Abstract
In the human malaria parasite Plasmodium falciparum, a member of the sirtuin family has been implicated in the epigenetic regulation of virulence genes that are vital to malaria pathogenesis and persistence. This eukaryotic sirtuin, PfSir2, is divergent in sequence from those characterized thus far and belongs to the phylogenetic class that contains primarily eubacterial and archaeal sirtuins. PfSir2 cofractionates with histones in blood-stage parasites, and the recombinant enzyme efficiently deacetylates the N-terminal tails of histones H3 and H4. In addition, PfSir2 can ADP-ribosylate both histones and itself, an activity that is minimal or absent in most sirtuins with significant deacetylase activity. Strikingly, the deacetylase activity of PfSir2 is dependent on its ADP-ribosylation. Finally, although PfSir2 is not affected by established sirtuin inhibitors, it can be completely inhibited by nicotinamide, a natural product of the sirtuin reaction. This study shows that PfSir2 has the appropriate characteristics to be a direct regulator of chromatin structure in P. falciparum. It also raises the significant possibility that both ADP-ribosylation and deacetylation of histones could be sirtuin-regulated modulators of chromatin structure in this species.
Class III deacetylases are a family of NAD+-dependent deacetylase enzymes that are widely conserved from archaea to humans (21). They are also termed “sirtuins” after the founding member of the family, Saccharomyces cerevisiae ‘silencing information regulator 2’ (Sir2). ScSir2 plays a central role in epigenetic transcriptional control, acting as a histone deacetylase (HDAC) to establish regions of silent heterochromatin at subtelomeres, mating-type loci, and ribosomal DNA within the budding yeast genome (23). At least one of the seven sirtuins in humans, HsSirT1, appears to have a similar role in establishing gene silencing in human cells (61), suggesting that this function has been conserved through evolution. Many other sirtuins do not, however, act efficiently on histone substrates, including two additional nuclear sirtuins in humans (45), while others act on substrates in the cytoplasm or mitochondria. The roles of these sirtuins are generally not completely understood, but various members of the family have been assigned to processes such as longevity control (reviewed reference 26), DNA repair (22, 48), and the control of metabolic enzymes (27, 28).
All the sirtuin proteins crystallized so far have the same basic structure: a large NAD+-binding “Rossmann fold” domain and a smaller domain consisting of a zinc-binding module plus a variable, primarily helical, module (17, 46, 66). Sirtuins have a complex reaction mechanism compared to NAD+-independent (“class I and II”) deacetylases, involving the breakdown of an NAD+ cofactor to produce nicotinamide and O-acetyl-ADP-ribose as well as the deacetylated lysine product. This mechanism allows sirtuins to act as both deacetylases and ADP-ribosyltransferases (51). In fact, both ScSir2 and a human enzyme, HsSirT2, were initially shown to have ADP-ribosyltransferase activity rather than deacetylase activity: ScSir2 ADP-ribosylates both itself and its histone substrates, albeit at a very low level (20, 56). It was subsequently demonstrated, however, that a second, more significant activity, the deacetylation of histones H3 and H4, correlated with the effects of ScSir2 on transcriptional silencing and life span extension (34). It now appears that both deacetylation and ADP-ribosylation by different sirtuins are of biological importance, with certain sirtuins, such as ScSir2, being biased towards deacetylation and others, such as HsSirT6 and HsSirT4, towards ADP-ribosylation (27, 40).
In several eukaryotic microbial pathogens, including Plasmodium spp., variantly expressed gene families encoding virulence determinants are located in potentially heterochromatic, subtelomeric regions of the genome. This observation suggests that there may be a common sirtuin-mediated mechanism for controlling antigenic variation in these pathogens (43). In the human malarial parasite Plasmodium falciparum, a sirtuin homolog, PfSir2, was recently shown to be involved in the epigenetic transcriptional control of a large number of subtelomeric genes that are vital for virulence (15). These included the multigene families var and rifin, both of which encode variantly expressed antigens exposed on the surface of infected erythrocytes during a blood-stage malarial infection (11, 38, 54). When the PfSIR2 gene was experimentally disrupted, mutually exclusive expression of var genes was abolished, and the expression of many var genes (and also some rifin genes) was simultaneously upregulated (15). In a complementary study, semiquantitative chromatin immunoprecipitation showed that H4 was more highly acetylated within an active subtelomeric var gene than within a silent one and that this hyperacetylation was mutually exclusive with the presence of PfSir2 (19). All of this evidence points to the idea that heterochromatic silencing, mediated by PfSir2, could directly control the expression of var virulence genes in P. falciparum.
In this study we have characterized the PfSir2 protein, establishing it as a genuine HDAC with the ability to act on Plasmodium histones. In P. falciparum, the protein was found in the parasite nucleus, where it cofractionated with histones. Furthermore, PfSir2 could partially complement a sir2 mutant in the heterologous species Schizosaccharomyces pombe. Strikingly, in addition to deacetylase activity, PfSir2 had robust ADP-ribosyltransferase activity. PfSir2 therefore possesses all the necessary characteristics to contribute to the epigenetic control of virulence genes in P. falciparum.
MATERIALS AND METHODS
Recombinant protein preparation from P. falciparum.
P. falciparum was cultured in human O+ erythrocytes using standard procedures (59). The PfSIR2 gene was amplified by PCR from genomic DNA (P. falciparum 3D7 strain) and cloned into an expression vector under control of the PfHsp86 promoter (65) with a C-terminal His6 tag. Transformation into parasites was carried out as previously described (14). Cytoplasmic and nuclear extracts of transformed parasites were made as described previously (63), using mixed-stage cultures at 3 to 5% parasitemia. For Ni-nitrilotriacetic acid (Ni-NTA) purification of PfSir2, parasite extracts were made as described previously (63) but using a modified buffer with sodium phosphate buffer replacing HEPES. All soluble fractions were then pooled and bound to Ni-NTA resin (Novagen, Madison, WI). The resin was extensively washed with 50 mM Na2HPO4-300 mM NaCl-20 mM imidazole, and the protein was eluted in the same buffer with 250 mM imidazole.
Recombinant protein preparation from Escherichia coli.
The PfSIR2 gene was cloned into the pET-21a vector (Novagen) between the BamHI and XhoI sites. BL21(DE3)/pLysS E. coli (Stratagene, La Jolla, CA) was transformed with this plasmid, grown to an optical density of ∼0.5, and induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight at room temperature. Cells were then lysed with “Bugbuster” reagent (Novagen) containing 5 U/ml Benzonase (Novagen) and “Complete” protease inhibitors (Roche, Basel, Switzerland). Protein was purified as described previously (49). Protein-containing fractions were concentrated to ∼2 mg/ml on Amicon Ultra 5-kDa-cutoff columns (Millipore, Billerica, MA) and then exchanged into 50 mM Tris-HCl (pH 8.0)-100 mM NaCl-0.1 mM dithiothreitol-0.01% sodium azide with the addition of 20% glycerol for storage at −20°C.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
SDS-10% polyacrylamide gels and 0.45-μm pore-size nitrocellulose were used for Sir2-His6 blots; 15% gels and 0.2-μm-pore-size nitrocellulose were used for histone blots. All samples were reduced with dithiothreitol before electrophoresis. Primary antibodies were as follows: mouse anti-tetra-His (QIAGEN, Valencia, CA), 1:1,000; mouse anti-T7 tag (Novagen), 1:10,000; rabbit anti-H3 (Abcam Inc, Cambridge, MA), 1:1,000; rabbit anti-AcH3 (Lys9,14) (Upstate, Charlottesville, VA), 1:500; and rabbit anti-AcH4 (Lys5,8,12,16) (Upstate), 1:500. Secondary horseradish peroxidase-linked anti-mouse and anti-rabbit antibodies (sheep) were from Amersham, Piscataway, NJ, and detection was via ECL (Pierce, Rockford IL). Bands were quantitated by using AlphaEase FC densitometry software (Alpha Innotech, San Leandro, CA).
Deacetylase assays.
Assays using substrate A, ZMAL, were carried out as described previously (31), using 2.5 to 7.5 μg PfSir2. Reaction mixtures were incubated at 37°C for 16 h, and fluorescence was read in an fmax plate reader (Molecular Devices) (355 nm excitation/460 nm emission). All assays were performed in triplicate. Assays were carried out identically using substrate B (41), substituting 0.54 μM of the acetylated peptide for ZMAL. Product lysis was achieved with thermolysin instead of trypsin, and fluorescence was read at 540 nm/585 nm.
Assays testing the effects of treatment with nicotinamide or other drugs (splitomicin, sirtinol, and resveratrol) were carried out using substrate A and 5 μg PfSir2, as described above. The drug was dissolved in dimethyl sulfoxide and added at between 25 μM and 10 mM (constant final concentration of 1% dimethyl sulfoxide). The reaction rate was expressed as a percentage of the fluorescence from the uninhibited reaction.
Histone deacetylation assays were carried out as described previously (57), using 500 μM NAD+, 5 μg Sir2, and 5 μg histones (either from sodium-butyrate-treated P. falciparum or from sodium butyrate-treated HeLa cells [Upstate]).
P. falciparum histone preparation.
Histones were prepared from trophozoite-stage cultures of P. falciparum. For deacetylation assays, parasites were subjected to 2 h of pretreatment with 5 mM sodium butyrate before collection. Nuclear extraction was carried out as described previously (63), and then the remaining insoluble material containing DNA and histones was treated twice with 0.5 M ice-cold HCl for 1 h. The acid-extractable protein was precipitated with trichloroacetic acid (TCA), washed in ice-cold acetone, and resuspended in double-distilled water.
Schizosaccharomyces pombe complementation assay.
The complementation assay was carried out as described previously (18). PfSIR2 was amplified from 3D7 genomic DNA and cloned into the an S. pombe nmt+ thiamine-repressible vector, pREP-1 (42). Wild-type S. pombe and a sir2Δ strain, SPY48, carrying the URA4 gene inserted into a centromeric repeat region of the genome, were transformed via the lithium acetate method with either the PfSir2 construct or the empty vector, and 10-fold dilutions of the yeast were plated on EMM-leucine with 5-fluoroorotic acid (5-FOA) and 15 mM thiamine. Four to 5 days of growth was allowed at 30°C before imaging the plates.
ADP-ribosylation assay.
The ADP-ribosylation assay was carried out as described previously (56), with the addition of EDTA-free “Complete” protease inhibitors (Roche). Each reaction used ∼5 μg PfSir2 or Hst2, 5 μg P. falciparum histones and/or 5 μg bovine serum albumin (BSA), and 3μCi [32P]NAD+, giving a final concentration of 375 μM NAD+. Directly before TCA precipitation, 5 μg BSA was added to all reaction mixtures lacking BSA, to act as a carrier. For high resolution of histones, Tris-Tricine gels were used and stained with SYPRO Ruby protein stain (Invitrogen) instead of Coomassie blue. For phosphodiesterase treatment, 5 μg phosphodiesterase (Sigma) was added to a completed reaction in 100 mM Tris-HCl (pH 8.9), 100 mM NaCl, and 15 mM MgCl2 and then incubated for 30 min at 37°C before TCA precipitation. For the repurification of PfSir2 after such a treatment, 60 μg PfSir2 was incubated with 60 μg phosphodiesterase for 2 h at 37°C in the above buffer, and the mixture was diluted five times in 150 mM NaCl and then bound to Ni-NTA and eluted as before.
RESULTS
PfSir2 is a putative NAD+-dependent HDAC.
PfSir2 is one of the smallest eukaryotic sirtuins, having only 273 residues; it is smaller than any of the S. cerevisiae or Homo sapiens sirtuins (357 to 562 and 310 to 747 residues, respectively). In fact, PfSir2 more closely resembles the Archaeoglobus fulgidus sirtuin Af1 in size and overall sequence similarity. Moreover, a classification of sirtuins from across the evolutionary spectrum into four classes by molecular phylogeny places PfSir2 into class III, the class containing Af1, many eubacterial sirtuins, and only a single human homolog, the mitochondrial protein SirT5 (21).
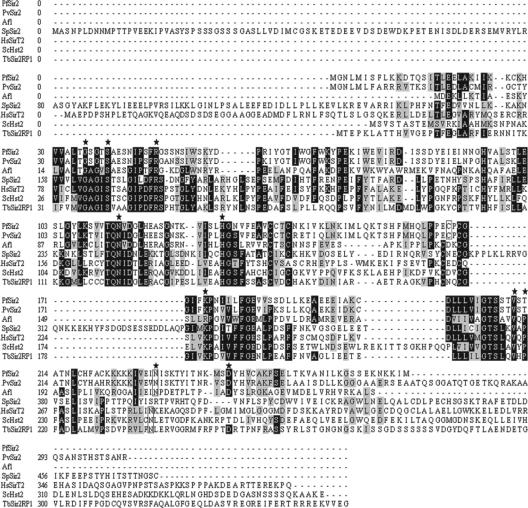
Figure 1 shows the predicted protein sequence of PfSir2 aligned with sirtuin sequences from archaea, yeast, and human for which the encoded proteins have been crystallized and structures solved. The alignment also includes a sirtuin from Plasmodium vivax and the recently characterized sirtuin from Trypanosoma brucei, TbSIR2RP1. The core sirtuin domains of PfSir2 are relatively poorly conserved with the human, yeast, and Archeoglobus sirtuins (23 to 35%), but there is nevertheless identity or similarity between PfSir2 and the extensively studied S. cerevisiae sirtuin Hst2 at all but one of the residues known to be important for catalytic activity (36) (Fig. 1). For example, the two pairs of cysteines required for zinc binding (Cys140, Cys143, Cys168, and Cys170 in PfSir2) remain conserved, and it has previously been noted that considerable diversity is tolerated within the zinc-binding module outside of these residues (17). PfSir2 is also broadly conserved at the most invariant sirtuin motifs, the residues of which form an NAD+-binding pocket in the Af2 crystal structure (2). Here, it has only three substitutions: GAGXS/GSGXS, GIPXFR/NIPXFR, and TQNIDXL/TQNVDXL.
FIG. 1.
PfSir2 aligned with a selection of crystallized sirtuins from other species. The predicted sequence of PfSir2 is aligned with the sequences of A. fulgidus Af1, S. cerevisiae Hst2, S. pombe Sir2, H. sapiens SirT2, T. brucei SIR2RP1, and the predicted homolog in P. vivax, PvSir2. Strictly conserved and conserved residues are shaded in black and gray, respectively. Starred residues are essential for catalytic activity in Hst2 (36), and where necessary, essential residues with slightly different spacings in different sirtuins are boxed to indicate their presence.
PfSir2 has several changes at residues implicated in inhibition of the deacetylation reaction by nicotinamide. Of the residues that form a nicotinamide-binding pocket in Hst2 (50), Glu is changed to Arg62 and Phe to Gly65 in PfSir2. This phenylalanine is also altered in the Af1 and Af2 proteins, to alanine, possibly pointing to an altered potential for nicotinamide inhibition among all these class III enzymes. Similarly, most of the residues which contact an acetyl-lysine substrate in a ternary complex of Hst2, substrate, and product (67) remain conserved in PfSir2, although there are two conservative substitutions, Val to Ile179 and Leu to Val185, as well as one nonconservative change, Pro to Thr214. This could influence any binding of PfSir2 to acetyl-lysine substrates, altering the efficiency or specificity of this putative HDAC enzyme. Interestingly, most but not all of the alterations described here are also present in the P. vivax sirtuin homolog, which shares 80% similarly with PfSir2.
Outside of the core enzymatic domain, the N and C termini of the larger eukaryotic sirtuins are thought to mediate protein-protein interactions and/or regulate activity. For example, the N- and C-terminal domains in Hst2 have both been assigned functional roles: the C-terminal helices self-interact with the NAD+-binding site, causing autoinhibition, and this region also bears a nuclear export signal which keeps the enzyme primarily cytoplasmic and limits its nuclear deacetylase activity. The N-terminal seven residues of Hst2 mediate trimerization by interacting with, and apparently blocking, the substrate-binding cleft of an adjacent molecule (64, 66). Figure 1 shows that PfSir2 has a small N-terminal extension similar in length to that of Hst2, which may therefore be sufficient to play a functional role. The C terminus of PfSir2, however, is almost as short as that of the very minimal A. fulgidus sequence.
In order to support some of this in silico analysis with experimental evidence, we sought to express a recombinant PfSir2 protein.
Production and characterization of recombinant PfSir2.
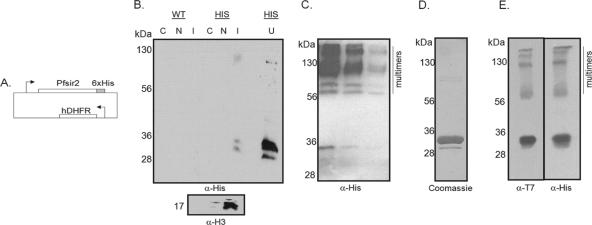
The PfSIR2 gene was cloned from P. falciparum genomic DNA into a vector containing a C-terminal His6 tag and a strong P. falciparum promoter, the Pfhsp86 promoter (65) (Fig. 2A). This was transfected as an episome into the 3D7 strain of P. falciparum, and biochemical fractionation of transgenic parasites showed that tagged PfSir2 was in the insoluble chromatin-containing fraction together with the great majority of histones (Fig. 2B). This suggests that PfSir2 has the appropriate location for an HDAC enzyme and is bound to chromatin (or possibly another insoluble nuclear component), either directly or via protein-protein interactions. PfSir2 was resistant to extraction with 800 mM KCl, possibly pointing to a hydrophobic protein-protein interaction. Limited amounts of soluble PfSir2 could, however, be released using modified buffer conditions, and when this protein was purified from parasite extract via Ni-NTA, it eluted as a series of multimers that were at least partially resistant to SDS-PAGE (Fig. 2C).
FIG. 2.
Expression and purification of PfSir2. (A) Schematic of the construct used to express His6-tagged Sir2 in P. falciparum. (B) Western blots of extracts from mixed-stage parasites expressing His6-tagged PfSir2 (HIS) or from nonrecombinant 3D7 parasites (WT). Parasites were fractionated to give cytosolic proteins (C), soluble nuclear proteins (N), and proteins from the remaining insoluble material (I). PfSir2 (α-His) cofractionates with histones (α-H3) in the insoluble, chromatin-containing fraction. Unfractionated parasites are also shown (U). (C) Western blot of His-tagged PfSir2 from P. falciparum, purified via Ni-NTA. Successive elution fractions with 250 mM imidazole are shown. The recombinant protein (α-His) appears as a series of multimers. (D) Coomassie blue-stained SDS-polyacrylamide gel showing recombinant PfSir2 produced in E. coli (eluate from Ni-NTA). (E) Western blots of the Ni-NTA eluate shown in panel D, using antibodies against the His6 tag or T7 tag.
In order to produce greater quantities of recombinant PfSir2, the gene was expressed in E. coli with a C-terminal His6 tag and an N-terminal T7 tag. The recombinant protein was produced in a soluble form with the expected size of ∼30 kDa, purified via Ni-NTA, and eluted at high purity (judged from SDS-PAGE and Coomassie blue staining) (Fig. 2D). Western analysis and mass spectrometry confirmed that the major, upper band of this doublet was indeed full-length, N- and C-terminally tagged PfSir2 (Fig. 2E). Several peptides, including the His6-containing peptide, were not detected in the lower, minor band, and this may be a C-terminally truncated version of the protein, although since purification was via the C-terminal His tag, a truncated protein could be purified only via tight binding to a second, full-length molecule. Alternatively, one band of the doublet might represent a modified form, and data in Fig. 6 do indeed show that the upper band is ADP-ribosylated, as has also been observed in recombinant ScSir2 (56).
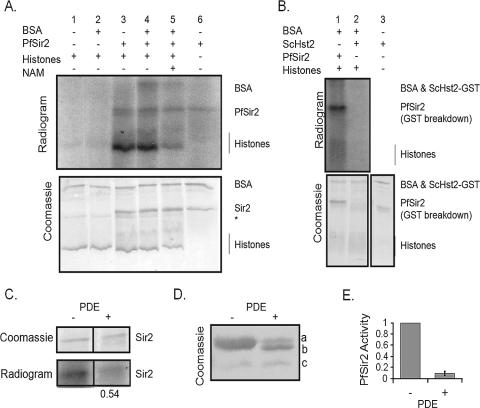
FIG. 6.
Recombinant PfSir2 has ADP-ribosyltransferase activity. (A) P. falciparum histones and/or BSA was treated with PfSir2 and [32P]NAD+ for 30 min at 37°C and then precipitated with TCA and subjected to SDS-PAGE. The Coomassie blue-stained SDS-polyacrylamide gel shows the positions of PfSir2, histones, BSA, and a minor breakdown product of BSA marked *. The autoradiogram of the gel shows ADP-ribosylated proteins. Lanes 1 and 2, substrates incubated with [32P]NAD+ in the absence of PfSir2. Lane 1, histones; Lane 2, histones plus BSA. Lanes 3 to 6, substrates incubated with [32P]NAD+ in the presence of PfSir2. Lane 3, histones; lane 4, histones plus BSA; lane 5, histones plus BSA plus 10 mM nicotinamide (NAM); lane 6, PfSir2 alone. (B) Histones and BSA were treated with either PfSir2 or GST-tagged Hst2, as for panel A. The Coomassie blue-stained SDS-polyacrylamide gel shows the positions of PfSir2, histones, BSA, and GST-Hst2. Lane 3 is included to show the size of GST-Hst2, which is very close to the size of BSA. The autoradiogram of the gel shows ADP-ribosylation by PfSir2 but not by GST-Hst2. (C) After incubation of PfSir2 with histones and [32P]NAD+ as described for panel A, 5 μg phosphodiesterase (PDE) was added to half of the reaction mixture and incubated for a further 30 min at 37°C before TCA precipitation. Radiolabel on the treated protein was calculated as a percentage of that on the nontreated protein, normalized to the amount of Coomassie blue-stained Sir2 in each lane. (D) Coomassie blue-stained SDS-polyacrylamide gel with extensive resolution, showing PfSir2 incubated for 2 h with PDE or with buffer alone (−) and then repurified on Ni-NTA. a, original upper band; b, PDE-modified upper band; c, original lower band, unaltered by PDE. (E) In vitro deacetylation assays with substrate A were carried out in duplicate using PfSir2 treated as described for panel D. Activity is expressed as a percentage of the fluorescence generated by untreated PfSir2. The error bar indicates standard deviation.
In addition to the ∼30-kDa band of recombinant PfSir2, Western analysis revealed several minor, higher-molecular-mass forms of the protein resembling the multimers observed when the protein was purified from P. falciparum (Fig. 2E). This suggested that PfSir2 had an inherent tendency to form dimers, tetramers, and larger multimers. Indeed, native PAGE and fast protein liquid chromatography confirmed that the protein existed as a very large multimer in solution (data not shown), and dynamic light scattering measurements estimated a uniform size for these multimers of ∼1,200 kDa (approximately 40 times the molecular mass of the monomer).
PfSir2 acts as a lysine deacetylase in vitro.
Two independent in vitro assays showed that recombinant PfSir2 from E. coli had NAD+-dependent deacetylase activity, as shown schematically in Fig. 3A. Both assays are based on synthetic acetylated substrates which, upon deacetylation, produce a compound that becomes fluorescent after trypsin treatment. Substrate A in Fig. 3B is an acetylated-lysine (AcLys) derivative termed ZMAL (30), and substrate B is a 19-residue synthetic peptide modeled on the tail of human p53, containing a single AcLys (41). In both assays PfSir2 showed deacetylase activity that was dependent upon NAD+ and about 7 times lower than the activity of a positive control, recombinant glutathione S-transferase (GST)-tagged Hst2 (data not shown). Hst2 has previously been shown to possess strong protein deacetylase activity (39). A non-HDAC protein prepared from E. coli and used as a negative control had no effect on substrate A (see Fig. S1 in the supplemental material), so the apparent deacetylase activity of PfSir2 cannot be attributed to nonspecific protease activities.
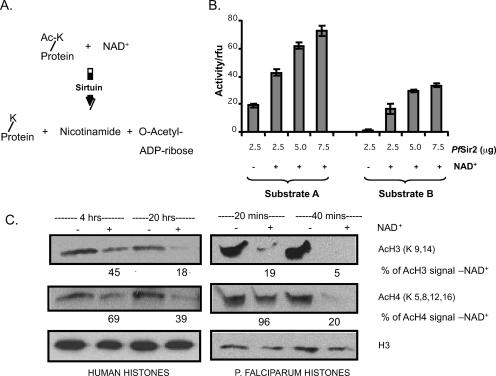
FIG. 3.
Recombinant PfSir2 has NAD+-dependent lysine deacetylase activity. (A) Reaction scheme for NAD+-dependent deacetylase enzymes. (B) Two independent in vitro deacetylase assays were carried out on increasing amounts of PfSir2, with or without 500 μM NAD+. Substrate A is a synthetic AcLys derivative termed ZMAL, and substrate B is a 19-residue synthetic peptide bearing a single AcLys. Assays were carried out in triplicate at 37°C for 16 h, and deacetylase activity is represented in relative fluorescence units (rfu). Error bars indicate standard deviations.(C) Western blots showing histones from sodium butyrate-treated HeLa cells or P. falciparum trophozoites, incubated with recombinant PfSir2 in the presence (+) or absence (−) of 500 μM NAD+ for the indicated times and then probed with antibodies against AcH3, AcH4, or the C-terminal portion of H3 (i.e., total H3). Ratios of acetylated histone signal in the NAD+ (+) reaction to that in the NAD+ (−) reaction are indicated at each time point. Each ratio is normalized to the ratio of total H3 signals in NAD+ and NAD+ lanes.
Having established that recombinant PfSir2 could deacetylate synthetic substrates, we wanted to determine whether it could also deacetylate histones, a probable native substrate. This was tested using commercially available histones purified from HeLa cells and also histones purified from P. falciparum itself. These were Western blotted and probed with antibodies specific to acetyl-lysine residues on H3 and H4.
The H3 and H4 tails are almost completely conserved between P. falciparum and H. sapiens (44), and PfSir2 effectively deacetylated H3 and H4 from either source (Fig. 3C). PfSir2 generally appeared to prefer H3 as a substrate over H4, and P. falciparum histones were deacetylated more efficiently, particularly in the case of H3, than commercial preparations of HeLa histones. Previous studies of other sirtuins have reported significant preferences for particular acetylated residues, especially Lys16 of H4 and Lys9 of H3, which may be important components of the histone code relating to epigenetic silencing. It was not possible to establish conclusively whether PfSir2 preferred a single residue in H3 because the antibody recognized both AcLys9 and AcLys14, while an antibody specific for AcLys14 does not react well with P. falciparum histones (44), perhaps due to a single residue change at position 12, Gly to Ala. However, since the overall AcH3 tail signal was readily reduced to near-undetectable levels, both Lys9 and Lys14 are clearly deacetylated efficiently.
In regard to H4, there are four potentially modified lysines in the N-terminal tail, Lys5, Lys8, Lys12, and Lys16. Again, it was not possible to conclusively determine a preference for a particular residue because only antibodies to AcLys8 and AcLys12 gave detectable signals. PfSir2 had no activity on AcLys8 and only a very limited activity on AcLys12 (data not shown). It may, however, act more strongly on AcLys5 or AcLys16.
PfSir2 is inhibited by nicotinamide but not by other established sirtuin inhibitors.
We next investigated whether the deacetylase activity of PfSir2 could be specifically inhibited with small molecules. Several inhibitors and activators of ScSir2 have been identified, including the inhibitors nicotinamide (5), sirtinol (25), and splitomicin (3) and the activators isonicotinamide (52) and resveratrol (33). All of these were tested on PfSir2 using the in vitro assay with substrate A.
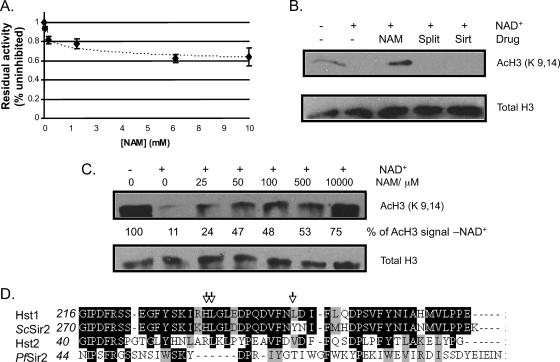
Nicotinamide is a natural product of the NAD+-dependent deacetylase reaction (Fig. 3A), which can promote a back reaction (“nicotinamide exchange”) at the expense of the forward reaction (53). This compound only partially inhibited PfSir2 in the synthetic-substrate assay, reaching a plateau at ∼65% of the uninhibited reaction rate (Fig. 4A). Strikingly, however, it inhibited the deacetylation of native histones much more effectively, with a high level of 10 mM being completely inhibitory (Fig. 4B) and even much lower levels (25 to 500 μM) showing substantial dose-dependent inhibition (Fig. 4C).
FIG. 4.
PfSir2 is inhibited by nicotinamide but not by other established sirtuin inhibitors. (A) The deacetylase assay using substrate A as for Fig. 3B was carried out in the presence of 25 μM to 10 mM nicotinamide. Residual activity in nicotinamide-treated reactions is expressed as a percentage of the fluorescence released in the uninhibited reaction. The graph shows the averages from three assays, each carried out in triplicate. Error bars indicate standard deviations. (B) Western blots as in Fig. 3C, showing P. falciparum histones incubated with recombinant PfSir2 in the presence (+) or absence (−) of 500 μM NAD+, with 10 mM nicotinamide (NAM), 50 μM splitomicin (Split), or 50 μM sirtinol (Sirt). (C) Western blots showing P. falciparum histones incubated for 1 h with recombinant PfSir2 in the presence (+) or absence (−) of 500 μM NAD+ and then with NAD+ and also increasing levels of nicotinamide (25 to 10,000 μM). Densitometric quantification of the AcH3 signal in the absence of NAD was set at 1, and all subsequent signals were compared to this. Each value was corrected for the ratio of total H3 signal in each lane to total H3 signal in the NAD+ (−) lane. Quantification of Western blot signals is linear only over a limited range, however, so values are at best semiquantitative over the whole range shown on this blot. (D) Alignment of the splitomicin target region of ScSir2 with the splitomicin-sensitive sirtuin Hst1 and the insensitive sirtuins Hst2 and PfSir2. Arrows mark residues which can be mutated to confer splitomicin resistance in ScSir2. None of these are conserved in PfSir2.
By contrast, splitomicin and sirtinol failed to inhibit PfSir2 in either assay (Fig. 4B and data not shown). Both drugs were tested at 50 μM, a concentration reported to reduce the reaction rate of a sensitive sirtuin, HsSirT1, by ∼40% for splitomicin and ∼70% for sirtinol (5). Indeed, both drugs at this concentration substantially inhibited ScSir2 in the same assay (see Fig. S2 in the supplemental material). Even at 400 μM, splitomicin did not inhibit PfSir2 (data not shown); the same test cannot be carried out with sirtinol due to its precipitation in aqueous solutions. The failure of splitomicin to affect PfSir2 may be due to the lack of conservation between PfSir2 and ScSir2 (the sirtuin for which splitomicin was initially discovered) in the region that is thought to contact the drug (Fig. 4D). Hst2, which also lacks homology in this region, is similarly insensitive to splitomicin (3). No target region has been reported for sirtinol, but it may likewise be effective only against the sirtuins most closely related to its initial target, ScSir2.
Finally, PfSir2 was also unaffected by the activators isonicotinamide and resveratrol (data not shown). Isonicotinamide can apparently bind to the nicotinamide-binding site of ScSir2 and specifically inhibit nicotinamide exchange (53), but this compound had little effect on the reaction rate of PfSir2. Likewise, resveratrol failed to activate the enzyme, a finding which may not be surprising as it has been reported that the in vitro “activation” of sirtuins by this drug is related specifically to the use of the Fluor-de-Lys synthetic substrate (35).
PfSir2 partially complements a fission yeast sir2 mutant.
To test whether PfSir2 could serve as an active HDAC in vivo as well as in vitro, it was used to complement a sir2 mutant strain of the fission yeast Schizosaccharomyces pombe. This strain carries the URA4 marker gene in a pericentromeric locus under Sir2-mediated silencing control. In the mutant, the URA4 gene cannot be silenced and a toxic prodrug, 5-FOA, can therefore be metabolized, killing the yeast. The addition of a functional sirtuin rescues this strain by silencing the URA4 gene; such successful cross-complementation was previously demonstrated when ScSir2 was expressed in S. pombe (18).
A PfSir2 construct was transformed into both wild-type and sir2 mutant S. pombe strains. When the yeast was grown on plates containing no thiamine, the overexpression of PfSir2 was toxic, an effect also observed with high levels of ScSir2 (18). Under conditions of thiamine repression and more moderate PfSir2 expression, however, the complemented strain grew 5- to 10-fold better on 5-FOA than an empty-vector control (Fig. 5).
FIG. 5.
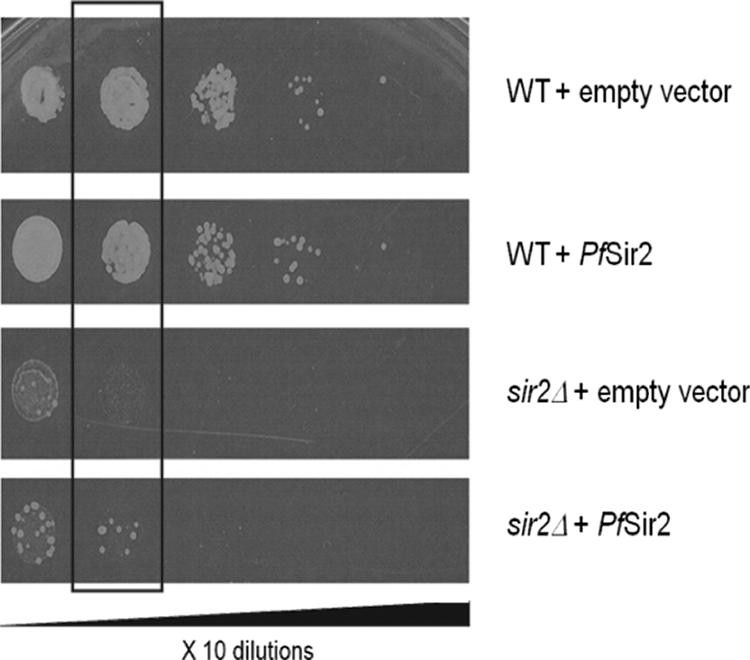
PfSir2 complements an S. pombe sir2 mutant. Wild-type (WT) or sir2Δ S. pombe was transformed with either the empty p-REP1 vector or with a construct containing the PfSir2 gene. Transformed cells were plated in 10-fold dilutions on EMM-Leu with 5-FOA. The highlighted dilution demonstrates better growth of the sir2Δ mutant carrying PfSir2 than of the mutant carrying only empty vector.
PfSir2 has ADP-ribosyltransferase in addition to HDAC activity.
Most of the sirtuins characterized thus far have either deacetylase or ADP-ribosyltransferase activity but are not significantly bifunctional. A sirtuin from Trypanosoma brucei, however, was recently reported to perform both functions robustly, ADP-ribosylating both histones and BSA, as well as itself, and also deacetylating histones (22). This sirtuin, TbSIR2RP1, falls into phylogenetic class I, and it is not very closely related to PfSir2 in sequence (Fig. 1); it is, however, another sirtuin from a protozoan parasite which, like P. falciparum, has relatively few sirtuin genes (4).
When PfSir2 was tested for the ability to ADP-ribosylate either BSA or Plasmodium histones using radioactive NAD+, significant activity was detected on both substrates (Fig. 6A, lanes 3 and 4). This was genuinely dependent on PfSir2, since a high level of nicotinamide inhibited the activity (Fig. 6A, lane 5), and substrate proteins were not significantly modified in the absence of the sirtuin (Fig. 6A, lanes 1 and 2). PfSir2 also ADP-ribosylated itself and was able to do this in the absence of other proteins (Fig. 6A, lane 6). Self-modification was, however, promoted by the presence of additional protein substrates, since the sirtuin became more strongly labeled when incubated in the presence of BSA and histones than when incubated alone. All four major histones became modified (see Fig. S3 in the supplemental material) but there was some preference for ADP-ribosylation of H2A, H2Bv, and/or H3 over H2B and H4. Figure 6B shows that the ADP-ribosyltransferase activity of PfSir2 was very robust compared to that of another sirtuin, Hst2 (which nevertheless has robust deacetylase activity). Finally, treatment with phosphodiesterase, which cleaves the pyrophosphate bond in ADP-ribose, was used to confirm that the labeling of PfSir2 was definitely due to the presence of [32P]ADP-ribose on the protein. This treatment did indeed reduce the intensity of the signal on radiolabeled PfSir2 (Fig. 6C).
Interestingly, prolonged treatment of recombinant PfSir2 with phosphodiesterase followed by repurification via Ni-NTA reduced the size of the major band of this protein. The size reduction was small but clearly visible on a highly resolved SDS-polyacrylamide gel (Fig. 6D), and concomitant with this, deacetylase activity was reduced by a factor of 10 (Fig. 6E). ADP-ribosylation of the major, upper band of recombinant PfSir2 therefore occurs when it is produced in E. coli, and the enzyme's activity depends on this modification. It remains to be established whether this is because ADP-ribosylation is actually required to activate PfSir2—an unprecedented finding among sirtuins—or because artificial cleavage of the ADP-ribose moiety at the pyrophosphate bond is somehow inhibitory to deacetylase activity.
DISCUSSION
This report provides the first detailed characterization of an apicomplexan sirtuin, PfSir2. It is also one of the few reports published so far examining a sirtuin from phylogenetic class III, since most of the extensively studied human and yeast sirtuins fall into class I. Class III sirtuins appear to be a very ancient group, as they are the most frequently occurring sirtuins in both eubacteria and archaea (21). They have been maintained in some although not all eukaryotic species; humans, for example, possess a single class III sirtuin, SirT5. SirT5 is found in mitochondria, consistent with a possible endosymbiotic origin, and it is presently without an established function or substrates.
In silico analysis of the protein sequence of PfSir2 suggested that it was probably a functional sirtuin, possessing the great majority of residues known to be important for catalytic activity in the yeast sirtuin Hst2 and having broadly conserved NAD-binding motifs. P. falciparum also contains a second, larger sirtuin gene in phylogenetic class IV (21), but this gene remains unstudied and it is unclear whether the two sirtuins, both of which are predicted to be nuclear, could perform all of the functions shared between multiple sirtuins in several locations in other higher eukaryotes. Apicomplexans diverged very early in evolution, and other species in the phylum, including Plasmodium spp. and also Toxoplasma gondii, similarly have only two sirtuin genes, orthologous to those in P. falciparum (Fig. 1). Some sirtuin functions, such as tubulin deacetylation or the regulation of metabolic enzymes, may have been lost in the apicomplexan lineage, but specialized partner proteins may also have evolved to adapt the apicomplexan sirtuins to a number of different roles. ScSir2, for example, is able to act in at least two distinct genomic locations via its binding to different protein complexes (24, 57).
PfSir2 formed large multimers in solution, and a similar property has been observed in ScSir2 (57), so this may be a physiological feature of certain sirtuins. ScSir2 spreads along stretches of DNA when establishing silenced heterochromatin, and the ability to multimerize could perhaps facilitate such spreading. Alternatively, the multimeric form may simply be an artifact of overexpression or of preparation in the absence of any physiological protein partners and/or substrates. ScSir2, HsSirT1, and HsSirT2 can all be produced under certain conditions as stable homotrimers, and Hst2 exists in an equilibrium between trimeric and monomeric states, with substrate binding specifically disrupting the trimer (66). ScSir2 also preferentially forms heterodimers and heterotrimers with its heterologous binding partners Sir4 and Sir3 (12, 47, 61). No evidence was found in the present study for preferential trimerization of PfSir2, a property which could be unique to class I sirtuins.
When His-tagged PfSir2 was expressed in P. falciparum, the protein was located in the insoluble chromatin-containing fraction of the parasite, together with the histones. PfSir2 may be bound here via hydrophobic interactions with a larger DNA-associated protein complex, perhaps containing many molecules of PfSir2 and being consistent with previous analyses of the large DNA-bound complexes containing ScSir2. ScSir2 does not itself have DNA-binding or nucleosome-binding activity (24), but it associates with chromatin via other proteins. The nuclear location of PfSir2 reported here is also consistent with a previous report that detected this protein in the parasite nucleus by immunocytochemistry (19). In the same report, regions of chromatin with highly acetylated histones were found to lack detectable PfSir2 and vice versa, pointing to a direct or indirect role for PfSir2 in histone deacetylation.
In this report, PfSir2 is directly shown to be an active HDAC. When its deacetylase activity was tested on two different synthetic substrates, PfSir2 clearly acted as an NAD+-dependent deacetylase, although rather slowly, requiring overnight incubations to produce significant signals. PfSir2 also deacetylated Plasmodium histones, a probable physiological substrate, and this reaction could be completed to the limit of detection within 1 h under appropriate conditions. These assays are not directly comparable, but any disparity between them may occur because the synthetic substrates only poorly mimic optimal, physiological substrates for PfSir2 (the synthetic peptide, for example, is modeled on human p53, which has no clear homolog in P. falciparum). PfSir2 may also require an absent binding partner(s) for its optimal activity, as is the case for ScSir2 (12, 57).
When deacetylating histones, PfSir2 acted most strongly on the N-terminal tail of H3, which is acetylated in this species by the acetyltransferase PfGCN5 (16). By contrast, all of the class I sirtuins ScSir2, SpSir2, HsSirT1 and HsSirT2 act primarily on H4Lys16, although they also deacetylate H3Lys9 to various extents (55, 57, 61, 62). It remains possible that H4Lys16 is specifically deacetylated by PfSir2 as well, though this did not reduce the overall acetylation of H4 very significantly whereas H3 could be deacetylated to the limit of detection. In any case, PfSir2 may still play a central role in transcriptional silencing because mutational studies in yeast have shown that the critical residues for heterochromatic silencing are H4 Lys16, H3 Lys9 and H3 Lys14 (9, 29, 58). Interestingly, there is some recent evidence that methylation of H3 Lys9 is associated with epigenetic silencing of virulence genes in Plasmodium falciparum (10).
PfSir2 could partially complement a sir2 mutant in the fission yeast S. pombe, despite the large evolutionary distance between the two species. This suggested that it supplied a bona fide HDAC activity within the S. pombe nucleus sufficient to partially repress transcription in a region normally silenced by SpSir2 (55), although it does remain possible that PfSir2 merely played a structural role, facilitating some degree of silencing in the absence of actual HDAC activity. If PfSir2 did supply a genuine HDAC activity, this may be undirected since the DNA elements and/or protein binding partners that would normally recruit SpSir2 to pericentromeric DNA are unlikely to be closely conserved between the two species. Alternatively, PfSir2 may recognize, via relatively conserved sirtuin domains, the repetitive DNA structure of the S. pombe centromere. P. falciparum itself has extensive repetitive DNA at its telomeres, which are heterochromatic (19). In support of this second theory, S. pombe can be complemented by ScSir2 as well as PfSir2 (18), despite the absence of S. pombe homologs for the ScSir2 protein partners Sir3 and Sir4 and despite the fact that ScSir2 does not normally silence pericentromeric DNA in its own genome.
PfSir2 was unusual among other characterized sirtuins in having robust ADP-ribosyltransferase activity as well as deacetylase activity, as shown schematically in Fig. 7. Only TbSIR2RP1 has previously been reported to act in this bifunctional way (22), and it is interesting to speculate that this is related to the relative paucity of sirtuins in both parasites, especially since TbSIR2RP1 and PfSir2 fall into different phylogenetic classes and are not closely homologous.
FIG. 7.
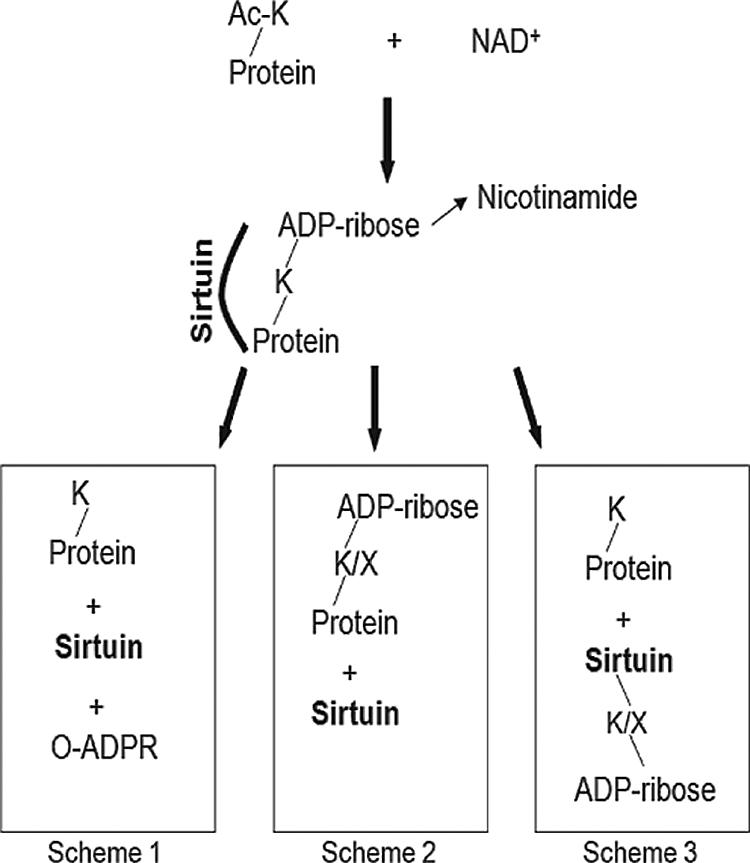
Model of reaction schemes showing possible ADP-ribosylation and deacetylation outcomes of the sirtuin reaction. Scheme 1 shows the classical deacetylation reaction, producing O-acetyl-ADP-ribose and a deacetylated lysine in the target protein. Scheme 2 shows an ADP-ribosylation reaction in which ADP-ribose is either left attached to the target lysine or subsequently transferred to another residue within the target protein. Scheme 3 shows the transfer of ADP-ribose to the sirtuin instead of to the target protein.
The ADP-ribosylation of histones is a well-established phenomenon in mammalian cells, where it is associated with replication (8) and with the response to DNA damage (1, 6, 7, 37). This has not, however, been traced directly to sirtuin activity, and it remains unclear what proportion of functional histone modification occurs via sirtuins and what via poly-ADP-ribose polymerase. We have found no obvious P. falciparum homolog of poly-ADP-ribose polymerase or of its counterpart poly-ADP-ribose glycohydrolase, so if a DNA damage response involving ADP-ribose does occur in this species, it might be mediated by ADP-ribosylation via PfSir2. TbSIR2RP1 certainly seems to be involved in DNA damage survival in T. brucei (22). Alternatively, or perhaps additionally, mono-ADP-ribosylation of histones could act like acetylation or methylation as a mark used to control chromatin structure, either for transcriptional control or to facilitate DNA repair. However, histones in mammalian cells can be modified on a range of different residues, and sirtuins would be limited to modifying lysine unless they can ultimately transfer ADP-ribose to another acceptor residue within a target protein (Fig. 7, scheme 2). No mechanism for such a reaction has been published, but the concept arises in a recent report showing that HsSIRT4 regulates mitochondrial glutamate dehydrogenase (27), which is regulated by cysteine-specific rather than lysine-specific ADP-ribosylation (32). If this is so, PfSir2 could potentially modify a range of acceptor residues within its target histones, widening its scope beyond the deacetylation of lysine.
The fact that ADP-ribosylation was detected on PfSir2 itself as well as on its substrates may be due to the trapping of a stable reaction intermediate in which ADP-ribose is bound to PfSir2, or it may by due to the secondary transfer of ADP-ribose onto other residues within the sirtuin (Fig. 7, scheme 3). The fact that phosphodiesterase, an enzyme which cleaves ADP-ribose, can reduce the size of freshly purified PfSir2 certainly suggests the latter, and intriguingly, this cleavage correlated with a severe reduction in deacetylase activity, suggesting a positive regulatory role for the modification. The very low level of ADP-ribosylation seen on recombinant ScSir2 has previously been proposed to play a regulatory role in vivo (56), and it was recently reported that the correct function and location of a Drosophila sirtuin depends upon poly-ADP-ribose glycohydrolase, perhaps because a regulatory ADP-ribose group(s) must be removed from this sirtuin for it to function properly (60).
Turning to the possibility of using inhibitors or activators to modify PfSir2 activity, all of the most commonly used compounds that affect class I sirtuins were either ineffective or only minimally effective on PfSir2, possibly due to the lack of conservation between PfSir2 and the S. cerevisiae sirtuin for which these drugs were first selected (3). The exception was nicotinamide, a universal product of the NAD+-dependent deacetylase reaction which can act as a competing nucleophile attacking the peptidyl-ADP-ribose reaction intermediate (53). The deacetylation of histones by PfSir2 could accordingly be completely inhibited by sufficiently high levels of nicotinamide and at least partially inhibited within the Ki range of the previously studied class I enzymes ScSir2 and Hst2 (110 μM and 180 μM, respectively) (50, 52). PfSir2 was much less responsive to nicotinamide when acting on a synthetic substrate, suggesting that the enzyme's configuration may be altered when it binds to an actual physiological substrate. This is not without precedent, as the sensitivity of ScSir2 to nicotinamide is also altered by its binding to two different protein complexes in vivo (57). Interestingly, nicotinic acid (metabolized to nicotinamide in an in vivo mouse model) has been shown to directly modulate the expression of virulence genes in a sirtuin-dependent manner in the fungal pathogen Candida glabrata (13). This raises the possibility of targeting PfSir2 as a means of altering virulence gene expression and pathogenesis in P. falciparum.
In conclusion, this study characterizes PfSir2, which is one of only two sirtuins in the most important human malarial parasite, P. falciparum. PfSir2 is a genuine HDAC with all of the properties required to act as a controller of the epigenetic silencing of subtelomeric virulence genes. It is also an ADP-ribosyltransferase, an activity that certainly merits further investigation, as it may reveal both regulatory mechanisms and secondary functions for this sirtuin that are either related or unrelated to epigenetic silencing. Little is currently known about sirtuins in phylogenetic class III, and further work may now clarify the precise targets of PfSir2 in vivo, the mechanisms of its regulation, and the identity of any potential binding partners, leading to a better understanding of class III sirtuins in general and of epigenetic silencing in P. falciparum in particular.
Supplementary Material
Acknowledgments
This work was funded by a Burroughs Wellcome Fund New Investigator in the Pathogenesis of Infectious Diseases Award to M.T.D.
We thank Danesh Moazed and members of his lab for help with S. pombe complementation and for providing the GST-Hst2 and GST-ScSir2 constructs, Wilhelm Haas and Steve Gygi for mass spectrometry, and Manfred Jung and Patrick Marcotte for providing the synthetic sirtuin substrates.
Footnotes
Published ahead of print on 7 September 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adamietz, P., and A. Rudolph. 1984. ADP-ribosylation of nuclear proteins in vivo. Identification of histone H2B as a major acceptor for mono- and poly(ADP-ribose) in dimethyl sulfate-treated hepatoma AH 7974 cells. J. Biol. Chem. 259:6841-6846. [PubMed] [Google Scholar]
- 2.Avalos, J. L., J. D. Boeke, and C. Wolberger. 2004. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol. Cell 13:639-648. [DOI] [PubMed] [Google Scholar]
- 3.Bedalov, A., T. Gatbonton, W. P. Irvine, D. E. Gottschling, and J. A. Simon. 2001. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. USA 98:15113-15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berriman, M., E. Ghedin, C. Hertz-Fowler, G. Blandin, H. Renauld, D. C. Bartholomeu, N. J. Lennard, E. Caler, N. E. Hamlin, B. Haas, U. Bohme, L. Hannick, M. A. Aslett, J. Shallom, L. Marcello, L. Hou, B. Wickstead, U. C. Alsmark, C. Arrowsmith, R. J. Atkin, A. J. Barron, F. Bringaud, K. Brooks, M. Carrington, I. Cherevach, T. J. Chillingworth, C. Churcher, L. N. Clark, C. H. Corton, A. Cronin, R. M. Davies, J. Doggett, A. Djikeng, T. Feldblyum, M. C. Field, A. Fraser, I. Goodhead, Z. Hance, D. Harper, B. R. Harris, H. Hauser, J. Hostetler, A. Ivens, K. Jagels, D. Johnson, J. Johnson, K. Jones, A. X. Kerhornou, H. Koo, N. Larke, S. Landfear, C. Larkin, V. Leech, A. Line, A. Lord, A. Macleod, P. J. Mooney, S. Moule, D. M. Martin, G. W. Morgan, K. Mungall, H. Norbertczak, D. Ormond, G. Pai, C. S. Peacock, J. Peterson, M. A. Quail, E. Rabbinowitsch, M. A. Rajandream, C. Reitter, S. L. Salzberg, M. Sanders, S. Schobel, S. Sharp, M. Simmonds, A. J. Simpson, L. Tallon, C. M. Turner, A. Tait, A. R. Tivey, S. Van Aken, D. Walker, D. Wanless, S. Wang, B. White, O. White, S. Whitehead, J. Woodward, J. Wortman, M. D. Adams, T. M. Embley, K. Gull, E. Ullu, J. D. Barry, A. H. Fairlamb, F. Opperdoes, B. G. Barrell, J. E. Donelson, N. Hall, C. M. Fraser, S. E. Melville, and N. M. El-Sayed. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416-422. [DOI] [PubMed] [Google Scholar]
- 5.Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves, and D. A. Sinclair. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099-45107. [DOI] [PubMed] [Google Scholar]
- 6.Boulikas, T. 1988. At least 60 ADP-ribosylated variant histones are present in nuclei from dimethylsulfate-treated and untreated cells. EMBO J. 7:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulikas, T. 1989. DNA strand breaks alter histone ADP-ribosylation. Proc. Natl. Acad. Sci. USA 86:3499-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulikas, T. 1990. Poly(ADP-ribosylated) histones in chromatin replication. J. Biol. Chem. 265:14638-14647. [PubMed] [Google Scholar]
- 9.Braunstein, M., R. E. Sobel, C. D. Allis, B. M. Turner, and J. R. Broach. 1996. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16:4349-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chookajorn, T., R. Dzikowski, M. Frank, F. Li, A. Z. Jiwani, D. L. Hartl, and K. W. Deitsch. 2007. Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. USA 104:899-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig, A., and A. Scherf. 2001. Molecules on the surface of the Plasmodium falciparum infected erythrocyte and their role in malaria pathogenesis and immune evasion. Mol. Biochem. Parasitol. 115:129-143. [DOI] [PubMed] [Google Scholar]
- 12.Cubizolles, F., F. Martino, S. Perrod, and S. M. Gasser. 2006. A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol. Cell 21:825-836. [DOI] [PubMed] [Google Scholar]
- 13.Domergue, R., I. Castano, A. De Las Penas, M. Zupancic, V. Lockatell, J. R. Hebel, D. Johnson, and B. P. Cormack. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308:866-870. [DOI] [PubMed] [Google Scholar]
- 14.Duraisingh, M. T., T. Triglia, S. A. Ralph, J. C. Rayner, J. W. Barnwell, G. I. McFadden, and A. F. Cowman. 2003. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 22:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duraisingh, M. T., T. S. Voss, A. J. Marty, M. F. Duffy, R. T. Good, J. K. Thompson, L. H. Freitas-Junior, A. Scherf, B. S. Crabb, and A. F. Cowman. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13-24. [DOI] [PubMed] [Google Scholar]
- 16.Fan, Q., L. An, and L. Cui. 2004. Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling. Eukaryot. Cell 3:264-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finnin, M. S., J. R. Donigian, and N. P. Pavletich. 2001. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 8:621-625. [DOI] [PubMed] [Google Scholar]
- 18.Freeman-Cook, L. L., E. B. Gomez, E. J. Spedale, J. Marlett, S. L. Forsburg, L. Pillus, and P. Laurenson. 2005. Conserved locus-specific silencing functions of Schizosaccharomyces pombe sir2+. Genetics 169:1243-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freitas-Junior, L. H., R. Hernandez-Rivas, S. A. Ralph, D. Montiel- Condado, O. K. Ruvalcaba-Salazar, A. P. Rojas-Meza, L. Mancio-Silva, R. J. Leal-Silvestre, A. M. Gontijo, S. Shorte, and A. Scherf. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25-36. [DOI] [PubMed] [Google Scholar]
- 20.Frye, R. A. 1999. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260:273-279. [DOI] [PubMed] [Google Scholar]
- 21.Frye, R. A. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273:793-798. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Salcedo, J. A., P. Gijon, D. P. Nolan, P. Tebabi, and E. Pays. 2003. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. EMBO J. 22:5851-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasser, S. M., and M. M. Cockell. 2001. The molecular biology of the SIR proteins. Gene 279:1-16. [DOI] [PubMed] [Google Scholar]
- 24.Ghidelli, S., D. Donze, N. Dhillon, and R. T. Kamakaka. 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20:4522-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grozinger, C. M., E. D. Chao, H. E. Blackwell, D. Moazed, and S. L. Schreiber. 2001. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 276:38837-38843. [DOI] [PubMed] [Google Scholar]
- 26.Guarente, L. 2005. Calorie restriction and SIR2 genes-towards a mechanism. Mech. Ageing Dev. 126:923-928. [DOI] [PubMed] [Google Scholar]
- 27.Haigis, M. C., R. Mostoslavsky, K. M. Haigis, K. Fahie, D. C. Christodoulou, A. J. Murphy, D. M. Valenzuela, G. D. Yancopoulos, M. Karow, G. Blander, C. Wolberger, T. A. Prolla, R. Weindruch, F. W. Alt, and L. Guarente. 2006. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126:941-954. [DOI] [PubMed] [Google Scholar]
- 28.Hallows, W. C., S. Lee, and J. M. Denu. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA 103:10230-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583-592. [DOI] [PubMed] [Google Scholar]
- 30.Heltweg, B., F. Dequiedt, E. Verdin, and M. Jung. 2003. Nonisotopic substrate for assaying both human zinc and NAD+-dependent histone deacetylases. Anal. Biochem. 319:42-48. [DOI] [PubMed] [Google Scholar]
- 31.Heltweg, B., J. Trapp, and M. Jung. 2005. In vitro assays for the determination of histone deacetylase activity. Methods 36:332-337. [DOI] [PubMed] [Google Scholar]
- 32.Herrero-Yraola, A., S. M. Bakhit, P. Franke, C. Weise, M. Schweiger, D. Jorcke, and M. Ziegler. 2001. Regulation of glutamate dehydrogenase by reversible ADP-ribosylation in mitochondria. EMBO J. 20:2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howitz, K. T., K. J. Bitterman, H. Y. Cohen, D. W. Lamming, S. Lavu, J. G. Wood, R. E. Zipkin, P. Chung, A. Kisielewski, L. L. Zhang, B. Scherer, and D. A. Sinclair. 2003. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425:191-196. [DOI] [PubMed] [Google Scholar]
- 34.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 35.Kaeberlein, M., T. McDonagh, B. Heltweg, J. Hixon, E. A. Westman, S. D. Caldwell, A. Napper, R. Curtis, P. S. DiStefano, S. Fields, A. Bedalov, and B. K. Kennedy. 2005. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 280:17038-17045. [DOI] [PubMed] [Google Scholar]
- 36.Khan, A. N., and P. N. Lewis. 2006. Use of substrate analogs and mutagenesis to study substrate binding and catalysis in the Sir2 family of NAD-dependent protein deacetylases. J. Biol. Chem. 281:11702-11711. [DOI] [PubMed] [Google Scholar]
- 37.Kreimeyer, A., K. Wielckens, P. Adamietz, and H. Hilz. 1984. DNA repair-associated ADP-ribosylation in vivo. Modification of histone H1 differs from that of the principal acceptor proteins. J. Biol. Chem. 259:890-896. [PubMed] [Google Scholar]
- 38.Kyes, S. A., J. A. Rowe, N. Kriek, and C. I. Newbold. 1999. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 96:9333-9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liszt, G., E. Ford, M. Kurtev, and L. Guarente. 2005. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280:21313-21320. [DOI] [PubMed] [Google Scholar]
- 41.Marcotte, P. A., P. L. Richardson, J. Guo, L. W. Barrett, N. Xu, A. Gunasekera, and K. B. Glaser. 2004. Fluorescence assay of SIRT protein deacetylases using an acetylated peptide substrate and a secondary trypsin reaction. Anal. Biochem. 332:90-99. [DOI] [PubMed] [Google Scholar]
- 42.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 43.Merrick, C. J., and M. T. Duraisingh. 2006. Heterochromatin-mediated control of virulence gene expression. Mol. Microbiol. 62:612-620. [DOI] [PubMed] [Google Scholar]
- 44.Miao, J., Q. Fan, L. Cui, J. Li, J. Li, and L. Cui. 2006. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene. 369:53-65. [DOI] [PubMed] [Google Scholar]
- 45.Michishita, E., J. Y. Park, J. M. Burneskis, J. C. Barrett, and I. Horikawa. 2005. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16:4623-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Min, J., J. Landry, R. Sternglanz, and R. M. Xu. 2001. Crystal structure of a SIR2 homolog-NAD complex. Cell 105:269-279. [DOI] [PubMed] [Google Scholar]
- 47.Moazed, D., A. Kistler, A. Axelrod, J. Rine, and A. D. Johnson. 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 94:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mostoslavsky, R., K. F. Chua, D. B. Lombard, W. W. Pang, M. R. Fischer, L. Gellon, P. Liu, G. Mostoslavsky, S. Franco, M. M. Murphy, K. D. Mills, P. Patel, J. T. Hsu, A. L. Hong, E. Ford, H. L. Cheng, C. Kennedy, N. Nunez, R. Bronson, D. Frendewey, W. Auerbach, D. Valenzuela, M. Karow, M. O. Hottiger, S. Hursting, J. C. Barrett, L. Guarente, R. Mulligan, B. Demple, G. D. Yancopoulos, and F. W. Alt. 2006. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124:315-329. [DOI] [PubMed] [Google Scholar]
- 49.North, B. J., B. Schwer, N. Ahuja, B. Marshall, and E. Verdin. 2005. Preparation of enzymatically active recombinant class III protein deacetylases. Methods 36:338-345. [DOI] [PubMed] [Google Scholar]
- 50.Sanders, B. D., K. Zhao, J. T. Slama, and R. Marmorstein. 2007. Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes. Mol. Cell 25:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sauve, A. A., I. Celic, J. Avalos, H. Deng, J. D. Boeke, and V. L. Schramm. 2001. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40:15456-15463. [DOI] [PubMed] [Google Scholar]
- 52.Sauve, A. A., R. D. Moir, V. L. Schramm, and I. M. Willis. 2005. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol. Cell 17:595-601. [DOI] [PubMed] [Google Scholar]
- 53.Sauve, A. A., and V. L. Schramm. 2003. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry 42:9249-9256. [DOI] [PubMed] [Google Scholar]
- 54.Scherf, A., R. Hernandez-Rivas, P. Buffet, E. Bottius, C. Benatar, B. Pouvelle, J. Gysin, and M. Lanzer. 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 17:5418-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shankaranarayana, G. D., M. R. Motamedi, D. Moazed, and S. I. Grewal. 2003. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 13:1240-1246. [DOI] [PubMed] [Google Scholar]
- 56.Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz, and D. Moazed. 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99:735-745. [DOI] [PubMed] [Google Scholar]
- 57.Tanny, J. C., D. S. Kirkpatrick, S. A. Gerber, S. P. Gygi, and D. Moazed. 2004. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell. Biol. 24:6931-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, J. S., X. Ling, and M. Grunstein. 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369:245-247. [DOI] [PubMed] [Google Scholar]
- 59.Trager, W., and J. B. Jenson. 1978. Cultivation of malarial parasites. Nature 273:621-622. [DOI] [PubMed] [Google Scholar]
- 60.Tulin, A., N. M. Naumova, A. K. Menon, and A. C. Spradling. 2006. Drosophila poly(ADP-ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics. 172:363-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaquero, A., M. Scher, D. Lee, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16:93-105. [DOI] [PubMed] [Google Scholar]
- 62.Vaquero, A., M. B. Scher, D. H. Lee, A. Sutton, H. L. Cheng, F. W. Alt, L. Serrano, R. Sternglanz, and D. Reinberg. 2006. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 20:1256-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voss, T. S., T. Mini, P. Jenoe, and H. P. Beck. 2002. Plasmodium falciparum possesses a cell cycle-regulated short type replication protein A large subunit encoded by an unusual transcript. J. Biol. Chem. 277:17493-17501. [DOI] [PubMed] [Google Scholar]
- 64.Wilson, J. M., V. Q. Le, C. Zimmerman, R. Marmorstein, and L. Pillus. 2006. Nuclear export modulates the cytoplasmic Sir2 homologue Hst2. EMBO Rep. 17:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, Y., C. D. Sifri, H. H. Lei, X. Z. Su, and T. E. Wellems. 1995. Transfection of Plasmodium falciparum within human red blood cells. Proc. Natl. Acad. Sci. USA 92:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, K., X. Chai, A. Clements, and R. Marmorstein. 2003. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 10:864-871. [DOI] [PubMed] [Google Scholar]
- 67.Zhao, K., X. Chai, and R. Marmorstein. 2003. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure 11:1403-1411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.