Abstract
Understanding the function of histone modifications across inducible genes in mammalian cells requires quantitative, comparative analysis of their fate during gene activation and identification of enzymes responsible. We produced high-resolution comparative maps of the distribution and dynamics of H3K4me3, H3K36me3, H3K79me2 and H3K9ac across c-fos and c-jun upon gene induction in murine fibroblasts. In unstimulated cells, continuous turnover of H3K9 acetylation occurs on all K4-trimethylated histone H3 tails; distribution of both modifications coincides across promoter and 5′ part of the coding region. In contrast, K36- and K79-methylated H3 tails, which are not dynamically acetylated, are restricted to the coding regions of these genes. Upon stimulation, transcription-dependent increases in H3K4 and H3K36 trimethylation are seen across coding regions, peaking at 5′ and 3′ ends, respectively. Addressing molecular mechanisms involved, we find that Huntingtin-interacting protein HYPB/Setd2 is responsible for virtually all global and transcription-dependent H3K36 trimethylation, but not H3K36-mono- or dimethylation, in these cells. These studies reveal four distinct layers of histone modification across inducible mammalian genes and show that HYPB/Setd2 is responsible for H3K36 trimethylation throughout the mouse nucleus.
Keywords: chromatin, histone methyltransferases, histone modifications, HYPB/Setd2, transcription elongation
Introduction
Histone modifications, which include acetylation, phosphorylation, methylation and ubiquitination, play key roles in gene regulation (reviewed in Shilatifard, 2006; Berger, 2007; Bernstein et al, 2007; Kouzarides, 2007; Li et al, 2007). They are studied by chromatin immunoprecipitation (ChIP) to define their location, by genetic and biochemical analyses to identify enzymes involved and by identification of binding proteins or domains to determine their molecular function. ChIP is widely used to map histone modifications across genes and chromosomes, but their quantification and interpretation is contentious (reviewed in Clayton et al, 2006). In particular, high-resolution quantitative comparative studies in synchronised mammalian cells tracking changes in histone modification during transcriptional activation are currently unavailable.
The immediate-early (IE) genes c-fos and c-jun are rapidly induced in mammalian cells via ERK or p38 MAP kinase cascades. These kinases activate the downstream kinase MSK1/2, which phosphorylates histone H3 and HMGN1 (reviewed in Clayton and Mahadevan, 2003). Phosphorylated histone H3 is subject to continuous dynamic acetylation, creating phosphoacetylated H3 (Barratt et al, 1994), which is seen at c-fos and c-jun upon gene induction (Cheung et al, 2000; Clayton et al, 2000). Although they are colocalised at these genes, these two modifications are independently regulated by distinct mechanisms (Thomson et al, 2001). More recently, we reported that all K4-trimethylated H3 in these cells is also subject to extremely dynamic, continuous turnover of acetylation (Hazzalin and Mahadevan, 2005). ChIP with phosphoacetylated (S10phK9ac) or K4me3-specific antibodies and re-ChIP experiments showed that all three modifications can occur on the same nucleosomes (Hazzalin and Mahadevan, 2005). Because K4me3 was continuously detectable, whereas K9ac and S10ph were transient, we favoured a model whereby K4me3 was the key pre-existing modification, which attracts both dynamic acetylation and stimulus-dependent S10 phosphorylation (reviewed in Clayton et al, 2006). Such complex models can only be confirmed by detailed quantitative comparative mapping of these modifications across genes. H3 methylated at lysines 4, 36 or 79 is generally associated with active/permissive chromatin, whereas lysine 9 or 27 methylation is associated with repression (reviewed in Shilatifard, 2006). Within genes, K4me3, K9ac and K14ac occur around start sites of active genes (Liang et al, 2004; Bernstein et al, 2005; Pokholok et al, 2005; Barski et al, 2007; Guenther et al, 2007; Mikkelsen et al, 2007). K4me2 has been reported at start sites of active genes in metazoans (Liang et al, 2004; Schneider et al, 2004), but is spread throughout coding regions in yeast (Pokholok et al, 2005). In contrast, K4me1 is reported towards the 3′ end of active genes (Kouskouti and Talianidis, 2005; Pokholok et al, 2005), as have both K36me2 and me3 (Bannister et al, 2005; Farris et al, 2005; Morillon et al, 2005; Pokholok et al, 2005; Vakoc et al, 2006; Barski et al, 2007; Guenther et al, 2007; Mikkelsen et al, 2007). An exception to this is a transient post-initiation peak of K36me2 and me3 at the yeast MET16 promoter (Morillon et al, 2005). In Drosophila, K79me2 occurs throughout coding regions of active genes (Schübeler et al, 2004). Likewise, in yeast, K79me3 was found in coding regions, but there was no correlation with gene activity (Pokholok et al, 2005). At active mammalian genes, K79me2 and me3 are reported to peak towards the 5′ end of the coding region (Kouskouti and Talianidis, 2005; Vakoc et al, 2006), although another study found K79me2 at both promoter and coding regions of active genes (Im et al, 2003).
Complications to all such studies include quantitation and comparability of ChIP recovery using different antibodies and the possibility of microheterogeneity at any position, whereby distinct histone modifications, deriving from different cells within a population, are detectable at the same position along a gene. An obvious source of microheterogeneity is the transient dynamic nature of some histone modifications, for example, H3 acetylation described above. A second relates to the fact that some histone-modifying enzymes travel with elongating RNA polymerase II (Pol II), producing elongation-dependent variation across coding regions. In yeast, the K4 methyltransferase SET1/COMPASS complex (Briggs et al, 2001; Krogan et al, 2002) coimmunoprecipitates with Pol II phosphorylated at Ser 5 in its CTD (Ng et al, 2003), a form associated with early elongating complexes (Komarnitsky et al, 2000). In contrast, Set2, the yeast K36 methyltransferase (Strahl et al, 2002), associates with Pol II phosphorylated at Ser 2 in its CTD (Li et al, 2002; Krogan et al, 2003; Schaft et al, 2003; Xiao et al, 2003; Kizer et al, 2005), a form of Pol II found increasingly towards the 3′ end of transcribed genes (Komarnitsky et al, 2000). K36me then recruits a histone deacetylase (HDAC) complex via interaction with the chromodomain of Eaf3 (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005), which is required for suppression of internal transcript initiation in the STE11 and FLO8 genes (Carrozza et al, 2005). In higher eukaryotic cells, the location of K36me at 3′ coding regions of active genes (Bannister et al, 2005; Carrozza et al, 2005), the observation that it is dependent upon the CTD kinase p-TEFb (Zhou et al, 2004; Eissenberg et al, 2007) and sensitive to the transcriptional inhibitor DRB (Vakoc et al, 2006) all suggest an elongation-dependent mechanism similar to that in yeast. Potential enzymes include nuclear receptor-binding SET domain-containing protein 1 (NSD1), Smyd2 and the Huntingtin-interacting protein HYPB/SETD2, all of which methylate K36 in vitro (Rayasam et al, 2003; Sun et al, 2005; Brown et al, 2006), but questions of which enzyme is relevant in vivo and whether it catalyses mono-, di- and/or trimethylation in intact cells are unanswered.
Here, we have comprehensively mapped the distribution of histone H3 K9ac, K4me3, K36me3 and K79me2 across c-fos and c-jun at high resolution, and demonstrate rapid elongation-dependent changes in K4me3 and K36me3 within the coding regions upon gene activation. These studies reveal four distinct layers of histone modification that may be distinguished across these genes. Using siRNA to knock down the relevant enzymes, we show here that Setd2, the mouse homologue of human Huntingtin-interacting protein HYPB, is responsible for virtually all detectable K36me3, but not K36me2 or K36me1, throughout the nucleus. Consistent with its proposed interaction with RNA Pol II, this enzyme mediates EGF-induced transcription-dependent increase of H3K36 trimethylation at the 3′ ends of c-fos and c-jun. This represents the first unequivocal demonstration in mammalian cells that a single histone methyltransferase (HMTase) enzyme may be entirely responsible for the totality of a specific state of methylation, that is, trimethylation but not mono- or dimethylation, throughout the nucleus.
Results
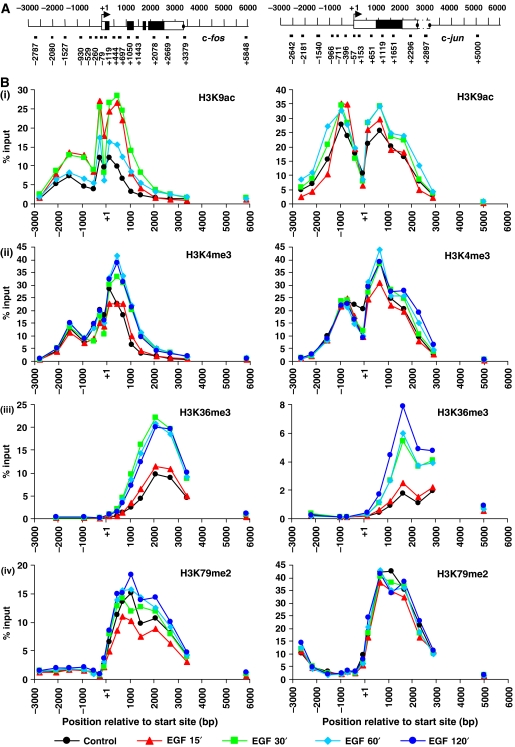
We first carried out a comprehensive mapping study to determine the distribution of histone modifications across c-fos and c-jun in control, EGF-stimulated (Figure 1) or anisomycin-stimulated (Supplementary Figure S1) mouse fibroblasts. A modified ChIP protocol, using MNase instead of sonication to generate predominantly mononucleosomal chromatin (MacDonald et al, 2005) was used to produce high-resolution quantitative comparative maps of the distribution and dynamics of H3K4 trimethylation (K4me3), K36 trimethylation (K36me3), K79 dimethylation (K79me2) and K9 acetylation (K9ac) across these genes (Figure 1 and Supplementary Figure S1). Each antibody was first titrated in ChIP to arrive at concentrations that recovered virtually all available epitopes, leaving little or none in the unbound fraction as monitored by western blotting. Peaks of distribution of each modification across these genes are shown graphically (Figure 1B and Supplementary Figure S1B) and as bar charts with error bars (Figure 1C and Supplementary Figure S1C). This yielded highly reproducible maps with striking differences in the amounts of each modification recovered. Their interpretation is extremely complex because of the continuous dynamic turnover of acetylation at these genes, the fact that methylation may occur transiently with the traverse of RNA pol II, and the phenomenon of microheterogeneity of histone modifications at any single position. Evaluating these maps in overview, common trends applying to both c-fos and c-jun emerge. First, K9ac (Figure 1B, panel i) and K4me3 (panel ii) have remarkably similar overlapping distributions across the start site of both genes. For both modifications, there is a striking dip precisely at the start site of both genes, and the positions of peaks of modification largely coincide. The stability of this colocalisation at all points analysed is consistent with sequential ChIP assays showing these modifications coexist on the same nucleosomes at these positions (Hazzalin and Mahadevan, 2005). The sharp dip in K4me3 and K9ac at the start site on both genes appears to be due to this position lacking a nucleosome and being more accessible, as indicated by MNase sensitivity maps across these genes (Figure 2 and Supplementary Figure S2). These show that the start sites of c-fos and c-jun are considerably more sensitive to MNase digestion with <5% (c-fos −79, c-jun −57) and <10% (c-jun −396) of the corresponding genomic DNA signal remaining after digestion (Figure 2 and Supplementary Figure S2).
Figure 1.
EGF-stimulated distribution of acetylated and methylated histone H3 across c-fos and c-jun. (A) Schematic representation of c-fos and c-jun showing positions analysed by real-time PCR. Primer positions indicate the 5′ position of the forward primer relative to the transcription start site. Exons are represented by boxes, unfilled for untranslated regions and filled for translated regions. Termination sites are shown as filled circles. (B) Quiescent cells were unstimulated (control) or stimulated with EGF (50 ng/ml) for 15, 30, 60 or 120 min and formaldehyde crosslinked mononucleosomes were prepared for ChIP. Aliquots of chromatin were immunoprecipitated with H3K9ac- (i), H3K4me3- (ii), H3K36me3- (iii) or H3K79me2- (iv) specific antibodies. Recovered DNA sequences were quantified by real-time PCR. Average % input recoveries from two independent experiments are plotted.
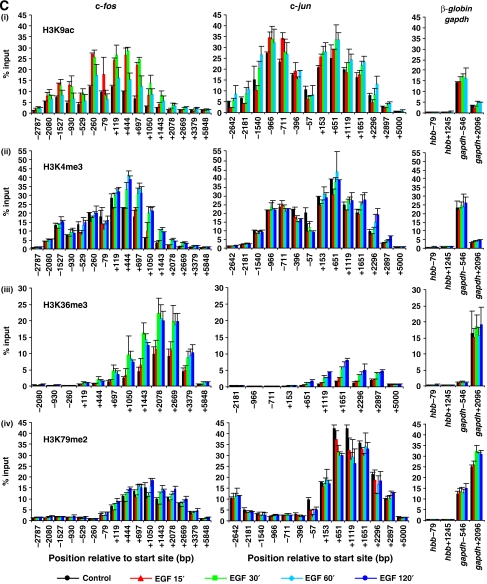
Figure 1.
(C) Data shown for c-fos and c-jun are the same as in (B) but plotted in bar chart format with error bars representing the standard deviation (s.d.) of the mean of the two independent experiments. For each ChIP, primers spanning two regions of the inactive β-globin (hbb) and the constitutively expressed gapdh genes were also analysed (right-hand side graphs) for comparison.
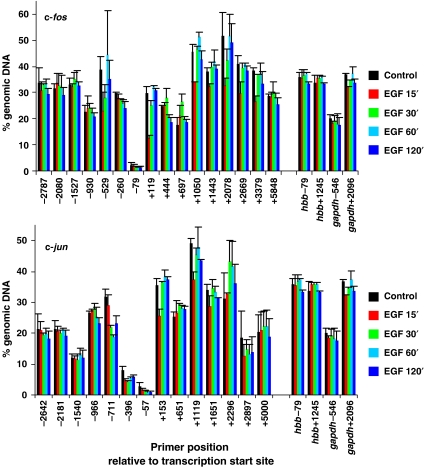
Figure 2.
MNase sensitivity across c-fos and c-jun in quiescent and EGF-stimulated mouse cells. Quiescent cells were unstimulated (control) or stimulated with EGF (50 ng/ml) for 15, 30, 60 or 120 min and formaldehyde crosslinked mononucleosomes were prepared. Equivalent amounts of DNA were isolated from each sample (the DNA is from the same (input) chromatin samples used for the ChIP assays shown in Figure 1 and Supplementary Figure S1) and specific sequences were quantified by real-time PCR. MNase sensitivity is expressed as % of amplifiable DNA sequence in the chromatin sample relative to that in an equivalent amount of intact genomic DNA (more digestion within a region is reflected by fewer intact template molecules that are amplifiable by PCR). Average values from two independent experiments are plotted with s.d. Primers spanning two regions of the hbb and gapdh genes were also analysed and the data are included on both graphs for comparison (c-fos upper panel, c-jun lower panel).
In contrast, K36me3 and K79me2 are found exclusively within the coding regions of c-fos and c-jun (Figure 1B, panels iii and iv). Although they do overlap, their precise distributions appear distinct. K36me3 peaks towards the 3′ end of the coding regions but drops off before the gene's terminus (Figure 1B, panel iii), whereas K79me2 peaks over the 5′ end of the coding region before declining towards the 3′ end (Figure 1B, panel iv). The presence of both these modifications in unstimulated control cells may be attributed either to their being deposited here in previous rounds of transcription prior to quiescence, or to their being part of a transcription-independent pattern of pre-existing, possibly epigenetic, mechanism; there is no evidence either way at present.
Finally, as a negative control, we monitored all four modifications at a site beyond the end of each gene (Figure 1B; c-fos +5848; c-jun +5000) and found none of these modifications were present there. Stimulation with EGF leads to rapid increases in H3K9ac on c-fos across promoter and coding region, peaking at 15 min over the promoter and 30 min over the coding region (Figure 1B, panel i left-side graph). These increases are transient and decrease substantially by 60 min (Figure 1B, panel i), correlating well with kinetics of c-fos transcription (Thomson et al, 1999). For c-jun, basal level of K9ac is much higher than for c-fos, and smaller increases are seen upon EGF stimulation, peaking at 60 min (Figure 1B, panel i, right-side graph). The elevated basal level of K9ac on c-jun may be due to a low background level of c-jun transcription in control cells (Hazzalin and Mahadevan, 2005). K4me3 and K36me3 also increase within the coding regions on c-fos and c-jun upon stimulation (Figure 1B, panels ii and iii). Compared to K9ac, these increases are slower and more sustained, remaining after 2 h of stimulation (Figure 1B, panels ii and iii). Increases in K4me3 on c-jun are much smaller than on c-fos (Figure 1B, panel ii).
The entire mapping study was repeated using a different stimulus, anisomycin, which activates these genes predominantly via stress-activated protein kinases (SAPKs) as opposed to the ERK MAP kinase pathway, which EGF utilises (reviewed in Hazzalin and Mahadevan, 2002). This yielded extremely similar MNase sensitivity maps and patterns of distribution and quantitative levels of these modifications across each gene (Supplementary Figures S1 and S2), testifying to the high reproducibility of the methodology used here.
Finally, we address a concern that activation of these genes may produce marked changes of nucleosomes occupancy across the gene, thereby skewing the ChIP recovery data provided at various time points after induction. First, there is no indication of a great change in MNase sensitivity at various time points after stimulation (Figure 2 and Supplementary Figure S2), suggesting that average nucleosomal occupancy at any position did not vary greatly upon gene activation. Second, we carried out ChIP using anti-H3 antibodies directed to the unmodified C terminus across these genes (Supplementary Figure S3A and B, panel i), and this too showed no major changes in recovery upon gene activation, confirming indications from MNase sensitivity maps. As expected, there was good correspondence between the relative H3 occupancy and the MNase sensitivity maps at all points compared across the genes (Supplementary Figure S3A and B, panels ii and iii; see for example, the dip across the start site), indicating that MNase sensitivity seen here is a good approximation of average nucleosomal occupancy.
EGF-induced increases in H3K4me3 and H3K36me3 within c-fos and c-jun coding regions are dependent on transcription elongation
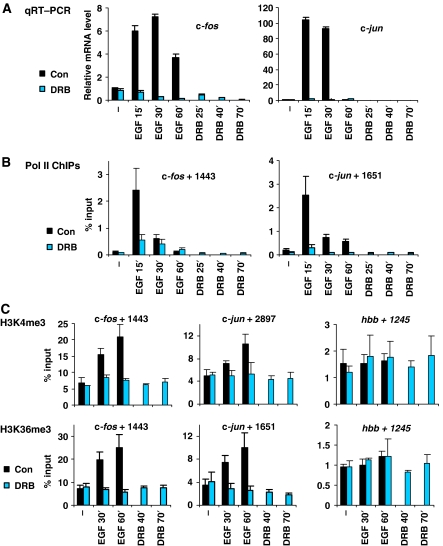
As shown in yeast (reviewed in Gerber and Shilatifard, 2003; Hampsey and Reinberg, 2003; Sims et al, 2004), stimulation-dependent increases in K4me3 and K36me3 across coding regions suggest a link with elongating Pol II, a possibility we tested with the transcriptional inhibitor DRB. Quantitative reverse transcription PCR (qRT–PCR) shows that DRB ablates induction of c-fos and c-jun (Figure 3A). This correlates with loss of Pol II across the coding region of both genes under these conditions, shown by ChIPs using anti-Pol II antibodies raised against its N terminus (Figure 3B). Similar results were obtained at several different positions within the coding regions of c-fos and c-jun (Supplementary Figure S4A), except for +444 of c-fos, which showed increased Pol II loading in response to DRB at later time points. This may be explained by the transcriptional pause site reported at this region (Mechti et al, 1991; Coulon et al, 1999).
Figure 3.
EGF-induced increases in H3K4me3 and H3K36me3 within c-fos and c-jun are transcription-dependent. (A) Quiescent cells were pretreated (10 min) with DRB (25 μg/ml) or untreated (con) prior to stimulation with EGF (50 ng/ml) for 15, 30 or 60 min or no-stimulation (−). Quiescent cells were also treated with DRB alone for 25, 40 or 70 min as controls, respectively. Total mRNA was isolated and relative levels of c-fos and c-jun mRNA were quantified by qRT–PCR with normalisation to gapdh mRNA. A representative experiment is shown. Error bars represent the s.d. from triplicate PCRs. (B) Formaldehyde crosslinked mononucleosomes were prepared from quiescent cells treated as in (A) and used for ChIPs with an anti-RNA-polymerase-II (Pol II) antibody. Recovery of c-fos and c-jun coding DNA sequences were quantified by real-time PCR. Average % input recoveries and s.d. from 3–4 independent experiments are plotted. (C) ChIPs and real-time PCR were performed as described in (B) using H3K4me3- (top panel) and H3K36me3- (bottom panel) specific antibodies. Amplicons analysed are indicated above each graph. Average % input recoveries and s.d. from 3–4 independent experiments are plotted.
EGF-induced increases in K4me3 and K36me3 within the coding regions of both genes were abolished by DRB pretreatment (Figure 3C, left and middle panels). Other coding region positions within c-fos and c-jun gave similar results (Supplementary Figure S4B and C). As expected, in the negative control hbb coding region, no significant change in either modification was observed in response to EGF or DRB (Figure 3C, right panel). Thus, EGF-induced increases in K4me3 and K36me3 within coding regions of these genes are dependent on transcription elongation.
Setd2 is non-redundantly responsible for H3K36 trimethylation in mouse fibroblasts
In Saccharomyces cerevisiae, Schizosaccharomyces pombe and Neurospora crassa, Set2 is an H3K36 methyltransferase responsible for mono-, di- and trimethylation (Strahl et al, 2002; Adhvaryu et al, 2005; Morris et al, 2005; Chu et al, 2006). In mammals, several candidates have been proposed. The first discovered, NSD1, has in vitro HMTase activity towards H3K36 and H4K20 (Rayasam et al, 2003). Second, a SET domain protein that interacts with the Huntington's Disease gene (HD) product, HYPB/SETD2, also methylates H3K36 in vitro and, via its C-terminal Pol II interaction domain, can bind to phosphorylated Pol II (Sun et al, 2005). Finally, Smyd2 has HMTase activity towards H3K36 in vitro, and interacts with Sin3 HDAC complexes (Brown et al, 2006). None of these studies addressed their ability to methylate H3K36 in vivo, or the specific states (mono, di or tri) of methylation for which they might be responsible. Dimethyl-H3K36 antibodies were used in the NSD1 and Smyd2 studies, but capacity for mono- or trimethylation was not analysed.
Using NCBI protein BLAST search, we identified mouse homologues of yeast Set2 (data not shown). Setd2 (the mouse homologue of human HYPB/SETD2) and NSD1 (among others) had significant homology. In view of this homology, and the fact that HYPB/SETD2 has a Pol II interaction domain (Sun et al, 2005), we investigated Setd2 and NSD1 as possible H3K36 methyltransferases in mouse fibroblasts.
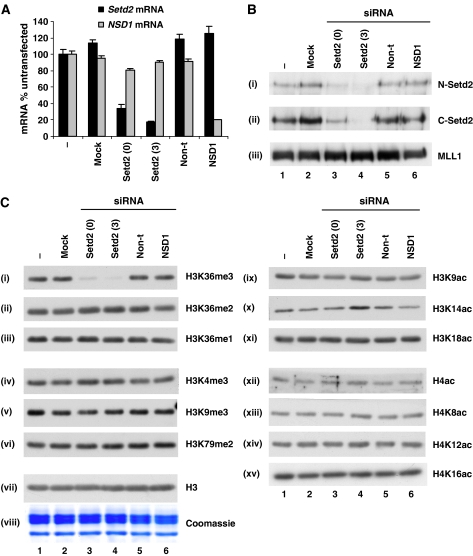
Two different siRNAs targeting Setd2 transcripts knocked down Setd2 mRNA levels by >60% (Setd2 0) and >80% (Setd2 3) (Figure 4A). siRNA targeting NSD1 transcripts knocked down NSD1 mRNA levels by >80% (Figure 4A). These were specific, as mock-transfected cells, or non-targeting siRNA, did not show similar effects (Figure 4A). Additionally, Setd2- or NSD1-specific siRNAs did not knock down mRNAs of other genes tested (Figure 4A and data not shown). We raised and characterised N- and C-terminal Setd2 antibodies to confirm that Setd2 protein levels were specifically diminished as a result of its mRNA knockdown (Figure 4B, panels i and ii; Supplementary Figure S5). We were not able to assess NSD1 protein levels, as a good antibody is currently unavailable. This became less relevant since the Setd2 knockdown produced a clear H3K36 trimethylation defect.
Figure 4.
Setd2 is a non-redundant H3K36-specific trimethyltransferase. (A) Cells were untransfected (−), mock transfected (no siRNA, mock) or transfected with Setd2 (0 and 3, two different siRNAs), NSD1 or non-targeting (non-t) siRNAs. Total mRNA was isolated 24 h later and relative levels of Setd2 and NSD1 mRNA were quantified by qRT–PCR and normalised to gapdh mRNA. A representative experiment is shown. Error bars represent the s.d. from triplicate PCRs. (B) Cells were transfected as in (A) and crude nuclear lysates were prepared 48 h later. Lysates were separated by 8% SDS–PAGE and transferred to PVDF membrane for immunoblotting with two different Setd2 antibodies (N-Setd2 (i) and C-Setd2 (ii)) and an MLL1 antibody (iii) as a control. (C) Crude nuclear lysates were prepared as in (B), separated by 15% SDS–PAGE and either Coomassie-stained (viii) or transferred to PVDF membrane for immunoblotting with various methyl (i–vi) or acetyl (ix–xv) modification-specific histone antibodies, as indicated in the figure. H4ac is an anti-histone H4 pan-acetyl antibody. Unmodified H3 (H3, vii) was used as a loading control.
Total cellular K36me3 was virtually absent in the Setd2 knockdowns (Figure 4C, panel i), whereas K36me2 and me1 levels remain unaffected (Figure 4C, panels ii and iii). In contrast, NSD1 knockdown had no effect on mono-, di- or trimethylation of K36 (Figure 4C, panels i–iii). Additionally, total levels of K4me3, K9me3 or K79me2 were unchanged in either Setd2 or NSD1 knockdowns (Figure 4C, panels iv–vi). Therefore, Setd2 appears to be a non-redundant methyltransferase specific for trimethylation of H3K36. This is the first demonstration in intact cultured cells of a H3K36-specific methyltransferase whose knockdown results in abrogation of virtually all trimethylation, but not mono- or dimethylation. It is not possible, however, to distinguish from these experiments if HYPB/Setd2 can catalyse all states of modification culminating in K36 trimethylation, or just the final conversion of dimethylated K36 to the trimethylated state. Since there are several recent reports of a link between H3K36 methylation and histone deacetylation in yeast (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005), we also analysed global levels of various H3 and H4 acetyl modifications in Setd2 knockdown cells. No significant reproducible changes were observed for any acetyl modification (Figure 4C, panels ix–xv).
Setd2 is responsible for H3K36 trimethylation at IE and housekeeping genes
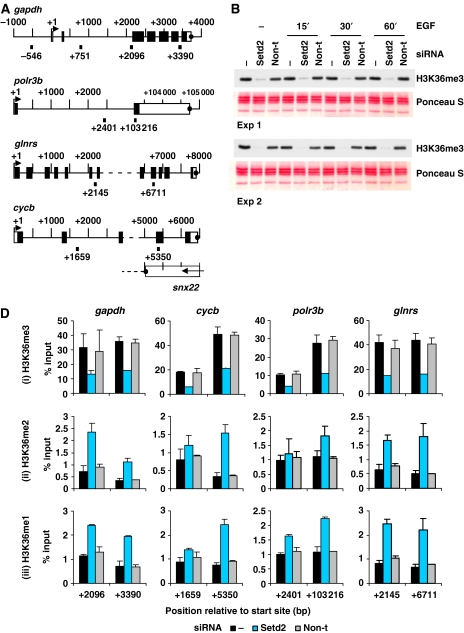
Next, we used ChIP to analyse all states of K36 methylation at c-fos- and c-jun coding regions (mid and 3′ end) after Setd2 knockdown. Four housekeeping genes were also analysed; glyceraldehyde-3-phosphate-dehydrogenase (gapdh), RNA-polymerase III subunit b (polr3b), glutaminyl-tRNA synthetase (glnrs) and cyclophilin b (cycb) (shown schematically in Figure 5A).
Figure 5.
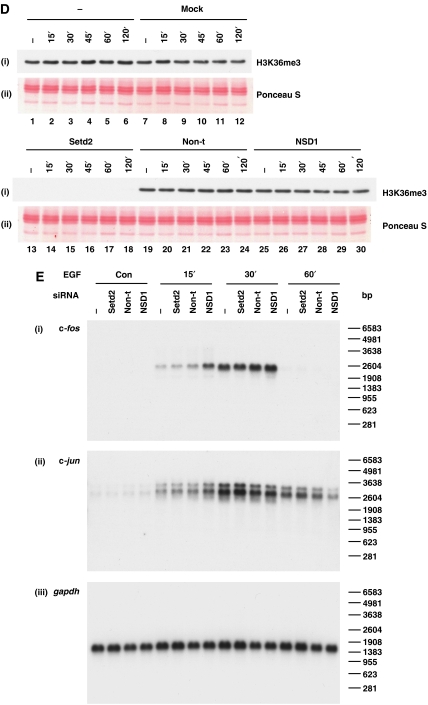
Setd2 is responsible for H3K36 trimethylation at IE and housekeeping genes. (A) Schematic representation of the gapdh, polr3b, glnrs and cycb housekeeping genes showing regions amplified by primers used for real-time PCR. Primer positions shown indicate 5′ position of the forward primer relative to the transcription start site. Exons are represented by boxes, unfilled for untranslated regions and filled for translated regions. Transcription termination sites are shown as filled circles. (B) Cells were untransfected (−) or transfected with Setd2 or non-targeting (non-t) siRNA. Cells were quiesced 24 h later, and after a further 24 h were unstimulated (−) or stimulated with EGF (50 ng/ml) for 15–60 min. Formaldehyde crosslinked mononucleosomes were prepared and aliquots of each sample were heated to reverse the crosslinks, separated by 15% SDS–PAGE, transferred to PVDF and immunoblotted with an H3K36me3-specific antibody. Membranes were stained with Ponceau S before immunoblotting to verify even loading. (D) Aliquots of formaldehyde crosslinked mononucleosomes from unstimulated cells prepared as in (B) were used in ChIP assays with H3K36me3- (i), H3K36me2- (ii) and H3K36me1- (iii) specific antibodies. Recovery of gapdh, cycb, polr3b and glnrs coding region sequences were quantified by real-time PCR. Average % input recoveries and s.d. from two independent experiments are plotted.
Trimethyl K36
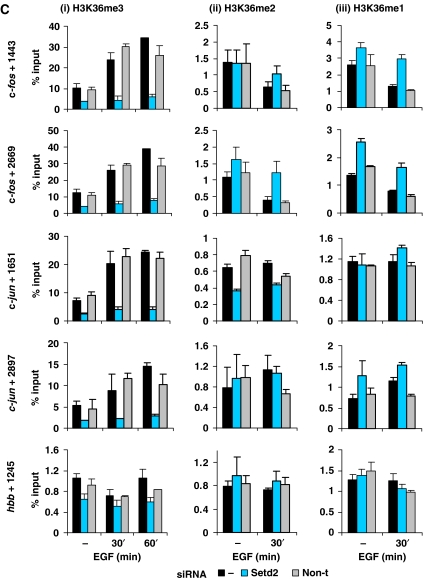
Setd2 knockdown produced almost complete loss of total cellular K36me3 in these cells (Figure 5B) and a marked reduction of K36me3 at c-fos- and c-jun coding regions (Figure 5C, panel i). Stimulation with EGF for 60 min resulted in a 2.7- to 3.4-fold increase in the K36me3 observed at these regions (black bars panel i), whereas in the Setd2 knockdown, induced levels of K36me3 were much smaller—1.3- to 1.8-fold (middle bars panel i). K36me3 was also decreased by >50% within the coding regions of gapdh, cycb, polr3b and glnrs after Setd2 knockdown (Figure 5D, panel i). Setd2 therefore appears to be responsible for both basal and inducible K36me3 at IE gene coding regions, and K36me3 levels at constitutively active genes.
Figure 5.
(C) Aliquots of formaldehyde crosslinked mononucleosomes prepared as in (B) were used in ChIP assays with H3K36me3- (i), H3K36me2- (ii) and H3K36me1- (iii) specific antibodies. Recovery of c-fos, c-jun and hbb coding region sequences were quantified by real-time PCR. Average % input recoveries and s.d. from two independent experiments are plotted. Regions analysed are indicated to the left of the panels.
Mono- and dimethyl K36
For all points tested, the recovery of nucleosomes with H3 bearing mono- and dimethyl K36 was very much lower (>10-fold) than that of trimethyl K36 (Figure 5C). In untransfected cells, there is no increase in mono- and dimethylated K36 upon activation of c-fos and c-jun (Figure 5C, panels ii and iii), indicating no correlation between these modifications and transcription. In Setd2 knockdown cells, there is no decrease in these low levels of K36me2 or me1 within the coding regions of c-fos and c-jun, with the exception of the c-jun +1651 region where there is a small decrease in K36me2 (Figure 5C, panel ii). Thus, in general Setd2 does not appear to be responsible for the low levels of mono- or dimethyl K36 at these regions.
As described for c-fos and c-jun, recovery of K36me1 and me2 was also very much lower than that of K36me3 at coding regions of gapdh, cycb, polr3b and glnrs (Figure 5D). In fact, these levels are very similar to that seen at the inactive hbb coding region (Figure 5C, panels ii and iii bottom graphs). Knockdown of Setd2 resulted in slightly higher levels of K36me2 and K36me1 at these positions (Figure 5D, panels ii and iii) but recoveries remain relatively very low. Thus, K36me2 and K36me1 levels do not seem to correlate with active transcription as K36me3 does, and Setd2 does not appear to be responsible for these modifications. By their nature, knockdowns do not produce the complete loss of any enzyme and low residual activity of the remaining HYPB/Setd2 enzyme may explain the incomplete loss of H3K36 sometimes seen.
Loss of H3K36me3 does not affect the kinetics of IE gene transcription, intragenic transcription or expression of constitutively active genes
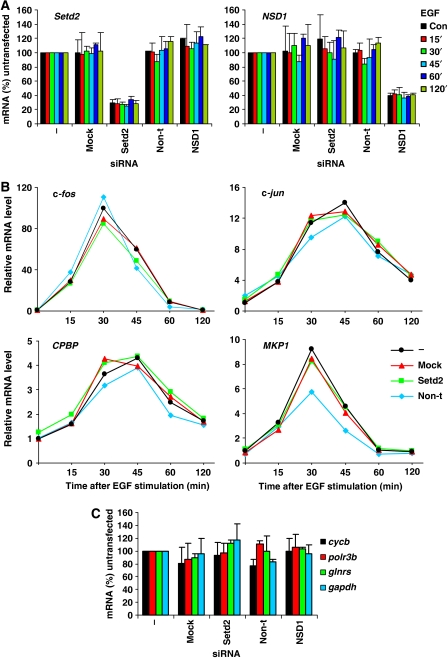
To investigate the potential role of K36me3 in transcriptional regulation, the effect of K36me3 loss on the transcription of IE and constitutively active genes was assessed by qRT–PCR (Figure 6). As shown previously, knock down of Setd2 mRNA (Figure 6A) led to virtually complete loss of all detectable K36me3 (Figure 6D). The gene induction kinetics of c-fos, c-jun and two other IE genes, core promoter element binding protein (CPBP) and MAP kinase phosphatase 1 (MKP1), was examined (Figure 6B). A recent report in yeast has shown that increased K36me leads to delayed induction of the HIS4 gene, whereas elimination of K36me accelerates HIS4 induction (Nelson et al, 2006). However, loss of K36me3 did not affect the kinetics of induction of any gene tested here (Figure 6B). Second, the steady-state mRNA levels of four constitutively active genes (cycb, polr3b, glnrs and gapdh) were assessed in the absence of K36me3, and no significant changes were observed for any transcript (Figure 6C). Furthermore, we did not observe any intragenic transcription from c-fos, c-jun or gapdh in K36me3-depleted cells (Figure 6E). This is in contrast to the observations in yeast where K36me appears to create a less permissive chromatin structure throughout yeast gene coding regions by recruiting HDAC complexes (Rpd3) via interaction with the chromodomain of Eaf3 (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005). Loss of K36me leads to increased acetylation, K4me3 and Pol II loading within specific gene coding regions and intragenic transcript initiation from the FLO8 and STE11 genes (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005; Kizer et al, 2005). Similar to the lack of any intragenic transcription in K36me3-depleted mouse fibroblasts (Figure 6E), we do not detect any increased H3 or H4 acetylation at 3′ regions of IE or housekeeping genes (Supplementary Figure S6), nor any change in Pol II occupancy at these genes (Supplementary Figure S7). These studies do not reveal a clear parallel at c-fos and c-jun in mammalian cells for the relationship between H3K36 methylation and histone deacetylation previously seen in yeast.
Figure 6.
Effects of H3K36me3 knockdown on gene expression levels: lack of intragenic transcription from IE genes or gapdh. (A) C3H 10T½ cells were untransfected (−), mock transfected (no siRNA, mock) or transfected with Setd2, non-targeting (non-t) or NSD1 siRNA. Cells were quiesced 24 h later, and after a further 24 h left untreated (con) or treated with EGF (50 ng/ml) for 15–120 min. Total RNA was isolated and relative levels of Setd2 and NSD1 mRNA were quantified by qRT–PCR and normalised to 18S rRNA. Average values and s.d. from two independent experiments are plotted. (B) Cells were transfected and stimulated and total RNA isolated as in (A). Kinetics of c-fos, c-jun, CPBP and MKP1 gene expression was assessed by qRT–PCR and normalised to 18S rRNA. Average values and s.d. from two independent experiments are plotted. (C) Cells were transfected as in (A), left unstimulated and levels of cycb, polr3b, glnrs and gapdh mRNA quantified by qRT–PCR, with normalisation to 18S rRNA. Average values and s.d. from two independent experiments are plotted.
Figure 6.
(D) Cells were transfected and stimulated as in (A) and cellular protein was isolated from the same TRIzol® reagent preparations used for RNA extraction. Protein was separated by 15% SDS–PAGE and transferred to PVDF membrane for immunoblotting with an H3K36me3 (i)-specific antibody. Ponceau S staining of histones was performed to verify even loading (ii). (E) C3H 10T½ cells were untransfected (−) or transfected with Setd2, non-targeting (non-t) or NSD1 siRNA. Cells were quiesced 24 h later, and after a further 24 h left untreated (con) or treated with EGF (50 ng/ml) for 15–60 min. Total mRNA was isolated and northern blotting carried out with 3′ c-fos (i) and 3′ c-jun (ii) probes and a full-length gapdh cDNA probe (iii).
Discussion
Four different layers of histone modifications can now be distinguished across inducible genes in mammalian cells. The first, which includes K4me3 across the start site as well as K79me2 and K36me3 in the coding region (Figure 1) are pre-existing, possibly epigenetic histone modifications detectable in quiescent G0 cells. The second relates to dynamic turnover of histone acetylation by the action of HATS and HDACs, which affects all K4me3-modified H3 in the mouse nucleus (Hazzalin and Mahadevan, 2005; reviewed in Clayton et al, 2006). Both of these are detectable in quiescent cells. Upon stimulation, a third signalling-dependent set, which includes H3 phosphorylation at IE genes (Chadee et al, 1999; Cheung et al, 2000; Clayton et al, 2000) or H4 acetylation at hsp70 (Thomson et al, 2004) becomes evident, also seen mostly across the start site. Finally, there are DRB-sensitive modifications restricted to coding regions of these genes that are dependent upon passage of Pol II, such as enhanced K4me3 and K36me3 (Figure 3C). Thus, a combination of epigenetic, dynamic, kinase- and Pol II-dependent histone modifications is required to fully explain their region and stimulus-specific appearance across these genes.
Setd2 mediates H3K36 trimethylation at the 3′ end of active gene coding regions
Enhancement of K36me3 peaks towards the 3′ end of the induced c-fos and c-jun genes. There is relatively low recovery of K36me2 and me1, and no increase upon stimulation. Further, mono- and dimethyl K36 states do not correlate with transcription as similar low levels (∼1% input) are detectable at active or inactive genes (Figure 5C hbb, and 5D, panels ii and iii). This emphasises the importance of considering quantitative aspects of ChIP when evaluating reports in yeast (Krogan et al, 2003; Morillon et al, 2005; Rao et al, 2005) and metazoa (Zhou et al, 2004; Bannister et al, 2005; Miao and Natarajan, 2005) showing that both di- and trimethylated K36 are associated with active genes.
The distinctive distribution of K4me3 (peaking 5′) and K36me3 (peaking 3′) across these active genes (Figure 1B and Supplementary Figure S1B, panels ii and iii) may result from selective methyltransferase recruitment to differentially phosphorylated Pol II, as shown in yeast (reviewed in Hampsey and Reinberg, 2003). MLL1, a mammalian H3K4-specific methyltransferase, is recruited to the 5′ end of active genes (Milne et al, 2002; Nakamura et al, 2002; Dou et al, 2005; Guenther et al, 2005). MLL1 or components of MLL1-containing complexes interact with Pol II phosphorylated at the Ser 5 position of its CTD (Hughes et al, 2004; Milne et al, 2005), the form of Pol II associated with 5′ coding regions (Komarnitsky et al, 2000). HYPB/SETD2 has been shown to coimmunoprecipitate with Ser 2-phosphorylated Pol II (Sun et al, 2005), suggesting that like Set2, it may ‘piggyback' with Pol II across the gene to direct co-transcriptional trimethylation of K36 at the 3′ coding regions of c-fos and c-jun. Attempts to use our N- and C-terminal antibodies to show association of HYPB/Setd2 with these regions by ChIP were unsuccessful (data not shown), but as we do not have a positive control for these experiments, they are not conclusive.
Selectivity and multi-enzyme systems for H3K36 methylation in mammals
In contrast to its homologue Set2, which is responsible for all three methyl states in S. cerevisiae, S. pombe and N. crassa (Adhvaryu et al, 2005; Morris et al, 2005; Chu et al, 2006), Setd2 is specifically responsible for K36 trimethylation in mouse cells (Figures 4C and 5B–D). This suggests a more complex multi-enzyme system for K36 methylation in higher eukaryotes, similar to that for K4 methylation. Set1 is the only enzyme shown to methylate K4 in yeast, whereas many enzymes exist in mammals, including hSet1, MLL1–4, Set7/9, SMYD2 and Meisetz (reviewed in Ruthenburg et al, 2007). Some of these are methyl-state specific. For example, Meisetz specifically catalyses trimethylation of K4 in vivo (Hayashi et al, 2005). Mouse NSD1 was previously shown to dimethylate K36 in vitro (Rayasam et al, 2003), but the mono- and trimethyl states were not analysed. In this study, NSD1 did not appear to be responsible for total nuclear mono-, di- or trimethyl K36 (Figure 4C, panels i–iii) nor that at c-fos and c-jun (data not shown). Other potential K36 mono- and/or dimethyltransferases include the closely related NSD2 and NSD3 proteins, or the Smyd2 protein recently found to dimethylate K36 in vitro (Brown et al, 2006). This study indicates an unexpectedly strict relationship between a specific state of H3K36 methylation and a single methyltransferase enzyme, a finding that may eventually be found relevant for other HMTases in mammals.
Materials and methods
Materials
Anisomycin, TSA and DRB were supplied by Sigma and rhEGF by Promega. siRNAs were synthesised by Dharmacon. PCR primers and 5′ FAM 3′ TAMRA probes were synthesised by Eurogentec.
Antibodies and peptides
H3K36me3 (ab9050), H3K36me1 (ab9048), H3K18ac (ab1191) and unmodified H3 (ab7834 for western and ab1791 for ChIP assays) antibodies were from Abcam. H3K36me2 (07-369), H3K9me3 (07-442), H3K14ac (07-353), H3K79me2 (07-366), H4K8ac (06-760), H4K12ac (06-761) and H4K16ac (06-762) antibodies were from Upstate, and Pol II antibody (sc-899) was from Santa Cruz. Anti-pan-acetyl H4 was a gift from Dr Tim Hebbes (University of Portsmouth). C-terminal MLL1 antibody was a gift from Dr James Hsieh (Washington University, St Louis).
In-house antibodies were raised by PTU/BS (Pentlands Science Park, Penicuik, Midlothian, UK), using peptides from Alta Bioscience (University of Birmingham). Rabbit anti-H3K9ac and anti-H3K4me3 antisera were raised against the peptides ARTKQTARK(ac)STGGKAPRKQLC and ARTK(me3)QTARKSGGC, respectively. N-Setd2 and C-Setd2 antisera were raised in rabbit and sheep, against the peptides MERRGKYTSKLERESKRTSEC and RGTKRPDDRYDTPTSKKKVRC, corresponding to amino acids 1–20 and 1560–1579 of mouse Setd2 (NCBI accession no.: XP_900231), respectively. All peptides were synthesised with C-terminal cysteine for coupling to keyhole limpet haemocyanin (KLH).
Cell culture
C3H 10T1/2 mouse fibroblasts were cultured in DMEM containing 10% (v/v) fetal calf serum (FCS) and 2 mM glutamine. Cells (70–90% confluent) were quiesced by transferring to DMEM with 0.5% (v/v) FCS and 2 mM glutamine for 24–28 h prior to stimulation and treatment as described.
Acid extraction of histones and western blotting
Extraction of histones, SDS–PAGE and western blotting were carried out essentially as described previously (Cano et al, 1995; Clayton et al, 2000). Setd2 and histones were resolved on 8 and 15% polyacrylamide gels, respectively. Antibodies were used at the following dilutions: H3K4me3 and H3K9ac, 1:10,000; H3K36me3, H3K36me2 and H3K79me2, 1:5000; H4K8ac, H4K12ac and H4K16ac, 1:4000; H3K14ac, MLL1, C-Setd2 and N-Setd2, 1:2500; H3K36me1, H3K18ac and unmodified H3, 1:2000; H3K9me3 and pan-acetyl H4, 1:1000.
Transfection with siRNA
Cells were cultured to 40–60% confluence before washing with, and transferring to OptiMEM® I medium (Invitrogen). Cells were transfected with siRNA (50 nM in final medium) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 5 h, an equal volume of DMEM medium containing 20% (v/v) FCS and 2 mM glutamine was added. Total RNA was extracted 24 or 48 h after transfection as described below. For protein analysis, cells were harvested in lysis buffer (0.2% (v/v) Triton X-100, 10 mM HEPES pH 8, 0.25 M sucrose, 1.5 mM MgCl2, 10 mM NaCl, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 100 μM sodium orthovanadate, 1 μM microcystin and protease inhibitors (Mahadevan and Bell, 1990)) 48 h after transfection. Crude nuclei were pelleted by centrifugation and resuspended in SDS–PAGE sample buffer. Cells were quiesced 24 h after transfection as described above. Crosslinked mononucleosomes were prepared 48 h after transfection as detailed below.
RNA extraction, qRT–PCR and northern blotting
Total RNA was extracted using TRIzol® reagent (Invitrogen) according to the manufacturer's instructions. For qRT–PCR, contaminating DNA was removed using the TURBO DNA-free™ kit (Ambion), and 0.5 μg of RNA was reverse-transcribed using the SuperScript™ III First-Strand Synthesis kit (Invitrogen). cDNA was then quantified by real-time PCR. For northern analysis, 5 μg of RNA was resolved on formaldehyde/agarose gels, transferred to nylon membrane (Hybond-N+, Amersham) and hybridised with probes as detailed in Hazzalin et al (1997). c-fos (+2091 –3109) and c-jun (+1853 –2920) 3′ gene fragments for probes were amplified by PCR from cDNA, labelled by random priming (Amersham RediPrime II kit) and sequentially hybridised to membranes.
Preparation of crosslinked mononucleosomes and ChIP
Formaldehyde crosslinked mononucleosomes were prepared as described in Macdonald et al (2005). Antibodies (10 μl H3K36me3/Pol II or 5 μl H3K36me2/H3K36me1/H3K79me2/H3K4me3 serum/H3K9ac serum/Pan-acetyl H4 serum) were added to chromatin aliquots (∼25–30 μg DNA) and rotated for 2 h at 4°C. Protein A-sepharose beads (∼5 mg) pre-blocked with 10 μg sonicated herring sperm DNA and BSA (0.02% final concentration after addition to chromatin) were then added and samples rotated for a further 2 h at 4°C. Beads were recovered by centrifugation, and unbound supernatant chromatin was retained. Beads were washed sequentially with 3 × RIPA buffer (Clayton et al, 2000) without phosphatase or protease inhibitors, 3 × RIPA containing 500 mM NaCl and 2 × TE (10 mM Tris–HCl pH 7.5, 1 mM EDTA). Input and antibody-bound chromatins were incubated at 65°C overnight to reverse crosslinks. Samples were treated with RNase A (100 μg/ml, 1 h, 37°C) followed by proteinase K (100 μg/ml, 3 h, 37°C) before phenol/chloroform extraction of DNA. Extracted DNA was ethanol precipitated and redissolved in 10 mM Tris–HCl pH 8.0, 0.1 mM EDTA. The presence of specific gene sequences was then quantified by real-time PCR.
Real-time PCR
Real-time PCR was performed using the ABI Prism 7000 sequence detection system (Applied Biosystems). All primers were designed with Primer Express 2.0 software (Applied Biosystems). All c-fos, c-jun, gapdh and hbb amplicons were detected by the 5′ nuclease (Taqman) assay using 5′ FAM 3′ TAMRA-labelled probes and qPCR Mastermix Plus (Eurogentec). All other amplicons were detected by the SYBR® Green assay using SYBR® Green PCR Master Mix (Applied Biosystems). Sequences of primers and probes and detailed conditions are available on request. PCR reactions were carried out in triplicate or duplicate.
For ChIP assays, standard curves (serially diluted sonicated genomic DNA) were included in every plate to ensure bound and input sample dilutions were within the linear range. Data were analysed by the CT method and plotted as % input recovered in the bound fraction for each amplicon, calculated by the following formula: % input recovery=[100/(input fold dilution/bound fold dilution)] × 2(input CT - bound CT), where no fold dilution=1. For qRT–PCR, data were analysed by relative quantification using the standard curve method (see Applied Biosystems user bulletin 2). All raw data from micrococcal nuclease sensitivity and ChIP experiments are available upon request.
Note added in proof
During review of this manuscript, Bell et al (2007) reported that in Drosophila, dHypb is responsible for the trimethylated state of H3K36, in agreement with our findings in mammalian cells. However, the global increase in H4K16 acetylation reported upon dHypb knockdown in Drosophila is not seen in mammalian cells used here (Figure 4C).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Information
Acknowledgments
We thank Drs Fiona Myers and Alan Thorne (University of Portsmouth) for advice on real-time PCR in the early stages of this work, and Dr James Hsieh (Washington University, St Louis) for the MLL1 antibody. We thank all members of the Nuclear Signalling Laboratory for help and discussions throughout the course of this work. JWE is supported by an MRC:CASE award with Syngenta. This work was funded by a Wellcome Trust Programme Grant (065373/Z/01/Z).
References
- Adhvaryu KK, Morris SA, Strahl BD, Selker EU (2005) Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot Cell 4: 1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2005) Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736 [DOI] [PubMed] [Google Scholar]
- Barratt MJ, Hazzalin CA, Cano E, Mahadevan LC (1994) Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc Natl Acad Sci USA 91: 4781–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, Macalpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, Garza D, Peters AH, Schübeler D (2007) Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J (advance online publication, 15 November 2007, doi:10.1038/sj.emboj.7601926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, Schreiber SL, Lander ES (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES (2007) The mammalian epigenome. Cell 128: 669–681 [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15: 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Sims RJ III, Gottlieb PD, Tucker PW (2006) Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer 5: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano E, Hazzalin CA, Kardalinou E, Buckle RS, Mahadevan LC (1995) Neither ERK nor JNK/SAPK MAP kinase subtypes are essential for histone H3/HMG-14 phosphorylation or c-fos and c-jun induction. J Cell Sci 108: 3599–3609 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Chadee DN, Hendzel MJ, Tylipski CP, Allis CD, Bazett-Jones DP, Wright JA, Davie JR (1999) Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J Biol Chem 274: 24914–24920 [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD (2000) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 5: 905–915 [DOI] [PubMed] [Google Scholar]
- Chu Y, Sutton A, Sternglanz R, Prelich G (2006) The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol Cell Biol 26: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AL, Hazzalin CA, Mahadevan LC (2006) Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell 23: 289–296 [DOI] [PubMed] [Google Scholar]
- Clayton AL, Mahadevan LC (2003) MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett 546: 51–58 [DOI] [PubMed] [Google Scholar]
- Clayton AL, Rose S, Barratt MJ, Mahadevan LC (2000) Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J 19: 3714–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon V, Veyrune JL, Tourkine N, Vie A, Hipskind RA, Blanchard JM (1999) A novel calcium signaling pathway targets the c-fos intragenic transcriptional pausing site. J Biol Chem 274: 30439–30446 [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG (2005) Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121: 873–885 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A, Dorokhov N, Michener DE (2007) Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment. Mol Genet Genomics 277: 101–114 [DOI] [PubMed] [Google Scholar]
- Farris SD, Rubio ED, Moon JJ, Gombert WM, Nelson BH, Krumm A (2005) Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A. Z J Biol Chem 280: 25298–25303 [DOI] [PubMed] [Google Scholar]
- Gerber M, Shilatifard A (2003) Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem 278: 26303–26306 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA (2005) Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci USA 102: 8603–8608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D (2003) Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113: 429–432 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y (2005) A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378 [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Cuenda A, Cano E, Cohen P, Mahadevan LC (1997) Effects of the inhibition of p38/RK MAP kinase on induction of five fos and jun genes by diverse stimuli. Oncogene 15: 2321–2331 [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Mahadevan LC (2002) MAPK-regulated transcription: a continuously variable gene switch? Nat Rev Mol Cell Biol 3: 30–40 [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Mahadevan LC (2005) Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol 3: e393 (doi:10.1371/journal.pbio.0030393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, Kay GF, Hayward NK, Hess JL, Meyerson M (2004) Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 13: 587–597 [DOI] [PubMed] [Google Scholar]
- Im H, Park C, Feng Q, Johnson KD, Kiekhaefer CM, Choi K, Zhang Y, Bresnick EH (2003) Dynamic regulation of histone H3 methylated at lysine 79 within a tissue-specific chromatin domain. J Biol Chem 278: 18346–18352 [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD (2005) A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 25: 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskouti A, Talianidis I (2005) Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J 24: 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A (2002) COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753–10755 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP (2002) Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 277: 49383–49388 [DOI] [PubMed] [Google Scholar]
- Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, Jones PA (2004) Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci USA 101: 7357–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, Moggs JG, Orphanides G, Thomson S, Edmunds JW, Clayton AL, Endicott JA, Mahadevan LC (2005) Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3. Mol Cell 20: 199–211 [DOI] [PubMed] [Google Scholar]
- Mahadevan LC, Bell JC (1990) Phosphate-labelling studies of receptor tyrosine kinases. In Receptors: a Practical Approach, Hulme EC (ed), pp 181–201. Oxford: IRL press [Google Scholar]
- Mechti N, Piechaczyk M, Blanchard JM, Jeanteur P, Lebleu B (1991) Sequence requirements for premature transcription arrest within the first intron of the mouse c-fos gene. Mol Cell Biol 11: 2832–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao F, Natarajan R (2005) Mapping global histone methylation patterns in the coding regions of human genes. Mol Cell Biol 25: 4650–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL (2005) MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci USA 102: 14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J (2005) Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell 18: 723–734 [DOI] [PubMed] [Google Scholar]
- Morris SA, Shibata Y, Noma K, Tsukamoto Y, Warren E, Temple B, Grewal SI, Strahl BD (2005) Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot Cell 4: 1446–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 10: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Nelson CJ, Santos-Rosa H, Kouzarides T (2006) Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126: 905–916 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Rao B, Shibata Y, Strahl BD, Lieb JD (2005) Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol 25: 9447–9459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, Lerouge T, Hager GL, Chambon P, Losson R (2003) NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J 22: 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Schaft D, Roguev A, Kotovic KM, Shevchenko A, Sarov M, Neugebauer KM, Stewart AF (2003) The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res 31: 2475–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2004) Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 6: 73–77 [DOI] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A (2006) Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Belotserkovskaya R, Reinberg D (2004) Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z (2005) Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 280: 35261–35271 [DOI] [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Mahadevan LC (2001) Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol Cell 8: 1231–1241 [DOI] [PubMed] [Google Scholar]
- Thomson S, Hollis A, Hazzalin CA, Mahadevan LC (2004) Distinct stimulus-specific histone modifications at hsp70 chromatin targeted by the transcription factor heat shock factor-1. Mol Cell 15: 585–594 [DOI] [PubMed] [Google Scholar]
- Thomson S, Mahadevan LC, Clayton AL (1999) MAP kinase-mediated signalling to nucleosomes and immediate-early gene induction. Semin Cell Dev Biol 10: 205–214 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA (2006) Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 26: 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 17: 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Deng L, Lacoste V, Park HU, Pumfery A, Kashanchi F, Brady JN, Kumar A (2004) Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J Virol 78: 13522–13533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Information