Abstract
Homogenates from rat spleen and lung could modify nitrotyrosine-containing BSA. With incubation, nitrotyrosine-containing BSA lost its epitope to a monoclonal antibody that selectively recognized nitrotyrosine-containing proteins. In the presence of protease inhibitors, the loss of the nitrotyrosine epitope occurred without protein degradation and hydrolysis. This activity was found in supernatant but not particulate fractions of spleen homogenates. The factor was heat labile, was sensitive to trypsin treatment, and was retained after passage through a membrane with a 10-kDa retention. The activity was time- and protein-concentration dependent. The activity increased about 2-fold in spleen extracts with endotoxin (bacterial lipopolysaccharide) treatment of animals, suggesting that the activity is inducible or regulatable. Other nitrotyrosine-containing proteins also served as substrates, while free nitrotyrosine and some endogenous nitrotyrosine-containing proteins in tissue extracts were poor substrates. Although the product and possible cofactors for this reaction have not yet been identified, this activity may be a “nitrotyrosine denitrase” that reverses protein nitration and, thus, decreases peroxynitrite toxicity. This activity was not observed in homogenates from rat liver or kidney, suggesting that there may also be some tissue specificity for the apparent denitrase activity.
Keywords: nitric oxide/peroxynitrite/protein nitration/denitrase
Since the discovery of the first biological effects of nitric oxide more than two decades ago (1–3), the pursuit of evidence of the ubiquitous involvement of this simple free radical in cell signaling and regulation has grown at a phenomenal rate (4–6). Nitric oxide can be produced from exogenous sources, such as nitrovasodilators, or endogenously from l-arginine by one of several isoforms of nitric oxide synthase. Nitric oxide may participate in the regulation of diverse physiological processes such as smooth muscle relaxation, platelet aggregation, neurotransmission, cytotoxicity, etc. (4–6). While many of the effects of nitric oxide are mediated through guanylyl cyclase activation and cyclic GMP accumulation (1–6), some of its effects are cyclic GMP-independent.
Nitric oxide can react with various functionalities and molecules, including thiol groups, transition metals, and other free radicals. The interaction of nitric oxide with superoxide is a rapid reaction, almost diffusion-limited, that forms the very reactive species peroxynitrite (7). It has been proposed that some of the cytotoxic effects of nitric oxide are the result of formation of peroxynitrite, which can form several reactive species and can nitrate free and protein-associated tyrosine residues (7–9). The formation of nitrotyrosine-containing proteins has been observed in some inflammatory disorders, atherosclerotic lesions, and neurodegenerative diseases (9–13).
While protein nitration has been used as an index of peroxynitrite formation, tyrosine nitration can also occur through peroxynitrite-independent mechanisms (12). The nitration of proteins may alter a protein’s conformation and structure, catalytic activity, and/or susceptibility to protease digestion (14–16). It has also been shown that tyrosine nitration can diminish a protein’s effectiveness as a substrate for tyrosine kinases (16–18). Thus, the present view is that protein nitration is associated with some toxic or deleterious effects of nitric oxide and with interruption of cell signaling processes.
Although some investigators have viewed protein nitration as an irreversible event, we thought that tyrosine nitration in proteins might be a reversible process catalyzed by an enzyme. If indeed protein nitration is a reversible and perhaps regulatable process, such protein modifications could also function in cellular signaling events analogous to protein phosphorylation and dephosphorylation. Without an enzyme to reverse tyrosine nitration such as a “denitrase,” to use protein nitration in cellular signaling processes that are reversible, de novo protein synthesis would be required to restore the function of nitrated proteins.
We report here the presence of an activity in rat spleen and lung extracts that modifies nitrotyrosine-containing BSA (nitrated BSA) and other nitrated proteins. The activity was assayed by using a selective antibody to nitrotyrosine-containing proteins. The antibody had decreased recognition of the epitope after incubation of nitrated BSA and other nitrated proteins with tissue extracts. Although the product(s) and possible cofactors of the reaction are yet to be identified, this activity appears to be a “nitrotyrosine denitrase.”
Incubation of nitrated BSA with samples in the presence of a mixture of protease inhibitors resulted in a loss of the nitrotyrosine epitope without apparent protein hydrolysis and/or degradation. The activity was time- and concentration-dependent, labile to heating and trypsin treatment, found in soluble fractions of rat spleen homogenates, retained by filtration through a membrane that retained molecules >10 kDa, and used some but not all nitrated proteins examined as substrates. This putative “nitrotyrosine denitrase” activity is increased with endotoxin treatment, suggesting that the activity can be regulated and is perhaps inducible. In other studies (unpublished work), we found that many tissues and cell cultures possess nitrotyrosine-containing proteins on Western immunoblots. These nitrated proteins have dynamic increases and decreases when intact animals or cells are treated with endotoxin and/or some cytokines. These findings, coupled with the finding of an activity in tissue extracts that reverses protein nitration, support our hypothesis that nitration of tyrosyl residues in proteins and the reversal of this process participate in cellular signaling processes. The significance of these observations and the complete characterization of the activity require additional studies.
MATERIALS AND METHODS
Materials.
BSA, goat anti-mouse antibody peroxidase conjugate, and lipopolysaccharide (LPS; from Escherichia coli, serotype O111:B4) were purchased from Sigma. Enhanced chemiluminescence and enhanced chemifluorescence reagents for the detection of immunoreactive substances were obtained from Amersham. 3-Morpholinosydnonimine (SIN-1) was from Alexis Co. (San Diego, CA). Nitrated molecular weight standards and rabbit polyclonal anti-nitrotyrosine antibody were from Upstate Biotechnology (Lake Placid, NY). Male Sprague–Dawley rats were obtained from Charles River Breeding Laboratories and were cared for in accordance with the guidelines of the Institutional Animal Care and Use Committee and the National Institutes of Health guidelines on laboratory animal welfare.
Treatment of Rats with LPS and Preparation of Tissues.
Twenty-four hours after the intraperitoneal injection of 20 mg/kg LPS or vehicle, phosphate buffered saline [PBS; 20 mM sodium phosphate buffer (pH 7.4)/130mM NaCl], rats were sacrificed. Spleen, lung, liver, and kidney were removed and homogenized in 10 vol of 20 mM Tris⋅Cl buffer (pH 7.4) containing protease inhibitors (10 μg/ml soybean trypsin inhibitor/10 μg/ml benzamadine/0.005 trypsin inhibitor unit/ml aprotinin/10 μg/ml leupeptin/10 μg/ml pepstatin A/5 μg/ml antipain/200 μM phenylmethanesulfonyl fluoride/100 μM EDTA) (buffer A). After centrifugation at 1,000 × g for 15 min, the supernatant fractions were stored at −135°C.
Several samples (300 μl) were further processed by centrifugation at 100,000 × g for 60 min. The pellet was resuspended in buffer A (500 μl) and recentrifuged at 100,000 × g. The resulting pellet was finally resuspended in buffer A (300 μl) and kept frozen at −135°C. The high-speed supernatant fraction was further separated by membrane filtration using an Amicon 10 membrane to retain material greater than 10 kDa. The retained material (>10 kDa) was resuspended twice with buffer A and refiltered. The retained sample was assayed for activity as described. Protein concentrations were determined by the Bradford method with the Bio-Rad protein assay, using BSA as a standard (19).
Preparation of Nitrated BSA and Incubation with Tissue Samples.
BSA (100 mg) was solubilized in 10 ml of PBS and incubated for 2–4 h at 37°C with SIN-1 (10 mM), which generates NO and superoxide radicals and thus forms peroxynitrite (20). The nitrated BSA was dialyzed against PBS. The nitration procedure converted about 5% of the tyrosine residues in BSA to nitrotyrosine, as determined by the absorbance at 430 nm (20) and also by the nitrotyrosine and tyrosine content of hydrolyzed samples treated with 6 M HCl and analyzed with HPLC by using electrochemical detection (21, 22). BSA was also nitrated by the incubation with 10 mM tetranitromethane for 30 min, which resulted in the nitration of ≈10% of the tyrosine residues in BSA.
Tissue homogenates with protein concentration adjusted to 5 mg/ml, except when indicated, were incubated with equal volumes of nitrated BSA at 2 mg/ml for 10 min or for the times indicated. Reactions were stopped by boiling for 10 min in the presence of 2% SDS. In some experiments, homogenates were boiled for 10 min before assays. In other experiments, tissue homogenates were dialyzed against Buffer A without protease inhibitors and then incubated with trypsin (20 μg/ml) for 0–60 min at 37°C. After the addition of the soybean trypsin inhibitor (100 μg/ml), the samples were used in assays.
Detection and Estimation of Nitrated BSA.
Samples were separated by using PAGE with 7.5% gels (23). Separated samples were then transferred to nitrocellulose membranes and treated with 5% nonfat dry milk in Tris-buffered saline [20 mM Tris⋅Cl (pH 7.6)/130 mM NaCl/0.2% Tween 20] (buffer TBS-T). Membranes were incubated at 4°C overnight with a mouse monoclonal antibody developed against a nitrated peptide coupled to keyhole limpet hemocyanin. Membranes were washed with TBS-T and incubated with peroxidase-conjugated goat anti-mouse antibody, and chemiluminesence was used to identify nitrotyrosine-containing proteins according to the enhanced chemiluminescence system. Some membranes were sequentially incubated with a commercial rabbit polyclonal anti-nitrotyrosine antibody, fluorescein-linked anti-rabbit antibody, and alkaline phosphatase-conjugated anti-fluorescein antibody, according to the enhanced chemifluorescence system. Some gels or transferred membranes were stained with Coomassie brilliant blue to determine the amount of BSA and other proteins. The amount of nitrated BSA after PAGE was estimated from the area and density of each band, compared with the nitrated BSA control.
Statistical Analysis.
Results are expressed as means ± SD from representative experiments. For comparison of activities of tissue homogenates from LPS-treated and control rats, we used the Wilcoxon rank sum test and Dunnett’s multiple comparison test with the analysis of variance. A P value <0.05 was considered to be statistically significant.
RESULTS
Effects of Rat Tissue Homogenates on Nitrated BSA.
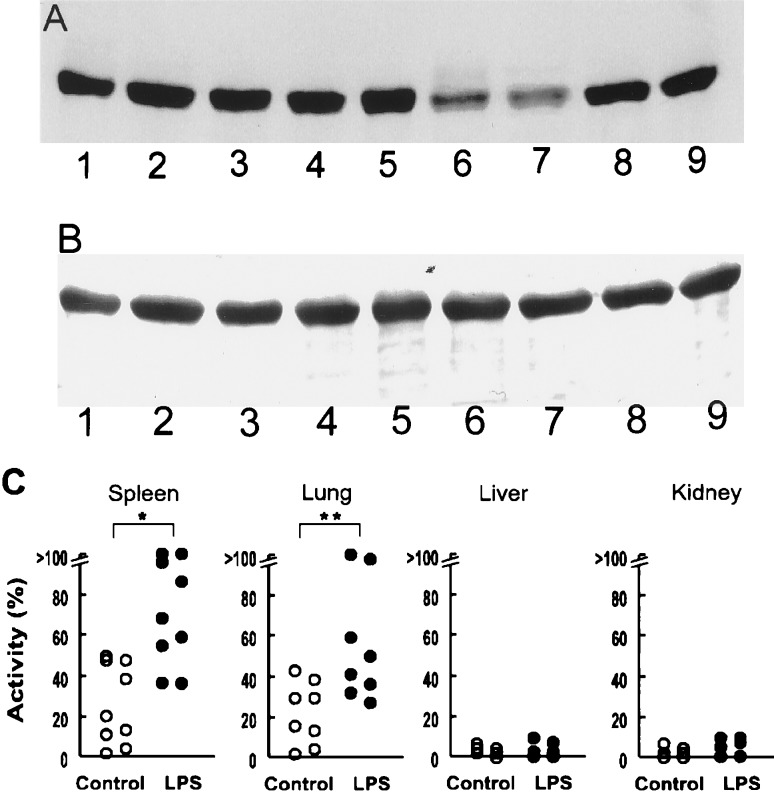
To test tissue homogenates for an activity that modified nitrated BSA, we examined tissues from control animals and rats treated with LPS (20 mg/kg intraperitoneally) for 24 h. We initially examined vascular-rich tissues such as spleen, lung, liver, and kidney. With our initial tissue homogenates from control rats, we found little or no activity that diminished the nitrotyrosine content of nitrated BSA (Fig. 1A). However, spleen and lung homogenates from animals treated with LPS decreased the nitrotyrosine content of nitrated BSA. The decrease in nitrated BSA was not associated with a decrease in Coomassie brilliant blue staining of the BSA band (Fig. 1B). These observations indicated that the decrease in nitrated BSA was not apparently because of proteolysis. In contrast, homogenates from liver and kidney did not decrease nitrated BSA levels (Fig. 1A). The amount of nitrated BSA in samples incubated with tissue homogenates was expressed as a percentage reduction of the nitrated BSA in control samples incubated without tissue homogenates (i.e., 100% was total loss of nitrated BSA). We found little or no activity in several homogenates from liver and kidney, whether or not animals were treated with LPS (Fig. 1C). However, the apparent activity in homogenates from spleen and lung was significantly greater in animals treated with LPS. Because the activity was greater in spleen homogenates, we used preparations from spleen for further characterization of the “denitrating activity.”
Figure 1.
Effects of LPS treatment on the activity of tissue homogenates to decrease nitrotyrosine content of nitrated BSA. (A) A representative Western immunoblot of nitrated BSA (1 mg/ml) that was incubated at 37°C for 10 min with tissue homogenates (2.5 mg/ml) of spleen (lanes 1, 6), lung (lanes 2, 7), liver (lanes 3, 8), and kidney (lanes 4, 9) from rats treated with LPS (20 mg/kg) for 24 h (lanes 6–9) or vehicle (control, lanes 1–4). Nitrated BSA was also incubated without tissue homogenate (lane 5). (B) Coomassie brilliant blue staining of a transferred membrane. Lanes are the same as in A. (C) Denitrating activity of tissues from (6–9) control rats (open symbols) and rats treated with LPS (solid symbols). Each symbol represents a different animal. Values are plotted as percent of maximal activity to decrease nitrated BSA. ∗, P < 0.01; ∗∗, P < 0.05, compared with control animals.
Time- and Concentration-Dependent Effects of Spleen Homogenates on Nitrated BSA.
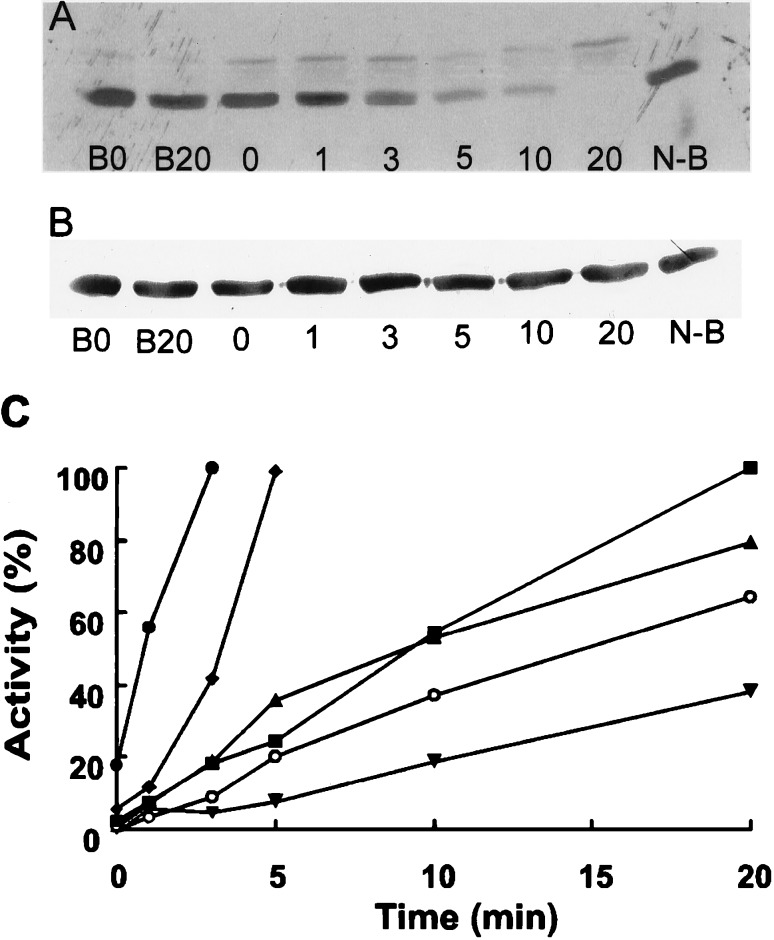
The effects of spleen homogenates in decreasing nitrated BSA as determined with Western immunoblots were time-dependent (Fig. 2A). The decrease in nitrated BSA was not associated with a decrease in BSA with Coomassie brilliant blue staining (Fig. 2B). The time-dependent decrease in nitrated BSA was observed with six different spleen homogenate preparations from control and LPS-treated rats (Fig. 2C). If the spleen homogenate was treated at 95°C for 10 min before incubation with nitrated BSA for 20 min, there was no decrease in nitrated BSA, indicating that the denitrating activity was heat labile (Fig. 2A).
Figure 2.
Effect of incubation time of spleen homogenate to decrease nitrated BSA. (A) Nitrated BSA was incubated for the designated times (0–20 min) with spleen homogenate from rats treated with LPS. Spleen homogenate was also boiled (B) before incubation for 0–20 min with nitrated BSA, as designated. Nitrated BSA (N-B) was also incubated without homogenate as a control. This is a representative experiment with six different preparations. (B) Coomassie brilliant blue staining of a transferred membrane. Lanes are the same as in A. (C) Time-dependent denitration of nitrated BSA with spleen homogenates from six different preparations representing six rats. Some rats were treated with LPS (solid symbols) while others (open symbols) received vehicle as controls.
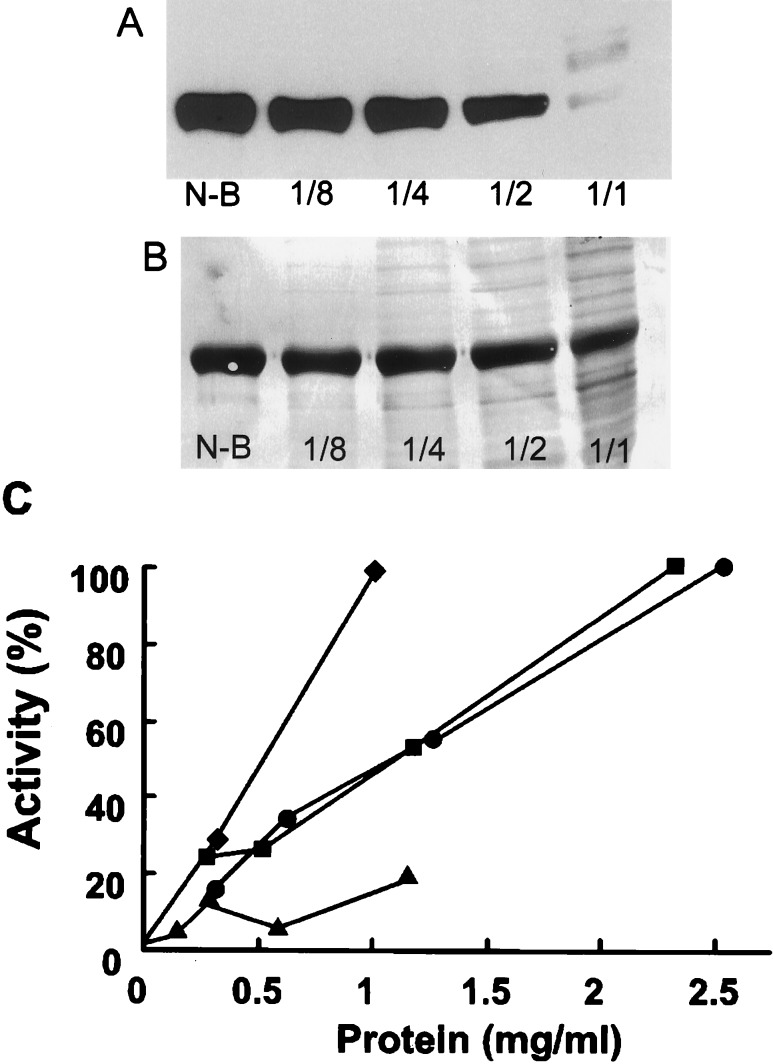
The denitrating activity in spleen homogenates diminished with decreasing protein concentration of the extracts during incubation (Fig. 3). The dependency of the activity on spleen homogenate concentration was observed with four different homogenate preparations (Fig. 3C).
Figure 3.
Effect of homogenate concentration on nitrated BSA. (A) Effect of a rat spleen homogenate at serial 2-fold dilutions incubated with nitrated BSA for 10 min. The protein concentration in the most concentrated homogenate (1:1) was 2.5 mg/ml. Control nitrated BSA is designated as N-B. This is a representative experiment with four different preparations. (B) Coomassie brilliant blue staining of a transferred membrane. Lanes are the same as in A. (C) Effect of protein concentration on denitration. Data from four different spleen homogenates from rats treated with LPS are presented.
After normalization of the relative activities, a linear regression analysis revealed that the correlation coefficients for time and protein dependency were 0.96 and 0.84, respectively.
Substrate Specificity of the Denitrating Activity.
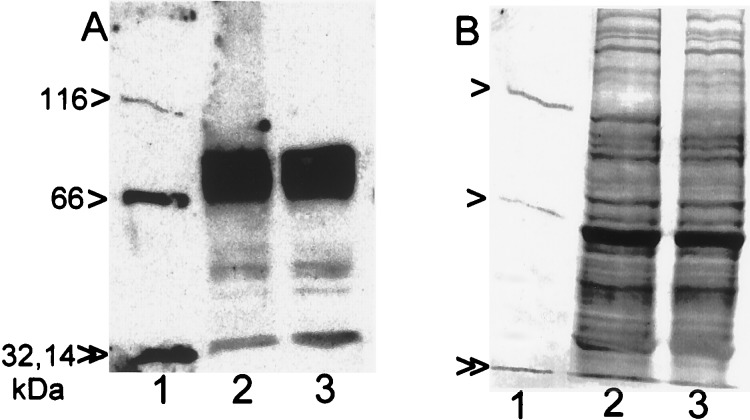
While the above experiments utilized SIN-1-nitrated BSA as substrate, similar data were obtained with BSA nitrated with tetranitromethane (data not shown). We also incubated spleen homogenates (50 μg of protein) with a mixture of nitrotyrosine-containing proteins (4.2 μg of total protein) (Fig. 4). Nitrotyrosine was coupled to β-galactosidase (116 kDa), BSA (66 kDa), carbonic anhydrase (32 kDa), and lysozyme (14 kDa). With incubation, all of these proteins lost their nitrotyrosine, as determined with Western immunoblots. However, the quantities of the corresponding proteins were not altered, as determined with Coomassie brilliant blue staining. Similar results were obtained when a commercial rabbit polyclonal anti-nitrotyrosine antibody was used for immunoblots (data not shown). Spleen homogenates also contained endogenous nitrotyrosine-containing proteins when greater quantities of extract were applied to gels. The endogenous nitrated proteins were 80, 40, and 30 kDa (Fig. 4A). They were not, however, substrates for the reaction because the intensity of the staining of these nitrotyrosine-containing proteins did not decrease with incubation (Fig. 4A).
Figure 4.
Substrate specificity of the spleen homogenate activity. (A) Nitrotyrosine-containing proteins were analyzed with Western immunoblotting after nitrated proteins (4.2 μg) (see text) were incubated without (lane 1) or with spleen homogenate (50 μg of protein) from a LPS-treated rat (lane 2). To examine endogenous nitrotyrosine-containing proteins in spleen homogenate, the spleen homogenate was also incubated without added nitrated proteins (lane 3). The arrowheads indicate the proteins with covalently coupled nitrotyrosine (116, 66, 32, and 14 kDa). (B) Coomassie brilliant blue staining of a duplicate gel. Lanes are the same as in A.
We also incubated free nitrotyrosine (0.5 mM) at 37°C for 20 min with increased concentrations of spleen homogenate (10 mg/ml). Under these conditions, 98 ± 5% (n = 3) of the added nitrotyrosine was recovered by HPLC analysis, indicating that the homogenate did not use free nitrotyrosine as substrate.
These studies indicated that other, but not all, nitrotyrosine-containing proteins could serve as a substrate for the activity. Presumably, the activity had some substrate specificity; this finding supported the view that the reaction was neither nonspecific nor nonenzymatic.
Other Properties of the Denitrating Activity in Spleen Homogenate.
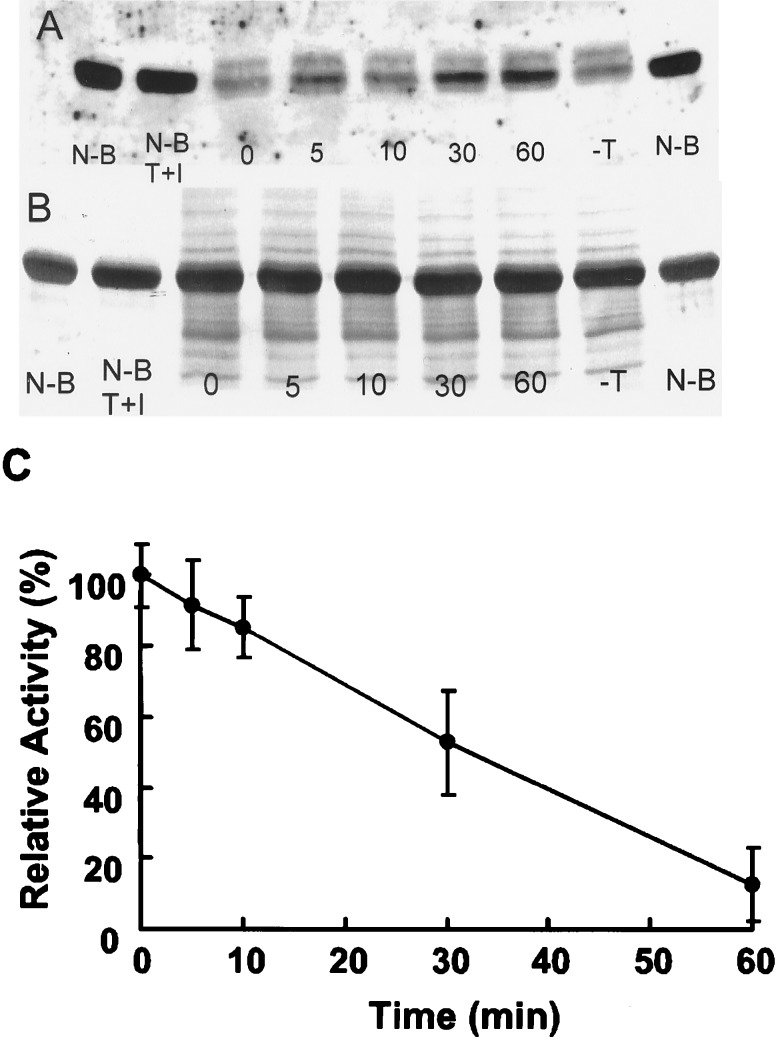
The activity in spleen homogenates was labile to heating (Fig. 2) and also to trypsin treatment. After dialysis of a spleen homogenate against buffer A without protease inhibitors, samples were treated with trypsin (20 μg/ml) for various times (0–60 min). Excess soybean trypsin inhibitor (100 μg/ml) was added to terminate digestions and samples were incubated with nitrated BSA (Fig. 5). While incubation of the dialyzed homogenate retained its activity (95 ± 7%, n = 5), incubation with trypsin produced a time-dependent decrease in the activity. The incubation of nitrated BSA with this low concentration of trypsin plus soybean trypsin inhibitor for 60 min resulted in no decrease in nitrated BSA.
Figure 5.
Effect of trypsin treatment on the activity in spleen homogenate. (A) Spleen homogenate (2 mg/ml) was treated with trypsin (20 μg/ml) at 37°C for the indicated times (min) or without trypsin for 60 min (-T). After addition of excess soybean trypsin inhibitor (100 μg/ml), samples were incubated with nitrated BSA for 10 min. Additional controls included nitrated BSA incubated without other additions (N-B) or with trypsin plus soybean trypsin inhibitor (N-B, T+I). (B) Coomassie brilliant blue stain of a transferred membrane from the experiment of A. Lanes correspond to those of A. (C) Effect of trypsin on the spleen denitrating activity as a function of time. Values are means ± SD of experiments with five different homogenates.
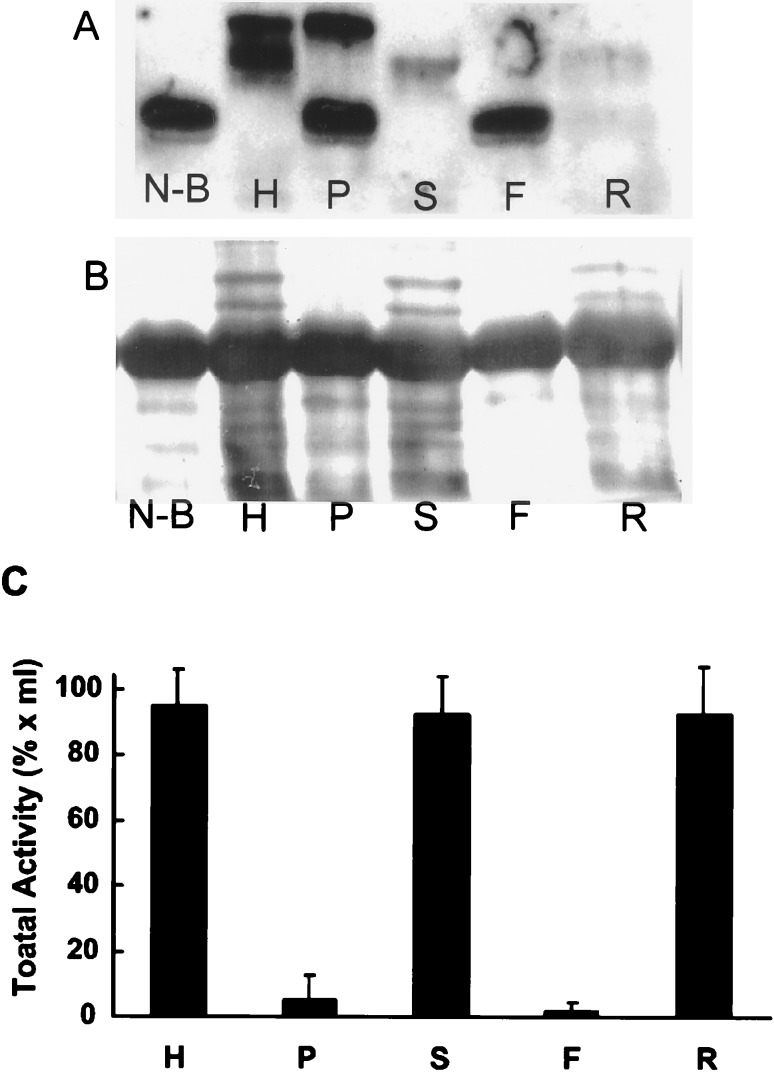
Spleen homogenates were also centrifuged at 100,000 × g for 60 min and the resulting supernatant and particulate fractions were tested for denitrating activity (Fig. 6). Most of the homogenate activity was recovered in the 100,000 × g supernatant fraction. When spleen homogenates were also subjected to membrane filtration by using Amicon 10 membranes to retain molecules >10 kDa, essentially all of the homogenate activity was recovered in the retained sample (Fig. 6). Furthermore, the retained samples with filtration were resuspended twice and recollected without apparent loss of enzyme activity. These latter experiments suggest that small cofactors are either not apparently required for activity or are tightly associated with the protein.
Figure 6.
Effects of centrifugation and membrane filtration on the activity in spleen homogenates. (A) The spleen homogenate (H) was separated into supernatant (S) and particulate (P) fractions with centrifugation at 100,000 × g. The supernatant fraction (S) was also subjected to membrane filtration by using an Amicon 10 membrane that retains 10,000-Da molecules. The retained material (R) and filtered material (F) were also examined for activity to denitrate the nitrated BSA (N-B). Incubated samples were examined for nitrated BSA with Western immunoblots. (B) Coomassie brilliant blue staining of a transferred membrane from the experiment in A. (C) Total enzyme activity in each of the fractions from eight different spleen homogenates was determined with appropriate dilutions. Columns and bars represent mean ± SD.
DISCUSSION
Post-translational modifications of proteins can participate in many regulatory processes. The phosphorylation, sulfation, adenylation, acylation, and other protein modifications can have dramatic effects on a protein’s structure, intracellular compartmentation, catalytic activity, or rate of degradation and turnover. It has been suggested that protein nitration increases the degradation of the protein (16). Presumably, protein nitration is but one of many possible post-translational modifications a protein may undergo. Often these modifications are reversible processes and, as noted above, they can have functional significance.
Thus, when protein nitration by peroxynitrite was described by Beckman and colleagues (7–9), we thought this could be a reversible and regulatable process that participated in cellular signaling by modifying the nitrated protein’s activity and/or function. To test this hypothesis, we approached the project from two directions. We examined cell culture and animal tissue extracts for nitrotyrosine-containing proteins on Western immunoblots after endotoxin and/or cytokine treatment to increase inducible nitric oxide synthase activity and nitric oxide formation. Endotoxin treatment has been used in models to mimic the effects of sepsis and to cause inflammation, processes that have been shown to be associated with the induction of nitric oxide synthase 2 and the increased formation of nitric oxide, peroxynitrite, and protein nitration. Interestingly, we found that many tissues and cells under control conditions have nitrotyrosine-containing proteins on Western immunoblots. In various tissue extracts, we found that the nitrotyrosine content of some proteins increased, decreased, or remained unchanged with endotoxin and/or cytokine treatment, indicating that protein nitration is a dynamic process. These studies will be reported elsewhere. As reported here, we also incubated nitrated BSA and other nitrated proteins with homogenates from tissues and cell cultures to examine for an enzyme activity that reversed protein nitration.
As described here, we used an antibody prepared to a nitrotyrosine-containing peptide coupled to keyhole limpet hemocyanin as the immunogen. The monoclonal antibody prepared has a high affinity for nitrotyrosine-containing proteins, is very specific for nitrotyrosine residues, and will recognize a variety of nitrated proteins with apparently similar affinity. The antibody does not recognize tyrosine or aminotyrosine. The properties of this antibody will be reported elsewhere. Preincubation of the antibody with nitrotyrosine or nitrotyrosine-containing proteins but not tyrosine, aminotyrosine, or aminotyrosine-containing proteins decreases the recognition of nitrotyrosine-containing proteins by the antibody (data not shown). We used this antibody to develop an assay for protein denitration by examining the loss of the nitrotyrosine epitope on Western immunoblots when nitrated proteins were incubated with tissue extracts. The assay proved to be sensitive and specific. We have also used the antibody to develop an ELISA method for nitrotyrosine-containing proteins. The method, to be reported elsewhere, will permit the detection of as little as 1–5 pmol of nitrated BSA.
Using Western immunoblots, we found that extracts from several rat tissues could decrease nitrated BSA on incubation. Homogenates of rat kidney, rat liver, and human erythrocytes could decrease nitrated BSA levels when samples were incubated in the absence, but not the presence, of various protease inhibitors (data not shown). The decrease in nitrated BSA on Western immunoblots was associated with the increase in smaller protein bands on Coomassie stained gels, presumably because of protein hydrolysis. With protease inhibitors present, the decrease in nitrated BSA was not observed with these tissue extracts. However, when homogenates from rat spleen or lung were used, nitrated BSA levels diminished without changes in the apparent BSA levels. The loss of the nitrated BSA epitope could result from modification of nitrotyrosine such as reduction to aminotyrosine, loss of the nitro functionality with a “denitrase,” protein hydrolysis, or possibly other modifications. Loss of the epitope without apparent decreases in BSA levels in incubations excludes protein hydrolysis. Because activity was retained after repetitive passage through a membrane with a 10,000-Da cutoff, the activity is presumably a macromolecule >10,000 Da, and no apparent small cofactors are evident. In other experiments (not shown), the enzyme activity was not decreased after gel filtration chromatography to remove small molecules and factors. These data suggest that a reductase activity is probably unlikely to explain our observations. Although the product(s) of the reaction is presently unknown, we believe that the activity will be a “denitrase” that removes the nitro group from nitrotyrosine in proteins. There is also some apparent substrate specificity for this activity. Although nitrated BSA and several nitrotyrosine-containing proteins could serve as substrate, free nitrotyrosine and some unknown endogenous nitrotyrosine-containing proteins were not effective substrates. These findings are consistent with our findings that endotoxin treatment of rats can increase or decrease the nitrotyrosine content of some select proteins (data to be reported elsewhere). The dynamic nature of these nitrotyrosine-containing proteins suggests that there may be some substrate selectivity for a “nitrotyrosine denitrase.” However, other interpretations are also possible. In our current studies, we have begun to purify the protein and scale up incubations to examine the product(s) of the reaction.
In rat spleen, the activity is increased 2- to 3-fold with endotoxin treatment, suggesting that the activity is regulatable and perhaps inducible. The increase in the activity with endotoxin and under conditions of nitric oxide synthase induction and increased peroxynitrite and nitrotyrosine formation is of interest. Perhaps the increased activity represents a regulatory mechanism to restore the function of the nitrated proteins with denitration and may protect the tissue from excess peroxynitrite formation.
The activity was found predominantly in 100,000 × g supernatant fractions of spleen homogenates, was time- and protein-concentration dependent, and was labile to heating or trypsin treatment, further supporting the view that the activity is a protein and probably an enzyme.
It has been shown that peptides and proteins that are nitrated on tyrosine residues are poor substrates for tyrosine kinases (16–18). Presumably the nitration of tyrosine on the 3-position and phosphorylation of the hydroxyl group on the 4-position interfere with each other. Thus, tyrosine nitration may compromise cell signaling via tyrosine kinase pathways. Therefore, an activity that removes the nitro functionality as a “nitrotyrosine denitrase” could have profound and important effects in cell signaling pathways. The denitration of proteins could permit the restoration of their function, allow them to become substrates for tyrosine kinases, and have important effects on cell signaling processes.
ABBREVIATIONS
- nitrated BSA
nitrotyrosine-containing BSA
- LPS
lipopolysacharide
- SIN-1
3-morpholinosydnonimine
References
- 1. Katsuki S, Arnold W, Mittal C K, Murad F. J Cyclic Nucleotide Res. 1977;3:23–35. [PubMed] [Google Scholar]
- 2.Arnold W P, Mittal C K, Katsuki S, Murad F. Proc Natl Acad Sci USA. 1977;74:3203–3207. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murad F, Mittal C K, Arnold W P, Katsuki S, Kimura H. Adv Cyclic Nucleotide Res. 1978;9:145–158. [PubMed] [Google Scholar]
- 4.Ignarro L, Murad F. In: Advances in Pharmacology. Ignarro L, Murad F, editors. Vol. 34. 1995. pp. 1–516. [Google Scholar]
- 5.Murad F. J Am Med Assoc. 1996;276:1189–1192. [Google Scholar]
- 6.Murad F. Rec Prog Horm Res. 1998;53:43–60. [PubMed] [Google Scholar]
- 7.Beckman J S, Beckman T W, Chen J, Marshall P A, Freeman B A. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckman J S, Ischiropoulos H, Zhu L, von der Woerd M, Smith C, Chen J, Harrison J, Martin J, Tsai M. Arch Biochem Biophys. 1992;298:438–445. doi: 10.1016/0003-9861(92)90432-v. [DOI] [PubMed] [Google Scholar]
- 9.Beckman J S, Ye Y Z, Anderson P G, Chen J, Accavitti M A, Tarpey M M, White C R. Biol Chem Hoppe-Seyler. 1994;375:81–88. doi: 10.1515/bchm3.1994.375.2.81. [DOI] [PubMed] [Google Scholar]
- 10.Kaur H, Halliwell B. FEBS Lett. 1994;350:9–12. doi: 10.1016/0014-5793(94)00722-5. [DOI] [PubMed] [Google Scholar]
- 11.Beckman J S, Carson M, Smith C D, Koppenol W. Nature (London) 1993;364:584. doi: 10.1038/364584a0. [DOI] [PubMed] [Google Scholar]
- 12.Eiserich J P, Hristova M, Cross C E, Jones A D, Freeman B A, Halliwell B, van der Vliet A. Nature (London) 1998;391:393–397. doi: 10.1038/34923. [DOI] [PubMed] [Google Scholar]
- 13.Bagasra O, Michaelis F H, Zheng Y M, Bobroski L E, Spistin S V, Fu Z F, Tawadros R, Koprowski H. Proc Natl Acad Sci USA. 1995;92:12041–12045. doi: 10.1073/pnas.92.26.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro L, Rodriguez M, Radi R. J Biol Chem. 1994;269:29409–29415. [PubMed] [Google Scholar]
- 15.Berlett B S, Friguet B, Yim M B, Chock P B, Stadtman E R. Proc Natl Acad Sci USA. 1996;93:1776–1780. doi: 10.1073/pnas.93.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gow A J, Duran D, Malcolm S, Ischiropoulos H. FEBS Lett. 1996;385:63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 17.Kong S K, Yim M B, Stadtman E R. Proc Natl Acad Sci USA. 1996;93:3377–3382. doi: 10.1073/pnas.93.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin B L, Wu D, Jakes S, Graves D J. J Biol Chem. 1990;265:7108–7111. [PubMed] [Google Scholar]
- 19.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Crow J P, Ischiropoulos H. Methods Enzymol. 1996;269:185–194. doi: 10.1016/s0076-6879(96)69020-x. [DOI] [PubMed] [Google Scholar]
- 21.Hensley K, Maidt M L, Pye Q N, Stewart C A, Wack M, Tabatabaie T, Floyd R A. Anal Biochem. 1997;251:187–195. doi: 10.1006/abio.1997.2281. [DOI] [PubMed] [Google Scholar]
- 22.Skinner K A, Crow J P, Skinner H B, Chandler R T, Thompson J A, Parks D A. Arch Biochem Biophys. 1997;342:282–288. doi: 10.1006/abbi.1997.0114. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]