Abstract
The previous characterization and structural analyses of Sfi1p, a Saccharomyces cerevisiae centrin-binding protein essential for spindle pole body duplication, have suggested molecular models to account for centrin-mediated, Ca2+-dependent contractility processes (S. Li, A. M. Sandercock, P. Conduit, C. V. Robinson, R. L. Williams, and J. V. Kilmartin, J. Cell Biol. 173:867-877, 2006). Such processes can be analyzed by using Paramecium tetraurelia, which harbors a large Ca2+-dependent contractile cytoskeletal network, the infraciliary lattice (ICL). Previous biochemical and genetic studies have shown that the ICL is composed of diverse centrin isoforms and a high-molecular-mass centrin-associated protein, whose reduced size in the démaillé (dem1) mutant correlates with defective organization of the ICL. Using sequences derived from the high-molecular-mass protein to probe the Paramecium genome sequence, we characterized the PtCenBP1 gene, which encodes a 460-kDa protein. PtCenBP1p displays six almost perfect repeats of ca. 427 amino acids (aa) and harbors 89 potential centrin-binding sites with the consensus motif LLX11F/LX2WK/R, similar to the centrin-binding sites of ScSfi1p. The smaller (260-kDa) protein encoded by the dem1 mutant PtCenBP1 allele comprises only two repeats of 427 aa and 46 centrin-binding sites. By using RNA interference and green fluorescent protein fusion experiments, we showed that PtCenBP1p forms the backbone of the ICL and plays an essential role in its assembly and contractility. This study provides the first in vivo demonstration of the role of Sfi1p-like proteins in centrin-mediated Ca2+-dependent contractile processes.
Centrins are EF-hand calcium-binding proteins conserved in all eukaryotes, where they localize as a discrete component at microtubule-organizing centers. Their implication in the duplication of the spindle pole body (SPB) in Saccharomyces cerevisiae (6, 41), of the centrosome in mammalian cells (6, 26, 37), and of basal bodies in ciliates and flagellates is well documented (21, 31, 39, 43, 45). In unicellular organisms, centrins are also associated with different organelles, like cilia (14, 15) and contractile vacuole pores (43), and are major constituents of diverse contractile organelles, such as the striated flagellar roots of Tetraselmis striata, where centrin was first characterized (34); the nucleus-basal body connector and the striated fibers of Chlamydomonas reinhardtii (35); the spasmoneme of Vorticella convallaria (2); and the infraciliary lattice (ICL) of Paramecium tetraurelia (11). Like calmodulin, centrins act as effectors of different Ca2+-mediated processes, and understanding their mode of action requires the identification of their partner(s).
Among the proteins associated with centrins, Sfi1p previously identified in S. cerevisiae (19) is of particular interest. Sfi1p interacts with the yeast centrin Cdc-31p and localizes to the half-bridge of the SPB. The study of different mutant alleles of SFI1 showed that this gene plays an essential role in SPB duplication both in the formation of the new half-bridge (19, 22) and in the processes allowing the separation of the new SPB from the old one (3, 44). Sfi1p, a 120-kDa polypeptide, presents 21 motifs with the consensus sequence AX7LLX3F/LX2WK/R, presently known as centrin-binding sites, and each repeat is capable of binding to centrin at a 1:1 molar ratio (19, 22). The coexpression of 15 repeats and Cdc-31p leads to the in vitro formation of filaments that can be observed by electron microscopy. However, in this in vitro system, Ca2+ has no effect on the length of the filaments (22).
By BLAST and motif searching for proteins with Sfi1 motifs, at least 23 Sfi1-like proteins in diverse organisms have been identified (22). The human homolog, HsSfi1p, contains 23 repeats, some of which have previously been shown to bind Cdc-31p as well as mammalian centrins (19). The HsSfi1p-green fluorescent protein (GFP) fusion localizes at the centrosome, suggesting the conservation of function between yeast and human protein centrin-binding sites (19).
The discovery of Sfi1p provides new clues to understand how centrins intervene in Ca2+-mediated contractility. An attractive model proposes that the association of Sfi1p and centrins forms contractile fibers (19, 33): upon Ca2+ influx, centrins would undergo a conformational change leading to the torsion of the Sfi1 molecule, to which they would remain attached, and, as a consequence, to the shortening of the fiber. This hypothesis fits the in vitro observation of the Ca2+-sensitive behavior of the centrosome and its filamentous associated matrix: calcium induces a decrease in the centriole diameter and a shortening of the intercentriolar link that connects centrioles (27). Similarly, Ca2+-induced basal body reorientation on an isolated basal apparatus of C. reinhardtii was observed previously (16).
While the validity of this model implying the contractility or the elasticity of Sfi1p-centrin complexes remains to be demonstrated for SPB or centrosome duplication, it can be experimentally tested in the case of the cytoskeletal contractile arrays present in unicellular organisms. The ICL of Paramecium is a Ca2+-associated contractile cytoskeletal network made of bundles of 4-nm filaments subtending the whole cell surface (1). In previous biochemical and genetic studies, we have identified, in addition to at least 10 centrin isoforms (12, 24), a large centrin-associated protein (L-CAP; >350 kDa) as a major component of the ICL (20). We show here that L-CAP, hereinafter referred to as PtCenBP1p (for P. tetraurelia centrin-binding protein 1), is indeed an Sfi1p-like protein which presents the same remarkable succession of centrin-binding sites over the whole length of the molecule. By using RNA interference (RNAi) and GFP fusion experiments, we demonstrated that PtCenBP1p is an essential constituent of the filamentous bundles that form the ICL and, thus, provide the first experimental demonstration of the models proposed by Salisbury (33) and Kilmartin (19) for the direct involvement of Sfi1p-like proteins in centrin-mediated Ca2+-dependent contractility.
MATERIALS AND METHODS
Strains and culture conditions.
Stock d4-2 of P. tetraurelia, the wild-type reference strain, was used in all experiments. Two mutants derived from the wild-type stock were also used: the démaillé (dem1) mutant, which contains a monogenic nuclear mutation affecting the organization and/or the dynamics of the ICL (20), and the Δ-PtCenBP1 cell line, described herein, which carries an epigenetically maintained deletion of the macronuclear PtCenBP1 gene (see below). Cells were grown at 27°C in a wheatgrass infusion (BHB; l'Arbre de Vie, Luçay Le Male, France) or wheatgrass powder (Pines International, Lawrence, KS) bacterized with Klebsiella pneumoniae and supplemented with 0.4 μg of β-sitosterol/ml according to standard procedures (42).
In vivo contractility assay.
Aminoethyldextran (AED) is a vital secretagogue that induces the exocytosis of trichocysts, the secretory vesicles docked beneath the plasma membrane (28). Exocytosis is accompanied by a transient intracellular Ca2+ influx (18), which provokes the contraction of the ICL, resulting in cell contraction. For in vivo observations, cells were transferred into a mixture of 10 mM Tris buffer, pH 7.3, and 40 μM CaCl2 containing 3 μM AED. Cell contraction, which persists for ca. 30 s, is easily monitored under a dissecting microscope.
Peptide sequencing.
One hundred micrograms of an ICL-enriched fraction (11) was subjected to limited trypsin proteolysis in a solution of 0.1 M ammonium carbonate, pH 8.0, and 5 mM CaCl2 (with 0.17-mg/ml trypsin from Boehringer) for 5 min at 25°C. Proteolysis was stopped by the addition of 0.05 mg of phenylmethylsulfonyl fluoride (Sigma)/ml. The digested fragments were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE), and the two heaviest major bands (45 and 40 kDa), generated by the proteolysis of the heaviest protein, were electroblotted onto Problott membranes (Applied Biosystems, CA) and sequenced on a 470A protein sequencer (ABI; Applied Biosystems, CA) using a Problott cartridge. Phenylthiohydantoin amino acids were identified using an online analyzer (ABI 120 A). For the determination of internal sequences, gel bands were digested by endoproteinase AspN according to the method described in reference 29. Peptides were purified by reverse-phase high-performance liquid chromatography and sequenced.
Gene silencing by feeding.
The gene sequences of interest were amplified by PCR and cloned into the L4440 feeding vector between two T7 promoters (46). For the CenBP1 gene, we amplified the region encompassing positions 2627 to 4768 with the following primers: 5′-ATTTGGAATCCATTATACGA-3′ (sense) and 5′-TGTGTGTATTCTTATGGTAT-3′ (antisense). The resulting plasmid was used for the transformation of HT115, an RNase III-deficient strain of Escherichia coli with an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 polymerase (38). Wild-type paramecia were incubated in double-stranded RNA-expressing bacteria, as previously described (9), and observed over successive days through daily transfer into freshly induced feeding bacteria. For negative controls, Paramecium cells were fed with HT115 carrying the L4440 plasmid without the insertion.
Expression vector for GFP fusion proteins and transformation.
The GFP synthetic gene sequence designed by E. Meyer and J. Cohen (unpublished data) was cloned at its 3′ extremity into the plasmid pPXV (32). The part of the PtCenBP1 gene encoding amino acids (aa) 59 to 1344 was amplified using primers 5′-TTAAGGTACCGGAGGAATGATCGAGAAAAAAATAGATGTCTC and 3′-TTTTTGGTACCTCAAATAATAGCTCTCTTCTTCAAAGC and cloned into the KpnI site (underlined) of pPXV-GFP. Plasmid DNA was prepared with a plasmid midi kit according to the protocol of the manufacturer (QIAGEN, Courtaboeuf, France). Microinjection of the DNA was carried out as previously described (13).
Fluorescence microscopy.
The immunostaining of cells was carried out as previously described (20). The monoclonal antibody 1A9, raised against the Paramecium ICL (7), was used at a dilution of 1:200; the polyclonal anticentrin antibody 26/14-1 (9), the monoclonal antitubulin antibody ID5 (47), and the secondary anti-mouse and anti-rabbit antibodies from Jackson ImmunoResearch Labs (West Grove, PA) were used at a dilution of 1:500. For the recording of GFP expression in living cells, cells were washed twice in Dryl's buffer (46) containing 0.2% bovine serum albumin and then transferred in a small drop onto a coverslip and overlaid with paraffin oil. Alternatively, GFP-labeled paramecia were fixed in 2.5% formaldehyde. Cells were observed under an Axioskop 2-plus fluorescence microscope equipped with a Roper Coolsnap-CF intensifying camera (Zeiss, Oberkochen, Germany) with GFP filters. Images were processed using the Metamorph software (Universal Imaging, Downington, PA).
Electron microscopy.
Cell pellets were fixed in 2% glutaraldehyde in 0.05 M cacodylate buffer for 90 min at 4°C. Fixed cells were washed three times in the same buffer and postfixed in 1% (wt/vol) osmium tetroxide in 0.05 M cacodylate buffer for 1 h at 4°C. After dehydratation, thin sections were contrasted with saturated ethanolic uranyl acetate and lead citrate and examined with a Philips CM10 microscope.
Computational sequence analysis.
Sequences were compared by ClustalW alignment (http://www.ch.embnet.org/software/ClustalW.html). Dot plots were established by Self-Matrix from DNA Strider (CEA, France). Sequence logos (8) was developed using WebLogo (http://weblogo.berkeley.edu/logo.cgi). The two-dimensional structure analysis was performed with PredictProtein (http://www.predictprotein.org). PROFsec or PHDsec (30) analyzes the α-helicoidal content of a protein. The probability of a coiled-coil structure was calculated according to the prediction programs Pepcoil and COILS, version 2.2, using the MTIDK matrix with different weights for positions a through g, as follows: a and d, 2.5, and b, c, e, f, and g, 1.0. The prediction was made using a scanning window of 28 residues (23).
Nucleotide sequence accession number.
The PtCenBP1 gene sequence was deposited in the EMBL database under accession number CR932274.
RESULTS
The ICL of P. tetraurelia forms a conspicuous cytoskeletal network whose Ca2+-dependent contractility can be observed in vivo upon Ca2+ influx (Fig. 1). Biochemical analysis of purified ICL fractions had previously established that the building blocks of the network are complexes of different centrin isoforms with a large, >350-kDa, centrin-associated protein and that these complexes are stable in the absence of Ca2+ (20). In vivo evidence for the role of L-CAP in the organization of the ICL was provided by the properties of the dem1 mutant, which displays a shorter L-CAP polypeptide and looser organization of the ICL (Fig. 2). The characterization of the corresponding gene led us to rename the L-CAP gene PtCenBP1 (for P. tetraurelia centrin-binding protein 1).
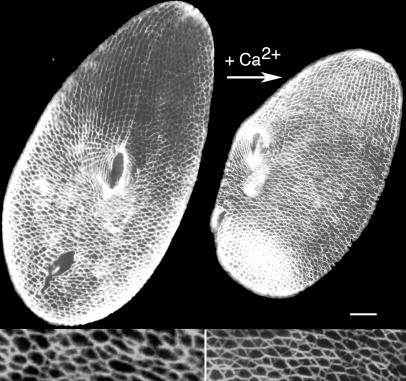
FIG. 1.
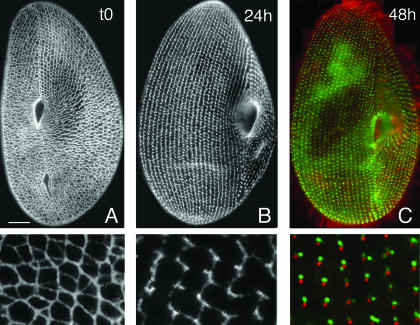
The Ca2+-dependent contractility of the ICL. Cells were immunolabeled with the monoclonal anticentrin antibody 1A9 (7). (Left panel) Ventral side of a wild-type resting cell. (Right panel) Cell labeled during the Ca2+ influx induced by AED treatment (see Materials and Methods), which triggers the contraction of the ICL, resulting in a general contraction of the cell. Scale bar, 10 μm.
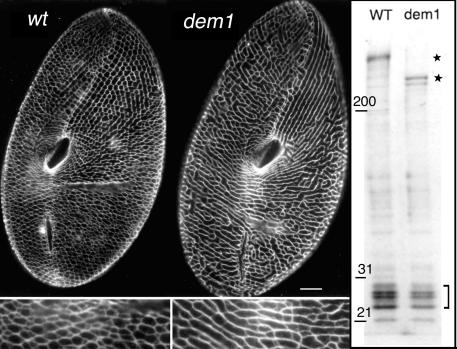
FIG. 2.
Phenotypical and biochemical features of the dem1 mutant. (Left panel) Wild-type (wt; left) and dem1 mutant (right) cells were immunolabeled with the monoclonal anticentrin antibody 1A9. Scale bar, 10 μm. The ICL of the dem1 mutant is characterized by a reduced number of meshes and an apparent deficit in branching. This phenotype is exhibited only by cells in growing populations, while cells in stationary phase reach a wild-type phenotype. (Right panel) Sodium dodecyl sulfate-PAGE pattern of purified ICL from wild-type and dem1 mutant cells. The gel is a 6 to 15% acrylamide gradient stained with Coomassie blue. A striking difference in the bands corresponding to the high-molecular-mass protein of wild-type and dem1 mutant cells, at 450 and 250 kDa (stars), respectively, is observed, whereas the same pattern for the low-molecular-mass bands (bracket), which correspond to centrins, is seen for both cell types. The presence of a doublet at the PtCenBP1-dem1p position may be due to proteolysis or to an unknown posttranslational modification.
Characterization of the PtCenBP1 genes from the wild type and the dem1 mutant.
To identify the gene encoding PtCenBP1p, the Paramecium genome sequence (4, 5) was probed by using BLAST with four sequences generated by tryptic proteolysis of the protein (Fig. 1 and Materials and Methods). As the BLAST search targeted two adjacent gene models (GSPATG00034434001 and GSPATG00034433001) corresponding to products whose sizes (714 and 1,233 aa, respectively) were not compatible with the observed size of the high-molecular-mass protein of more than 350 kDa, the genomic region was reassessed. The steps to the characterization of the wild-type gene organization and the characterization of the mutation present in the dem1 mutant PtCenBP1 allele are presented in the supplemental material. The wild-type PtCenBP1 gene turned out to be 11,862 bp long and to encode a 3,893-aa protein with a theoretical molecular mass of ca. 460 kDa. This protein contains six almost perfect repeats (P1 to P6), each with 427 aa, except for P1, with 428 aa, and P6, with 419 aa (Fig. 3). PtCenBP1p has a pI of 10.1 owing to a high content (18.5%) of the basic amino acids K and R. According to secondary-structure predictions, PtCenBP1p is an all-alpha protein that is ca. 86% α-helical. Coiled-coil arrangements were detected in the N-terminal part, between residues 210 and 245, and in the C-terminal part, between residues 3814 and 3867, with a high probability (>90%). These characteristics are compatible with a filamentous protein able to form homopolymers.
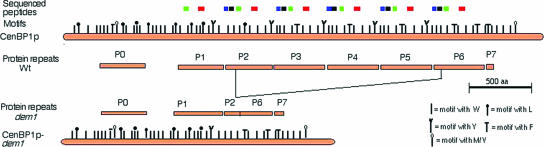
FIG. 3.
Analysis of the PtCenBP1p protein in the wild-type and dem1 mutant cells. The wild-type (Wt) PtCenBP1p sequence (orange rectangles) is composed of six repeats (P1 to P6) plus a divergent repeat (P0) and an incomplete repeat (P7). The mutant PtCenBP1-dem1p (CenBP1p-dem1) sequence derives from the wild-type sequence by the deletion of repeats P3 through P5. For both sequences, the positions of the sequenced peptides are indicated by colored boxes, red, black, blue, and green for the peptides KFERTLDILFRV/S(K)LKVSFDPLKE(I)YMXALNIK(T)M(G)LKKLF, DXAQALKKRAIXLMIKLQ, ETQLNKFTLXI, and DSNLRYFFMK, respectively. Vertical bars represent the centrin-binding motifs as defined by Kilmartin (19) and indicate whether the diagnostic hydrophobic amino acid is a tryptophan, a leucine, a tyrosine, a phenylalanine, or a methionine or valine.
We first looked for 23-aa centrin-binding motifs by using the consensus previously defined by Kilmartin, with a diagnostic tryptophan in position 22 (19). PtCenBP1p contains 44 such sites, with the consensus sequence KX4QXG/SX4A/TFX2WK/R (Fig. 4). This consensus is clearly similar to that of the Sfi1 proteins of S. cerevisiae, Schizosaccharomyces pombe, and H. sapiens previously described as being a centrin-binding domain and further demonstrated to act as a centrin-binding site in vitro (19). We then enlarged the search by allowing L, Y, F, M, or V in addition to W at the diagnostic position 22. This allowance led to the recognition of a total of 89 centrin-binding sites, which share a shorter consensus sequence, LLX11F/LX2WK/R (Fig. 4). Comparison of the PtCenBP1p centrin-binding sites with those of yeast and of a homologous protein found in the annotation of the Tetrahymena thermophila genome (corresponding to gene XM_001030914.1) shows that the most conserved residues are the C-terminal ones, F/LX2WK/R, which appear to be the true signature of the consensus. This finding is consistent with the crystal structures showing contacts between these conserved residues and helices VI and VII of the C-terminal centrin domain (22). Since the ICL is built up from a diverse multigene family of centrins, it is possible that different subclasses of these sites are specific for given ICL centrin subfamilies.
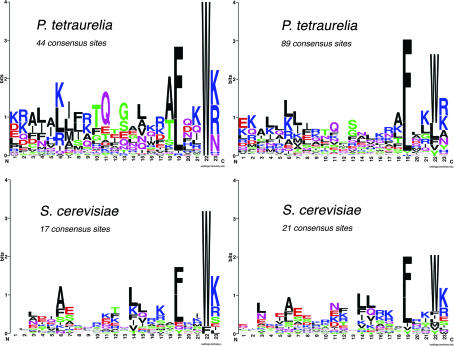
FIG. 4.
Sequence logos of the Sfi1 repeats in S. cerevisiae and homolog in P. tetraurelia. Among the potential centrin-binding sites identified in PtCenBP1p, 44 present a diagnostic W at position 22 in the 23-aa sequences defined as centrin-binding sites by Kilmartin (19). A less stringent definition of the consensus sequence, allowing L, Y, F, M, or V in addition to W at the diagnostic position 22, led to the identification of a total of 89 potential centrin-binding sites. The two sets of centrin-binding sites (44 and 89 consensus sites, respectively) were submitted to WebLogo. For comparision, logos corresponding to two sets of consensus sequences from S. cerevisiae are shown: a first set of 17, each with a W or F at position 22 (19), and a second set including all 21 centrin-binding sites recognized in Sfi1p (22) and showing more variability at position 22.
The mutation present in the dem1 strain was found to correspond to a large deletion of ca. 5,000 bp (see Fig. S1 in the supplemental material). The conceptual translation of the 6,738-bp dem1 mutant PtCenBP1 gene gives a 2,186-aa protein with a theoretical molecular mass of ca. 260 kDa and a pI of 10.1. The dem1 mutant protein PtCenBP1-dem1p lacks the central repeats (P3 through P5), retaining, however, 46 potential centrin-binding sites. Previous analyses (20) had shown that the patterns of centrin isoforms present in the ICL were the same in dem1 mutant and wild-type cells, consistent with the finding that only the number of each binding site, and not the specificity of the binding sites, is modified in the mutant.
Functional analysis of PtCenBP1p: RNAi experiments.
RNAi targeting the ICL1a gene, which codes for one of the constitutive centrins, was previously shown to cause the total disassembly of the ICL, without any other effect on cell growth or morphology (7). This dispensability of the ICL provides a favorable situation for its functional analysis. To investigate the role of PtCenBP1p, we silenced the corresponding gene in both wild-type and dem1 mutant cells by an RNAi approach using feeding (9). PtCenBP1-silenced cells grew normally without visible defects in swimming behavior, shape, or generation time. However, as early as two divisions after inoculation into the feeding medium, cells were no longer able to contract upon AED-induced Ca2+ influx (Fig. 1 and Materials and Methods), suggesting the disappearance of the ICL. Immunolabeling with anticentrin antibodies allowed us to monitor the disassembly of the network (Fig. 5). Although both wild-type and mutant cells continued to grow normally, disassembly was complete for 100% of the cells examined (i.e., 80 to 120 cells in each of five independent RNAi experiments) after three to four divisions for the wild type and slightly sooner for dem1 mutant cells. As shown in Fig. 5C, silenced cells displayed only ICL remnants, in the form of anticentrin-reactive spots close to each basal body, morphologically identical to the previously described ICL-organizing centers (ICLOCs) observed after RNAi in the ICL1a gene (7).
FIG. 5.
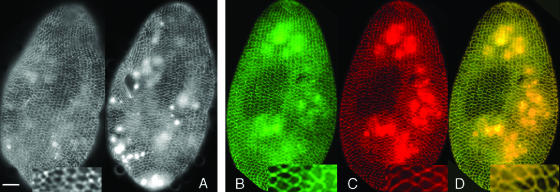
Disassembly of the ICL upon PtCenBP1 silencing by RNAi as visualized by using immunofluorescence. Disassembly triggered by PtCenBP1p depletion was monitored using the anticentrin antibody 1A9. (A) Ventral side of a wild-type cell before the start of the experiment. (B) Ventral side of a cell after 24 h (ca. three cell divisions) of gene silencing. The cell presents a disrupted ICL in which only the longitudinal part of the meshes is still present and the transversal branches are missing. (C) Ventral side of a cell after 48 h (ca. six to seven fissions) of CenBP1p depletion. The disassembly is complete. Double labeling with a polyclonal anticentrin antibody (26/14-1) and a monoclonal antitubulin antibody (ID5) reveals both basal bodies (red) and the remnants of the ICL (green), which form discrete masses of amorphous material, shown to be reactive to anticentrin antibodies and to be the sites of ICL reassembly, the ICLOCs (7). Enlargements of representative areas selected from panels A, B, and C are presented in the lower panels. Scale bar, 10 μm.
The RNAi effect could be reversed by transferring the cells back to normal growth medium. The reassembly of the network started from the ICLOCs, as previously described (7). After a few divisions, cells reacquired a complete contractile ICL. Conversely, a stable ICL-less condition can be established. When cells are subjected to RNAi during the nuclear reorganization accompanying sexual processes, some clones develop a new macronucleus (somatic nucleus) in which at least part of the target gene is deleted, although their micronulei (germinal nuclei) still retain the wild-type gene (10). Such clones carrying a deletion of PtCenBP1 in the macronucleus were obtained (data not shown) and were observed to be devoid of the ICL. These clones, designated Δ-PtCenBP1, could be maintained under normal culture conditions through cellular continuity over hundreds of cell divisions, and the phenotype was inherited maternally in crosses. Observations of a Δ-PtCenBP1 cell line by electron microscopy (Fig. 6B) confirmed that the meshes constitutive of the network were absent and that small masses of amorphous material corresponding to the ICLOC, as observed by anticentrin immunolabeling, were left near the basal bodies. Some small electron-dense triangular structures, the posts (1), normally located at the branching points of the meshes and possibly serving as anchoring points for the filament bundles in the native network, were observed within the ICL remnants (Fig. 6B). Spontaneous reversions to a contractile phenotype can occur during nuclear reorganization. Reversions always correlate with the restoration of wild-type copies of the PtCenBP1 gene within the maronucleus (data not shown). These experiments demonstrated that the assembly of the ICL, i.e., the organization of the residual masses of centrin-containing material into filaments, was directly dependent upon the availability of the PtCenBP1 gene product.
FIG. 6.
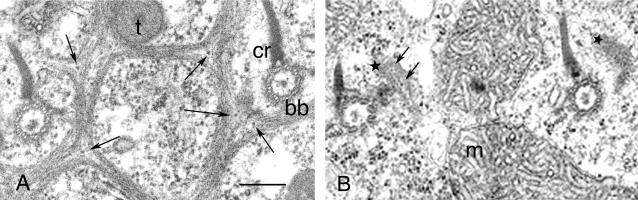
Disassembly of the ICL upon PtCenBP1 silencing by RNAi as shown by electron microscopy. Tangential thin sections of the cortex show the differences between the organization of the ICL in a control cell (A) and that in a cell after the inactivation of the PtCenBP1 gene (B). (A) The polygonal meshes surrounding basal bodies (bb) with their attached ciliary rootlet (cr) and secretory vesicle tip (t) are composed of branched filamentous bundles. At the branching points, note the presence of one or two dense structures (arrows) called posts (1). (B) In the inactivated cells, the meshes of the network are no longer present. Only small pieces of amorphous material (stars) are visible to the right of each basal body: the enclosed posts (arrows) ensure that they are genuine ICL remnants. Note the mitochondria (m), which are always found at the level of the proximal part of basal bodies in the absence of the ICL, while they are found deeper in the cytoplasm in wild-type cells when the ICL is present (our unpublished observations). Scale bar, 0.30 μm.
In vivo localization of GFP-PtCenBP1p.
In order to determine the localization of PtCenBP1p, a sequence encoding a GFP tag was fused to the part of the PtCenBP1 gene extending from positions 226 to 4160. The gene segment encoded a ca.-150-kDa polypeptide spanning aa 59 to 1344 and containing the presumed coiled-coil region of the N terminus and 30 consensus centrin-binding sites. The pPXV vector containing this construction was used to transform, by microinjection, both wild-type and Δ-PtCenBP1 cells. In the transformed wild-type cells, the GFP signal homogeneously localized to the preexisting network fibers as observed on living cells. GFP labeling of the ICL was detected as early as 3 h after microinjection (Fig. 7A). In living cells, in addition to the network labeling, strong background fluorescence throughout the cytoplasm was visible, most likely due to excess molecules in the cytosol and possibly also to the fact that the truncated GFP construct may have had lowered affinity for the network. Interestingly, when we processed the cells for labeling with anti-GFP antibodies in order to enhance the GFP signal, this signal was lost. However, if 1 mM Ca2Cl was added to the permeabilization-fixation buffers, the GFP signal was preserved and could be revealed by the anti-GFP antibodies. The Ca2+-dependent rescue of the GFP signal showed that the incorporation of the GFP fragment into the network was Ca2+ sensitive since the GFP signal was detected in vivo, destabilized in the standard EGTA-containing permeabilization-fixation buffer, and stabilized if the calcium concentration was raised. Under the latter conditions, as in vivo, the GFP label localized precisely with the centrin label along the extant ICL as shown in Fig. 7B to D.
FIG. 7.
GFP-tagged PtCenBP1p localizes at the ICL. Transformants expressing a GFP-tagged fragment of PtCenBP1p comprising aa 59 to 1344 were examined in vivo and after immunolabeling with anti-GFP antibodies. (A) Ventral and dorsal faces of a living cell showing the homogeneous fluorescence of the ICL. (B to D) Transformants after permeabilization and fixation in the presence of 1 mM Ca2+ and double labeling with a polyclonal anti-GFP antibody and the monoclonal anticentrin antibody 1A9. (B) Anti-GFP staining. (C) Anticentrin staining. (D) Merged images. Scale bar, 10 μm.
In Δ-PtCenBP1 cells transformed with pPXV, expressing GFP fused to the product of positions 226 to 4160 of the PtCenBP1 gene, the GFP signal specifically localized near basal bodies at the sites of the ICLOCs (data not shown), where ICL centrins localize even in the absence of the network. No rescue of the phenotype was observed, probably owing to the small size of the PtCenBP1 gene fragment used and/or the absence of the N- and C-terminal parts of the gene product.
DISCUSSION
We have used the ICL of P. tetraurelia to study the molecular mechanisms of Ca2+-mediated centrin-based contractility. We previously demonstrated that the building block of the ICL was a complex of different centrin isotypes and a high-molecular-mass protein, L-CAP (20). We report here the characterization of the L-CAP gene and its functional analysis. We showed that L-CAP, now renamed PtCenBP1p, is a 460-kDa, Sfi1p-like protein featuring 89 motifs of 23 aa, previously described by Kilmartin (19) as centrin-binding consensus sites. We demonstrated that PtCenBP1p forms the backbone of the contractile ICL by three converging experimental approaches and in vivo observations: (i) the monogenic recessive dem1 mutation, which causes a looser organization of the network (Fig. 2), corresponded to the deletion of 43 consensus centrin-binding sites; (ii) in cells depleted of PtCenBP1p following RNAi treatment, the ICL was totally disassembled but reassembled upon return to normal growth conditions; and (iii) a GFP-tagged fragment of PtCenBP1p comprising aa 59 to 1344 localized with the whole network in a Ca2+-dependent manner.
We will first examine to what extent the present findings account for all previous data on the biochemical composition, cytological organization, and contractile properties of the ICL. Then we will discuss how these results fit present models concerning the structure and function of Sfi1p-centrin complexes (19, 25).
PtCenBP1p as the backbone of the ICL.
In Paramecium, different centrins are present in different organelles and ensure different functions, not only in the ICL but also in basal body positioning (31) and in ciliary Ca2+ channels (14). While these diverse functions are also likely to involve centrin-binding proteins, our data show that the protein characterized here, PtCenBP1p, is specific to the contractile ICL, since its depletion results only in ICL disassembly without any effect on cell growth, morphology, or behavior. Conversely, most of the centrins of the ICL are specific to this structure: we have previously shown that ICL organization is not affected by the silencing of the basal body-specific centrins (31) and that different monoclonal antibodies against ICL centrins do not cross-react with basal bodies (7). Compared to the half-bridge of the SPB, the ICL stands out by virtue of its dimensions, which parallel the huge size of PtCenBP1p and the diversity of its constitutive centrins (20, 31). As first described by Allen (1), the ICL forms a subcortical, transcellular reticulum of polygonal meshes running around each basal body or pair of basal bodies and around the tips of trichocysts, the secretory vesicles docked at the cell surface (Fig. 6A). Each mesh corresponds to a bundle of 3- to 4-nm-diameter microfilaments. Filament bundles merge at one apex of the polygons, marked by a singular triangular structure, the post (12), which can be viewed as the branching or anchoring point of the network (Fig. 6A). The length of filament bundles varies from 0.3 to ca. 2 μm. In the images of the contracted network (as in Fig. 2), the meshes are 20 to 25% shorter. The molecular data presented here are in good agreement with these cytological data. The characterization of PtCenBP1p as a 3,893-aa protein of an essentially α-helical structure makes it reasonable to postulate that each molecule is ca. 600 nm long (a maximal value, calculated for a fully extended helix), and assuming that the numerous centrin isoforms known to colocalize to the ICL (7; D. Gogendeau et al., unpublished results) occupy their potential binding sites, the width of the PtCenBP1p molecule would be ca. 4 nm, as shown by the crystal structures of Sfi1p-centrin complexes (22). Furthermore, the predicted coiled-coil arrangement of the C and N termini of PtCenBP1p may be compatible with end-to-end interactions as shown for Sfi1p in the duplicating SPB (22). Clearly, the observed (3- to 4-nm) width of the unit filament within the ICL meshes in electron micrographs corresponds to the estimated width of the PtCenBP1p helix with its bound centrins, while the estimated length of the complex, 600 nm, falls within the size range of the shortest meshes in the resting network.
Biochemically, the unit complex (one PtCenBP1p molecule plus 89 centrin molecules) would have an estimated molecular mass of ca. 2,700 kDa. The size previously measured by the classical methods (liquid chromatography, ultracentrifugation, and PAGE) had led to a size estimate of more than 1,000 kDa for the elementary protein complex obtained by the dissociation of the purified ICL in the absence of Ca2+ (20).
PtCenBP1p would then form the backbone of the ICL, in agreement with the continuous and homogeneous GFP labeling of the network (Fig. 7). Finally, it is of interest to examine how the characteristics of the ICL in dem1 mutant cells (Fig. 2) can be accounted for by the reduced size of the mutant protein. With the length of mutant building blocks being half of that found in the wild type, the deletion of the central repeats may affect the flexibility of the bundles of filaments and lead to reduced or delayed branching activity. However, the question remains open as to the mechanism of formation of the branches and the nature of the posts.
Molecular mechanisms of centrin-based contractility.
Different models for the organization of Sfi1p and centrin have been proposed previously (19, 22, 25, 33, 41). Centrins bound to Sfi1p by their C-terminal moiety, but also by their free N-terminal moiety (41), are able to interact with other centrins located on the same Sfi1p molecule (22). Centrins may also interact with other centrins bound to a different Sf1p molecule (25), owing to their strong tendency to self-assemble (48). In all cases, the binding of Ca2+ onto centrins would induce conformational changes triggering or enhancing the interactions between centrins and resulting in the shortening of the filament. Moreover, because these filaments are highly α-helicoidal, it is possible that interactions between adjacent centrins bound along Sfi1p filaments (22) induce the supercoiling of the filament. This scenario corresponds to the idea first proposed in the case of the contraction of the rhizoplast of Platymonas subcordiformis, that centrin-based motility involves filament supercoiling (36).
In this conceptual framework, what information does the ICL contribute? For the general consensus that the association between the Sf1p-like protein and centrins is not Ca2+ dependent (17, 19, 25), the ICL provides experimental confirmation: the complete dissociation of the isolated network in the absence of Ca2+ (Ca2+ concentration, 10−8 M) could not dissociate centrins from the high-molecular-mass partner CenBP1p, as established by PAGE of high-speed pellets or exclusion chromatography. The CenBP1p-centrin association was quite strong, as a partial dissociation of centrins from the complex could be achieved only in the presence of 2 M urea (20).
Concerning the Ca2+ dependence of the interactions between centrins, both our in vitro and our in vivo observations indicate interactions between centrins bound to adjacent Sfi1p-like proteins. When the purified ICL has been completely dissociated in the absence of Ca2+, raising the Ca2+ concentration to 10−7 M (comparable to the resting-cell Ca2+ concentration) provokes the self-association of the elementary complexes into filamentous aggregates (12). However, association into ordered bundles of filaments was not observed in vitro. In vivo, the total disassembly of the network upon the depletion of the ICL1a centrin (7) shows that centrins contribute to the cohesion of the network and/or to the bundling of the filaments. Furthermore, we have shown here that the integration of GFP-tagged fragments of PtCenBP1p into the ICL filament bundles required a Ca2+ concentration at least equal to that in resting living cells (ca. 10−7 M). The range of calcium dependence observed for the ICL proteins, both in vitro (20) and in vivo as shown here, is similar to the values obtained previously by others measuring interactions between Sfi1p (one centrin binding site) and C. reinhardtii centrin at different Ca2+ concentrations (40). Evidence for interactions between centrins bound to different Sf1p-like proteins may not have general relevance, since the ICL architecture involves filament bundling and may have specific requirements for such lateral interactions. It is also possible that of the numerous centrin isotypes present in the ICL, some are more reactive than others for establishing intercomplex interactions. Our observations show, nevertheless, that such lateral interactions do occur and are likely to occur also in other systems since, in unicellular organisms, contractile organelles such as the distal connecting fibers in C. reinhardtii all consist of bundles of filaments.
As depicted in Fig. 8, we propose that in unstimulated Paramecium cells, PtCenBP1p-centrin molecules are assembled longitudinally in filaments with lengths that are multiples of 600 nm: only the high-affinity Ca2+-binding sites of the centrins are saturated, and there is no contraction. When the intracellular Ca2+ level rises, the lower-affinity sites are recruited and their binding provokes conformational changes leading to the supercoiling and thickening of the filaments and to the shortening of the bundles. This idea is in contrast to the model proposed for the contraction of the distal connecting fibers of Chlamydomonas (25), in which the contraction is proposed to occur perpendicular to the axis of the filaments. The large number of different centrins found in Paramecium as well as their diversity with respect to the numbers and positions of the functional Ca2+-binding sites (Gogendeau et al., unpublished) are likely to play a role in the fine regulation of the contractile strength of the ICL and its adjustment to the mosaic organization and dynamics of the cortex.
FIG. 8.
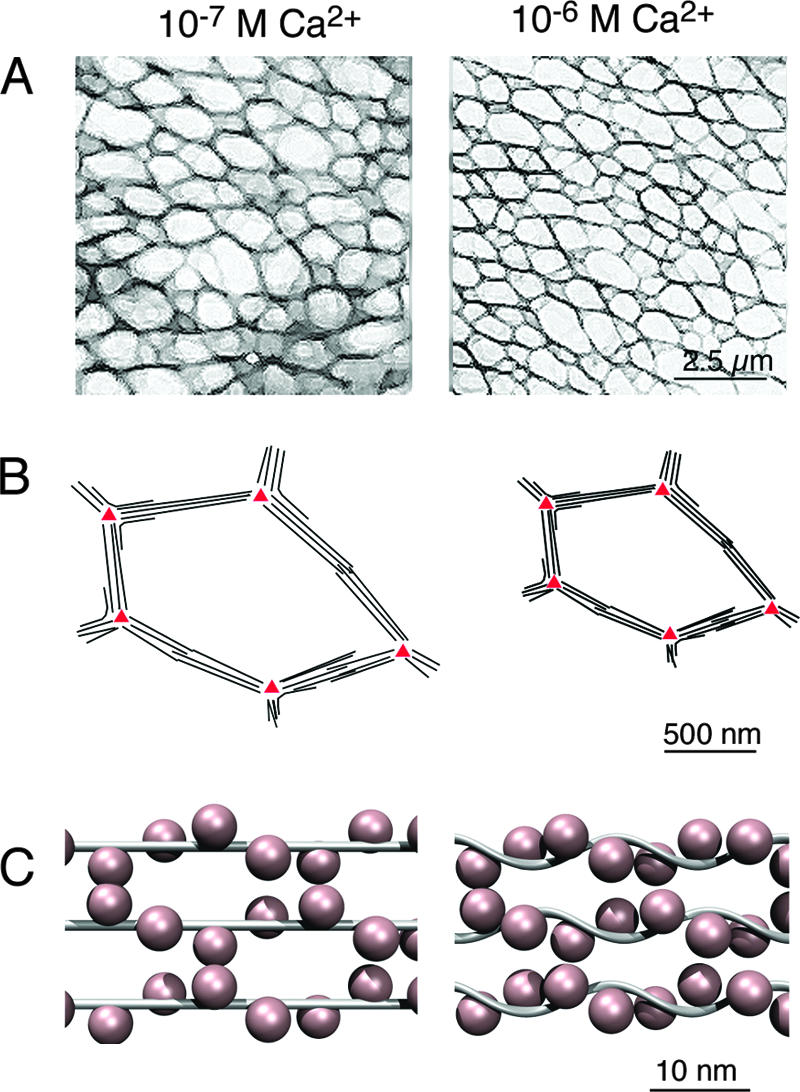
Role of CenBP1p in the Ca2+-dependent contractility of the ICL. (A) Schematic representation, based on Fig. 1, of the ICL. The geometry of the polygonal ICL meshwork at resting (10−7 M) and physiologically high (10−6 M) Ca2+ concentrations is shown. The filament bundles that constitute the edges of the polygons are on average 25% shorter in the presence of high levels of Ca2+. (B) Schematic representation of one ICL polygon. Under resting conditions, the unit filament consisting of one PtCenBP1p molecule with associated centrins is likely to be ∼0.3 to 0.5 μm in length (depending on whether or not the Sfi-like backbone is a completely extended α-helix). This is the distance between posts (red triangles) along the shortest polygon edges. The lateral association of unit filaments builds up the bundles. The N- and/or C-terminal coiled-coil domains may be involved in tethering the filament bundles to the posts. At high Ca2+ concentrations, the filament bundles shorten and the tension is relayed through the posts to yield a global reduction in the size of the ICL, and hence of the whole cell, with very little distortion of global cell shape (Fig. 1). (C) Schematic representation of a possible arrangement of PtCenBP1p and centrins. At resting Ca2+ levels, the ICL unit filament is probably a disordered helical array of ∼4-nm centrins (pink) wrapped around an essentially extended α-helical PtCenBP1p molecule (gray), as originally proposed by Kilmartin for yeast Sfi1p-Cdc31p complexes (19). The binding of Ca2+ to the centrin high-affinity sites at in vivo Ca2+ concentrations favors the lateral interactions necessary for filament bundling. At high Ca2+ levels, the occupation of centrin low-affinity Ca2+-binding sites leads to interactions between adjacent centrins along the length of the filament, as seen in the yeast Sfi1p-Cdc31p crystal structures (22). The consequence is the shortening of the filaments, probably owing to reduced distances between adjacent centrins and the consequent bending, twisting, or supercoiling of the Sfi1-like PtCenBP1p backbone.
The ICL still raises a number of intriguing questions concerning its elaborate cellular organization and the specific roles of its diverse centrins. However, the interactions between centrins and Sfi1p-like proteins, presumed to mediate Ca2+-dependent contractile processes in the SPB and centrosomes and in a variety of contractile organelles in unicellular eukaryotes, are likely to involve similar molecular mechanisms. The demonstration that CenBP1p forms the backbone of the ICL, a contractile network of Paramecium, provides the first direct evidence of the functional association of centrins and centrin-binding proteins to form contractile fibers, as proposed previously for the Sfi1p protein (22, 33).
Supplementary Material
Acknowledgments
We are particularly grateful to Michel Bornens for first drawing our attention to Kilmartin's publication on the properties of Sfi1p (19) and for critical reading of the manuscript. We are much indebted to Maud Sylvain for DNA sequencing, to Jean-Pierre Denizot (Institut A. Fessard, Gif-sur-Yvette, France) for electron microscopy facilities, and to R. Le Guyader for her participation in the artwork. We thank J. Wehland and J. Salisbury for the generous supply of antibodies.
This work was supported by the Centre National de la Recherche Scientifique and by grant ACI:IMPBIO 2004 no. 14 (L.S.). D.G. was supported by the Association de la Recherche Contre le Cancer (A06/3).
Footnotes
Published ahead of print on 3 August 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Allen, R. D. 1971. Fine structure of membranous and microfibrillar systems in the cortex of Paramecium caudatum. J. Cell Biol. 49:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amos, W. B., L. M. Routledge, and F. F. Yew. 1975. Calcium-binding proteins in a vorticellid contractile organelle. J. Cell Sci. 19:203-213. [DOI] [PubMed] [Google Scholar]
- 3.Anderson,V. E., J. Prudden, S. Prochnik, T. H. Giddings, and K. G. Hardwick. 2007. Novel sfi1 alleles uncover additional functions for Sfi1p in bipolar spindle assembly and function. Mol. Biol. Cell 18:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnaiz, O., S. Cain, J. Cohen, and L. Sperling. 2007. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 35(Database issue):D439-D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aury, J. M., O. Jaillon, L. Duret, B. Noel, C. Jubin, B. M. Porcel, B. Segurens, V. Daubin, V. Anthouard, N. Aiach, O. Arnaiz, A. Billaut, J. Beisson, I. Blanc, K. Bouhouche, F. Camara, S. Duharcourt, R. Guigo, D. Gogendeau, M. Katinka, A. M. Keller, R. Kissmehl, C. Klotz, F. Koll, A. Le Mouel, G. Lepere, S. Malinsky, M. Nowacki, J. K. Nowak, H. Plattner, J. Poulain, F. Ruiz, V. Serrano, M. Zagulski, P. Dessen, M. Betermier, J. Weissenbach, C. Scarpelli, V. Schachter, L. Sperling, E. Meyer, J. Cohen, and P. Wincker. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444:171-178. [DOI] [PubMed] [Google Scholar]
- 6.Baum, P., C. Furlong, and B. Byers. 1986. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. USA 83:5512-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beisson, J., J. C. Clerot, A. Fleury-Aubusson, N. Garreau de Loubresse, F. Ruiz, and C. Klotz. 2001. Basal body-associated nucleation center for the centrin-based cortical cytoskeletal network in Paramecium. Protist 152:339-354. [DOI] [PubMed] [Google Scholar]
- 8.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galvani, A., and L. Sperling. 2000. Regulation of secretory protein gene expression in paramecium. Role of the cortical exocytotic sites. Eur. J. Biochem. 267:3226-3234. [DOI] [PubMed] [Google Scholar]
- 10.Garnier, O., V. Serrano, S. Duharcourt, and E. Meyer. 2004. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol. Cell. Biol. 24:7370-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garreau de Loubresse, N., G. Keryer, B. Vigues, and J. Beisson. 1988. A contractile cytoskeletal network of Paramecium: the infraciliary lattice. J. Cell Sci. 90:351-364. [Google Scholar]
- 12.Garreau de Loubresse, N., C. Klotz, B. Vigues, B. Rutin, and J. Beisson. 1991. Ca2+-binding proteins and contractility of the infraciliary lattice in Paramecium. Biol. Cell 71:217-225. [Google Scholar]
- 13.Gilley, D., J. R. Preer, Jr., K. J. Aufderheide, and B. Polisky. 1988. Autonomous replication and addition of telomerelike sequences to DNA microinjected into Paramecium tetraurelia macronuclei. Mol. Cell. Biol. 8:4765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonda, K., A. Yoshida, K. Oami, and M. Takahashi. 2004. Centrin is essential for the activity of the ciliary reversal-coupled voltage-gated Ca2+ channels. Biochem. Biophys. Res. Commun. 323:891-897. [DOI] [PubMed] [Google Scholar]
- 15.Guerra, C., Y. Wada, V. Leick, A. Bell, and P. Satir. 2003. Cloning, localization, and axonemal function of Tetrahymena centrin. Mol. Biol. Cell 14:251-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi, M., T. Yagi, K. Yoshimura, and R. Kamiya. 1998. Real-time observation of Ca2+-induced basal body reorientation in Chlamydomonas. Cell Motil. Cytoskeleton 41:49-56. [DOI] [PubMed] [Google Scholar]
- 17.Hu, H., J. H. Sheehan, and W. J. Chazin. 2004. The mode of action of centrin. Binding of Ca2+ and a peptide fragment of Kar1p to the C-terminal domain. J. Biol. Chem. 279:50895-50903. [DOI] [PubMed] [Google Scholar]
- 18.Kerboeuf, D., and J. Cohen. 1990. A Ca2+ influx associated with exocytosis is specifically abolished in a Paramecium exocytosis mutant. J. Cell Biol. 111:2527-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilmartin, J. V. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162:1211-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klotz, C., N. Garreau de Loubresse, F. Ruiz, and J. Beisson. 1997. Genetic evidence for a role of centrin-associated proteins in the organization and dynamics of the infraciliary lattice in Paramecium. Cell Motil. Cytoskeleton 38:172-186. [DOI] [PubMed] [Google Scholar]
- 21.Koblenz, B., and K. F. Lechtreck. 2005. The NIT1 promoter allows inducible and reversible silencing of centrin in Chlamydomonas reinhardtii. Eukaryot. Cell 4:1959-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, S., A. M. Sandercock, P. Conduit, C. V. Robinson, R. L. Williams, and J. V. Kilmartin. 2006. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 173:867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lupas, A. 1996. Prediction and analysis of coiled-coil structures. Methods Enzymol. 266:513-525. [DOI] [PubMed] [Google Scholar]
- 24.Madeddu, L., C. Klotz, J. P. Le Caer, and J. Beisson. 1996. Characterization of centrin genes in Paramecium. Eur. J. Biochem. 238:121-128. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Sanz, J., A. Yang, Y. Blouquit, P. Duchambon, L. Assairi, and C. T. Craescu. 2006. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 273:4504-4515. [DOI] [PubMed] [Google Scholar]
- 26.Middendorp, S., T. Kuntziger, Y. Abraham, S. Holmes, N. Bordes, M. Paintrand, A. Paoletti, and M. Bornens. 2000. A role for centrin 3 in centrosome reproduction. J. Cell Biol. 148:405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moudjou, M., M. Paintrand, B. Vigues, and M. Bornens. 1991. A human centrosomal protein is immunologically related to basal body-associated proteins from lower eucaryotes and is involved in the nucleation of microtubules. J. Cell Biol. 115:129-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plattner, H., H. Matt, H. Kersken, B. Haake, and R. Stürzl. 1984. Synchronous exocytosis in Paramecium cells. A novel approach. Exp. Cell Res. 151:6-13. [DOI] [PubMed] [Google Scholar]
- 29.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 30.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz, F., N. Garreau de Loubresse, C. Klotz, J. Beisson, and F. Koll. 2005. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr. Biol. 15:2097-2106. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz, F., L. Vayssie, C. Klotz, L. Sperling, and L. Madeddu. 1998. Homology-dependent gene silencing in Paramecium. Mol. Biol. Cell 9:931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salisbury, J. L. 2004. Centrosomes: Sfi1p and centrin unravel a structural riddle. Curr. Biol. 14:R27-R29. [DOI] [PubMed] [Google Scholar]
- 34.Salisbury, J. L., A. Baron, B. Surek, and M. Melkonian. 1984. Striated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J. Cell Biol. 99:962-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salisbury, J. L., A. T. Baron, and M. A. Sanders. 1988. The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J. Cell Biol. 107:635-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salisbury, J. L., and G. L. Floyd. 1978. Calcium-induced contraction of the rhizoplast of a quadriflagellate green alga. Science 202:975-977. [DOI] [PubMed] [Google Scholar]
- 37.Salisbury, J. L., K. M. Suino, R. Busby, and M. Springett. 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12:1287-1292. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Selvapandiyan, A., A. Debrabant, R. Duncan, J. Muller, P. Salotra, G. Sreenivas, J. L. Salisbury, and H. L. Nakhasi. 2004. Centrin gene disruption impairs stage-specific basal body duplication and cell cycle progression in Leishmania. J. Biol. Chem. 279:25703-25710. [DOI] [PubMed] [Google Scholar]
- 40.Sheehan, J. H., C. G. Bunick, H. Hu, P. A. Fagan, S. M. Meyn, and W. J. Chazin. 2006. Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function. J. Biol. Chem. 281:2876-2881. [DOI] [PubMed] [Google Scholar]
- 41.Spang, A., I. Courtney, U. Fackler, M. Matzner, and E. Schiebel. 1993. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123:405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonneborn, T. M. 1970. Methods in Paramecium research, p. 241-339. In D. Prescott (ed.), Methods in cell physiology, vol. 4. Academic Press, New York, NY. [Google Scholar]
- 43.Stemm-Wolf, A. J., G. Morgan, T. H. Giddings, Jr., E. A. White, R. Marchione, H. B. McDonald, and M. Winey. 2005. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell 16:3606-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strawn, L. A., and H. L. True. 2006. Deletion of RNQ1 gene reveals novel functional relationship between divergently transcribed Bik1p/CLIP-170 and Sfi1p in spindle pole body separation. Curr. Genet. 50:347-366. [DOI] [PubMed] [Google Scholar]
- 45.Taillon, B. E., S. A. Adler, J. P. Suhan, and J. W. Jarvik. 1992. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 119:1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timmons, L., and A. Fire. 1998. Specific interference by ingested dsRNA. Nature 395:854. [DOI] [PubMed] [Google Scholar]
- 47.Wehland, J., and K. Weber. 1987. Turnover of the carboxy-terminal tyrosine of alpha-tubulin and means of reaching elevated levels of detyrosination in living cells. J. Cell Sci. 88:185-203. [DOI] [PubMed] [Google Scholar]
- 48.Yang, A., S. Miron, P. Duchambon, L. Assairi, Y. Blouquit, and C. T. Craescu. 2006. The N-terminal domain of human centrin 2 has a closed structure, binds calcium with a very low affinity, and plays a role in the protein self-assembly. Biochemistry 45:880-889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.