Muscle atrophy and bone loss after 90 days' bed rest and the effects of flywheel resistive exercise and pamidronate: results from the LTBR study. Bone 36: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R (1993) MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75: 1351–1359 [DOI] [PubMed] [Google Scholar]
- Ruff J, Wiesmann F, Hiller KH, Voll S, von Kienlin M, Bauer WR, Rommel E, Neubauer S, Haase A (1998) Magnetic resonance microimaging for noninvasive quantification of myocardial function and mass in the mouse. Magn Reson Med 40: 43–48 [DOI] [PubMed] [Google Scholar]
- Seynnes OR, de Boer M, Narici MV (2007) Early skeletal muscle hypertrophy and architectural changes in response to high-intensity resistance training. J Appl Physiol 102: 368–373 [DOI] [PubMed] [Google Scholar]
- Solomon AM, Bouloux PM (2006) Modifying muscle mass—the endocrine perspective. J Endocrinol 191: 349–360 [DOI] [PubMed] [Google Scholar]
- Vigoreaux JO (1994) The muscle Z band: lessons in stress management. J Muscle Res Cell Motil 15: 237–255 [DOI] [PubMed] [Google Scholar]
- Wiesmann F, Ruff J, Hiller KH, Rommel E, Haase A, Neubauer S (2000) Developmental changes of cardiac function and mass assessed with MRI in neonatal, juvenile, and adult mice. Am J Physiol Heart Circ Physiol 278: H652–H657 [DOI] [PubMed] [Google Scholar]
- Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C (2007) Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res 100: 456–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt CC, Burkart C, Labeit D, McNabb M, Wu Y, Granzier H, Labeit S (2006) Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. EMBO J 25: 3843–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt CC, Gerull B, Davies MJ, Centner T, Linke WA, Thierfelder L (2001) Hypercontractile properties of cardiac muscle fibers in a knock-in mouse model of cardiac myosin-binding protein-C. J Biol Chem 276: 5353–5359 [DOI] [PubMed] [Google Scholar]
- Witt CC, Ono Y, Puschmann E, McNabb M, Wu Y, Gotthardt M, Witt SH, Haak M, Labeit D, Gregorio CC, Sorimachi H, Granzier H, Labeit S (2004) Induction and myofibrillar targeting of CARP, and suppression of the Nkx2.5 pathway in the MDM mouse with impaired titin-based signaling. J Mol Biol 336: 145–154 [DOI] [PubMed] [Google Scholar]
- Witt SH, Granzier H, Witt CC, Labeit S (2005) MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. J Mol Biol 350: 713–722 [DOI] [PubMed] [Google Scholar]
- Yoo YA, Kim MJ, Park JK, Chung YM, Lee JH, Chi SG, Kim JS, Yoo YD (2005) Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1. Mol Cell Biol 25: 6603–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Chow A, Powers J, Fajardo G, Bernstein D (2004) Microarray analysis of gene expression after transverse aortic constriction in mice. Physiol Genomics 19: 93–105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Video 4
Supplementary Video 5
Supplementary Video 6
Supplementary Video 7
Supplementary Figure 8
Supplementary Table 9
Supplementary Figure 10
Supplementary Information