Abstract
A major mechanism of azole resistance in Candida albicans is overexpression of the genes encoding the ATP binding cassette transporters Cdr1p and Cdr2p due to gain-of-function mutations in Tac1p, a transcription factor of the zinc cluster family. To identify the Tac1p regulon, we analyzed four matched sets of clinical isolates representing the development of CDR1- and CDR2-mediated azole resistance by using gene expression profiling. We identified 31 genes that were consistently up-regulated with CDR1 and CDR2, including TAC1 itself, and 12 consistently down-regulated genes. When a resistant strain deleted for TAC1 was examined similarly, expression of almost all of these genes returned to levels similar to those in the matched azole-susceptible isolate. Using genome-wide location (ChIP-chip) analysis (a procedure combining chromatin immunoprecipitation with hybridization to DNA intergenic microarrays), we found 37 genes whose promoters were bound by Tac1p in vivo, including CDR1 and CDR2. Sequence analysis identified nine new genes whose promoters contain the previously reported Tac1p drug-responsive element (CGGN4CGG), including TAC1. In total, there were eight genes whose expression was modulated in the four azole-resistant clinical isolates in a TAC1-dependent manner and whose promoters were bound by Tac1p, qualifying them as direct Tac1p targets: CDR1, CDR2, GPX1 (putative glutathione peroxidase), LCB4 (putative sphingosine kinase), RTA3 (putative phospholipid flippase), and orf19.1887 (putative lipase), as well as IFU5 and orf19.4898 of unknown function. Our results show that Tac1p binds under nonactivating conditions to the promoters of its targets, including to its own promoter. They also suggest roles for Tac1p in regulating lipid metabolism (mobilization and trafficking) and oxidative stress response in C. albicans.
Candida albicans causes mucosal, cutaneous, and systemic infections, including oropharyngeal candidiasis, the most frequent opportunistic infection among patients with AIDS (25, 40). Azole antifungal agents have proven effective in the management of oropharyngeal candidiasis; however, with increased use of these agents, treatment failures that have been associated with the emergence of azole-resistant strains of C. albicans have occurred (47, 52, 56, 63, 82).
The azole antifungals target lanosterol demethylase (Erg11p), a key enzyme in the ergosterol biosynthesis pathway (38). Several mechanisms of resistance to the azole antifungal agents have been described for C. albicans, including increased expression of genes encoding multidrug efflux pumps (27, 28, 47, 67, 69, 80, 81). These include the gene encoding a transporter of the major facilitator superfamily (MDR1) and genes encoding two ATP binding cassette (ABC) transporters (CDR1 and CDR2) (27, 28, 47, 69, 80). Overexpression of these efflux pumps is presumed to prevent accumulation of effective concentrations of the azole antifungal agents within the fungal cell. Among studies examining multiple matched azole-susceptible and -resistant sets of isolates, some isolates overexpress only MDR1, whereas others overexpress only CDR1 and CDR2 (47, 56). These observations suggest that two distinct transcriptional pathways are involved in regulating these efflux pumps.
Previous studies have shown that a wild-type CDR1 promoter fused to a luciferase reporter construct becomes activated when placed in an azole-resistant isolate that overexpresses CDR1 and CDR2 (20). By use of this system, it was shown that a conserved DNA sequence element in the CDR1 and CDR2 promoters, named drug response element (DRE) (5′-CGGAA/TATCGG), was necessary for CDR1 constitutive as well as drug-inducible transcriptional activation (20). These results suggested that a gain-of-function mutation in a transcription factor was the cause of the CDR1 and CDR2 constitutive overexpression in azole-resistant isolates (20). The zinc cluster transcription factor Tac1p (for transcriptional activator of CDR genes) was recently identified and shown to be responsible for CDR1 and CDR2 transcriptional activation (15, 16). An amino acid change from asparagine to aspartic acid at position 977 (N977D) in Tac1p was able to confer increased expression of CDR1 and CDR2 accompanied by decreased azole susceptibility (15). It was also shown that the DNA-binding domain of Tac1p fused to the glutathione S-transferase protein binds in vitro to the CDR1 and CDR2 DRE (16).
Interestingly, increased azole resistance is observed when a gain-of-function mutation is present in both TAC1 alleles (15). The TAC1 locus is on chromosome 5. Homozygosity at the TAC1 locus occurs either through mitotic recombination between copies of chromosome 5 or through the presence of extra copies of chromosome 5 harboring a gain-of-function TAC1 allele and loss of chromosome 5 with the wild-type TAC1 allele (15). Selmecki et al. demonstrated that a specific segmental aneuploidy, consisting of an isochromosome composed of the two left arms of chromosome 5, is associated with azole resistance. Increases and decreases in azole resistance were found to be strongly associated with gain and loss of this isochromosome (73). Recent work by Coste et al. showed that the effect of specific mutations in azole resistance genes located on chromosome 5 (TAC1 and ERG11) can be enhanced further by loss of heterozygosity and/or addition of extra copies of chromosome 5 (14).
In addition to CDR1 and CDR2, other targets of Tac1p (RTA3, HSP12, and IFU5), all of which contain a putative DRE in their promoter region, have been identified previously (16). In a separate study, it was shown that PDR16, encoding a putative phosphatidylinositol transfer protein contributing to clinical azole resistance, is also a target of Tac1p (66, 85). In the present study, we used functional genomic approaches, namely, genome-wide expression and location profiling, to identify the Tac1p regulon. Our results provide a more comprehensive picture of the molecular effects of Tac1p mutations in azole-resistant clinical isolates and suggest other important cellular functions for Tac1p in C. albicans.
MATERIALS AND METHODS
Strains and growth media.
The C. albicans strains used in this study are listed in Table 1. The matched clinical isolate sets and strain SZY31 were grown in yeast-peptone-dextrose (YPD) broth (Sigma-Aldrich, St. Louis, MO) at 30°C. The CAI4 strain and its epitope-tagged derivatives were grown at 30°C in YPD or, under selective conditions, in synthetic complete medium lacking uracil (74) supplemented or not with 100 μg ml−1 of uridine and 1 mg ml−1 of 5-fluoroorotic acid (5-FOA) (Toronto Research Chemicals, Inc., Toronto, Ontario, Canada). The Escherichia coli DH5α bacterial strain was used for DNA cloning and maintenance of the plasmid constructs.
TABLE 1.
Strains used in this study
| Strain | Parental strain | Genotype or description | FLC MIC (μg/ml) | Reference or source |
|---|---|---|---|---|
| C43 (DSY294) | Azole-susceptible clinical isolate | 0.25a | 67 | |
| C56 (DSY296) | C43 | Azole-resistant clinical isolate | 128a | 67 |
| Gu2 | Azole-susceptible clinical isolate | 6.25b | 28 | |
| Gu5 | Gu2 | Azole-resistant clinical isolate | >100b | 28 |
| 3 | 2 | Azole-susceptible clinical isolate | 8c | 83 |
| 17 | 16 | Azole-resistant clinical isolate | >64c | 83 |
| 5457 | Azole-susceptible clinical isolate | ≤0.5d | 66 | |
| 5674 | 5457 | Azole-resistant clinical isolate | 32.0d | 66 |
| SZY31 | 5674 | tac1Δ::FRT/tac1Δ::FRTf | 0.5e | 85 |
| CAI4 | ura3Δ::imm434/ura3Δ::imm434 | 2.0e | 26 | |
| SZY51 | CAI4 | TAC1/TAC1::HA3-URA3-HA3 | NDg | This study |
| SZY63 | SZY51 | TAC1/TAC1::HA3 | 2.0e | This study |
| SZY103 | SZY63 | TAC1::HA3 | 2.0e | This study |
| SZY106 | SZY63 | TAC1::HA3 | 2.0e | This study |
| SZY91 | SC5314 | tac1Δ/TAC1(N972D)-MPAR-FLP | 16.0e | 85 |
FLC concentration yielding at least 90% growth inhibition after 24 h of growth compared with the growth of the control (69).
Lowest FLC concentration in which little or no growth was visually detected after 48 h of growth.
MIC80 according to the CLSI (formerly NCCLS) macrodilution reference method or a microdilution modification of the CLSI method (57, 83).
MIC50 value, determined as the first concentration of the azole drug able to reduce growth by 50% compared with that of control cells grown in the absence of drug.
MIC70 value (see Fig. S1 in the supplemental material).
FRT, FLP recombination target.
ND, not determined.
Construction of C. albicans expression microarrays.
The nucleotide sequences corresponding to 6,165 open reading frames (ORFs) for C. albicans were downloaded from the Galar Fungail European Consortium (assembly 6, http://www.pasteur.fr/Galar_Fungail/CandidaDB/). We set out to design two nonoverlapping probe sets targeting the 3′ 600-bp region of each ORF. Each probe set consisted of 13 perfect-match 25-bp probes and their corresponding mismatch control probes containing a single mismatch in the center of the oligonucleotide. For ORFs less than 600 bp in length, the sequence was divided in two equal segments for subsequent design procedures. Optimum probe sets were selected by the Affymetrix design team based on their model, which, among other things, considers probe hybridization quality and cross-hybridization potential. Consequently, in some cases, only one probe set was selected for a given ORF. For quality control and normalization purposes, we made two to three additional probe sets spanning the entire sequence of the C. albicans 18S rRNA gene (GenBank accession no. M60302) and genes encoding GAPDH (glyceraldehyde-3-phosphate dehydrogenase), actin, and Mdr1p (Bmr1p) in addition to the standard Affymetrix controls (BioBCD, cre, DAP, PHE, LYS, and THR). In the end, the GeneChip array contained 10,736 probe sets, including nine controls, 6,123 unique ORFs, and duplicate probe sets for 4,604 ORFs. The duplicate probe sets are made to distinct regions of the ORF, thereby allowing two independent measurements of the mRNA level for that particular gene. The C. albicans custom Affymetrix NimbleExpress arrays (CAN04a530004N) were manufactured by NimbleGen Systems (1) per our specification.
RNA preparation for microarrays.
The matched clinical isolate sets and strain SZY31 were grown in YPD broth at 30°C in a shaking incubator to mid-log phase as described previously (61). The cell pellets were frozen and stored at −80°C prior to RNA preparation. Experiments were repeated independently three times. Total RNA was isolated using a hot sodium dodecyl sulfate (SDS)-phenol method (70). Frozen cell pellets were suspended in 12 ml of 50 mM sodium acetate (pH 5.2), 10 mM EDTA at room temperature, after which 1 ml of 20% SDS and 12 ml of acid phenol (Fisher Scientific, Waltham, MA) were added. This mixture was incubated for 10 min at 65°C with vortexing each minute, cooled on ice for 5 min, and centrifuged for 15 min at 12,000 × g. Supernatants were transferred to new tubes containing 15 ml of chloroform, mixed, and centrifuged at 200 × g for 10 min. The aqueous layer was removed to new tubes, and RNA was precipitated with 1 volume isopropanol and 0.1 volume 2 M sodium acetate (pH 5.0) and then collected by centrifugation at 17,000 × g for 35 min at 4°C. The RNA pellet was suspended in 10 ml of 70% ethanol, collected again by centrifugation, and suspended in nuclease-free water.
cRNA synthesis and labeling.
Immediately prior to cDNA synthesis, the purity and concentration of RNA samples were determined from A260/A280 readings and RNA integrity was determined by capillary electrophoresis using an RNA 6000 Nano Laboratory-on-a-Chip kit and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) per the manufacturer's instructions. First- and second-strand cDNA was synthesized from 15 μg of total RNA by use of a SuperScript double-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA) and oligo-dT24-T7 primer (PrOligo; Sigma-Aldrich) according to the manufacturer's instructions. cRNA was synthesized and labeled with biotinylated UTP and CTP by in vitro transcription using the T7 promoter-coupled double-stranded cDNA as a template and a Bioarray HighYield RNA transcript labeling kit (ENZO Diagnostics, New York, NY). Double-stranded cDNA synthesized from the previous steps was washed twice with 70% ethanol and suspended in 22 μl of RNase-free water. The cDNA was incubated as recommended with reaction buffer, biotin-labeled ribonucleotides, dithiothreitol, RNase inhibitor mix, and T7 RNA polymerase for 5 h at 37°C. The labeled cRNA was separated from unincorporated ribonucleotides by being passed through a CHROMA SPIN-100 column (Clontech, Mountain View, CA) and ethanol precipitated at −20°C overnight.
Oligonucleotide array hybridization and analysis.
The cRNA pellet was suspended in 10 μl of RNase-free water, and 10 μg was fragmented by ion-mediated hydrolysis at 95°C for 35 min in 200 mM Tris-acetate (pH 8.1), 500 mM potassium acetate, 150 mM magnesium acetate. The fragmented cRNA was hybridized for 16 h at 45°C to the C. albicans NimbleExpress GeneChip arrays. Arrays were washed at 25°C with 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 0.01% Tween 20 followed by a stringent wash at 50°C with 100 mM MES [2-(N-morpholino)ethanesulfonic acid], 0.1 M NaCl, 0.01% Tween 20. Hybridizations and washes employed an Affymetrix Fluidics station 450 with the standard EukGE-WS2v5 protocol. The arrays were then stained with phycoerythrin-conjugated streptavidin (Molecular Probes/Invitrogen), and the fluorescence intensities were determined using a GCS 3000 high-resolution confocal laser scanner (Affymetrix, Santa Clara, CA). The scanned images were analyzed using software resident in GeneChip operating system v2.0 (GCOS; Affymetrix). Sample loading and variations in staining were standardized by scaling the average of the fluorescent intensities of all genes on an array to a constant target intensity. The signal intensity for each gene was calculated as the average intensity difference, represented by [Σ(PM − MM)/(number of probe pairs)], where PM and MM denote perfect-match and mismatch probes, respectively.
Expression microarray data analysis.
The scaled gene expression values from GCOS software were imported into GeneSpring 7.2 software (Agilent Technologies) for preprocessing and data analysis. Probe sets were deleted from subsequent analysis if they were called absent by the Affymetrix criterion and displayed an absolute value below 20 in all experiments. The expression value of each gene was normalized to the median expression of all genes in each chip as well as the median expression for that gene across all chips in the study. Pairwise comparison of gene expression was performed for each matched experiment.
Q-PCR for expression data.
An aliquot of the RNA preparations from the samples used in the microarray experiments was saved for quantitative real-time reverse transcription (RT)-PCR follow-up studies. First-strand cDNAs were synthesized from 2 μg of total RNA in a 21-μl reaction volume by using a SuperScript first-strand synthesis system for RT-PCR (Invitrogen) in accordance with the manufacturer's instructions. Quantitative real-time PCRs (Q-PCRs) were performed in triplicate using a 7000 sequence detection system (Applied Biosystems, Inc., Foster City, CA). Independent PCRs were performed using the same cDNA for both the gene of interest and the 18S rRNA gene with SYBR green PCR master mix (Applied Biosystems, Inc.). Gene-specific primers were designed for the gene of interest and the 18S rRNA gene by using Primer Express software (Applied Biosystems, Inc.) and an Oligo analysis and plotting tool (QIAGEN, Valencia, CA) and are shown in Table 2. The PCR conditions consisted of AmpliTaq Gold activation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. A dissociation curve was generated at the end of each PCR cycle to verify that a single product was amplified by using software provided with the 7000 sequence detection system. The change in fluorescence of SYBR green I dye in every cycle was monitored by the system software, and the cycle threshold (CT) above the background for each reaction was calculated. The CT value of the 18S rRNA gene was subtracted from that of the gene of interest to obtain a ΔCT value. The ΔCT value of an arbitrary calibrator (e.g., untreated sample) was subtracted from the ΔCT value of each sample to obtain a ΔΔCT value. The gene expression level relative to the calibrator was expressed as 2−ΔΔCT. Statistical analysis was performed using R software, version 2.5.0 (www.r-project.org). Enrichments (n-fold) were compared using Student's t test. The statistical significance threshold was fixed at α = 0.05.
TABLE 2.
Primers used for quantitative real-time PCR expression analysis
| Gene | Primer paira | Amplicon size (bp) |
|---|---|---|
| 18S rRNA | F, 5′-CACGACGGAGTTTCACAAGA-3′ | 135 |
| R, 5′-CGATGGAAGTTTGAGGCAAT-3′ | ||
| CDR1 | F, 5′-ATTCTAAGATGTCGTCGCAAGATG-3′ | 140 |
| R, 5′-AGTTCTGGCTAAATTCTGAATGTTTTC-3′ | ||
| CDR2 | F, 5′-TAGTCCATTCAACGGCAACATT-3′ | 76 |
| R, 5′-CACCCAGTATTTGGCATTGAAA-3′ | ||
| PDR16 | F, 5′-CTGCGGGACAAGATTCATTAGC-3′ | 62 |
| R, 5′-TTGAGTACCAACAGGATGTGCTTTA-3′ | ||
| TAC1 | F, 5′-TGGCAATGTATTTAGCAGATGAGG-3′ | 71 |
| R, 5′-TGCTTGAACTGAGGTGAATTTTG-3′ |
F, forward; R, reverse.
Generation of a hemagglutinin (HA)-tagged Tac1p-expressing strain.
A DNA fragment overlapping positions −402 to +937 of the C. albicans URA3 ORF (URA3 marker) was PCR amplified with Pfu DNA polymerase (Stratagene, La Jolla, CA) from plasmid pCaEXP (12) by using primers 5′-ATCATCTCTAGATAAGGATGGTATAAACGGAAAC (introduces an XbaI site, underlined) and 5′-GCCCCGGGAAGGACCACCTTTGATTG (introduces an SmaI site, underlined). The resulting fragment (1,362 bp) was digested with XbaI and SmaI and used to swap the equivalent XbaI-XhoI fragment of plasmid pMPY-3×HA (1,078 bp, containing the Saccharomyces cerevisiae URA3 marker) (72), generating plasmid pCaMPY-3×HA. A TAC1-tagging cassette was amplified from plasmid pCaMPY-3×HA by use of primers (forward) 5′-ttgattgataattcagctgtaaatttgccatttagtcaatttaataatttacccaattttttctttgacaataatttggggattAGGGAACAAAAGCTGG-3′ (the lowercase sequence corresponds to positions +2860 to +2943 of the TAC1 ORF) and (reverse) 5′-taggaaaaaatatatgaaacaataaatatttacaaagatatacattatacatcgctttcaccaattacaactcttttttaacccCTATAGGGCGAATTGG-3′ (the lowercase sequence corresponds to positions +2947 to +3030 of the TAC1 ORF), which anneal specifically to the in-frame pCaMPY-3×HA vector sequences PET-up and PET-down (uppercase sequences), as described previously (72). The resulting fragment (1,821 bp), containing the C. albicans URA3 marker flanked by direct repeats of the triple HA (HA3) epitope-encoding sequences and 84 bp of sequences homologous to the 3′ end of the TAC1 gene, was used to transform strain CAI4. Counterselection of the URA3 gene was carried out on plates containing 5-FOA as described previously (8), except that uracil was replaced with uridine.
C. albicans transformation.
C. albicans transformations were performed as described previously (35), with minor modifications. The cells were grown overnight in 15 ml of YPD medium, diluted to an optical density at 600 nm (OD600) of 0.1 in 100 ml of fresh YPD, and allowed to grow to an OD600 of 0.4. The cells were harvested, washed once with 10 ml of sterile water, and resuspended in 1 ml of 1× lithium acetate (LiAc) solution (35). Cells (100 μl) were transferred to a sterile tube containing 6 μg of gel-purified PCR fragment and 100 μg of denatured salmon sperm DNA as the carrier. A LiAc-40% polyethylene glycol solution (700 μl of 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, pH 8.0, 100 mM LiAc, 40% [wt/vol] polyethylene glycol 4000) was added, and the cell suspensions were incubated overnight at 30°C with gentle rotation. The cells were heat shocked at 42°C for 15 min and plated on synthetic complete medium lacking uracil.
Genomic DNA isolation and Southern blot analysis.
C. albicans genomic DNA was prepared as described previously for S. cerevisiae (62). For the Southern hybridization, genomic DNAs (500 ng) were digested to completion with HindIII and EcoRV, electrophoresed on a 1% agarose gel, and transferred to a nylon membrane (Hybond-N; Amersham Biosciences, Piscataway, NJ). Prehybridization, hybridization, and washing steps were carried out as previously described (66). The TAC1 probe used consists of a 32P-radiolabeled 927-bp PCR-amplified fragment from SC5314 genomic DNA overlapping positions +2020 to +2946 in the TAC1 ORF. The membrane was exposed to a FUJIFILM imaging plate screen. The signal was quantified using the Multi Gauge program, version 2.3 (FUJIFILM). The membrane was subsequently exposed to Kodak XAR film at −80°C.
Total protein preparation and Western blotting.
Total protein was prepared from 2 OD units of strains CAI4 and SZY63 grown overnight, as described previously for S. cerevisiae (62). Extracts were boiled for 5 min, and 35 μl (out of 100 μl total) was separated by electrophoresis on an SDS-8% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane with a Trans Blot SD semidry transfer apparatus (Bio-Rad, Hercules, CA) and analyzed with a mouse anti-HA monoclonal antibody [HA probe (F-7): sc-7392; Santa Cruz Biotechnology, Inc., Santa Cruz, CA] at a dilution of 1:2,000 by use of a chemiluminescence detection system under conditions recommended by the manufacturer (Pierce Biotechnology, Inc., Rockford, IL).
Chromatin immunoprecipitation (ChIP).
Three independent cultures (50 ml each) of strains CAI4 and SZY63 were grown in YPD medium to an OD600 of 0.8. The cultures were then treated with 1.4 ml of 37% formaldehyde (final concentration, 1%) to induce protein-DNA cross-links. Cells were incubated for 30 min at room temperature with agitation and the cross-linking stopped by adding 1.2 ml of 2.5 M glycine. Cells were harvested, washed three times with 40 ml of ice-cold TBS buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl), and snap-frozen in liquid nitrogen. Total cell extracts were prepared by adding 0.7 ml of lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 10 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin) and 0.5 ml of acid-washed glass beads (Sigma) and bead beating four times for 5 min at the highest settings in a mini-bead beater (Biospec Products). Preparation of soluble chromatin fragments was performed by sonicating the extracts four times during 20 s at power 2 to 3 (output power, 8 to 9 W) using a Sonic Dismembrator model 100 sonicator (Fisher Scientific). Immunoprecipitation was performed by incubating the sonicated extracts (from both the tagged SZY63 strain and the untagged parental CAI4 strain) overnight with monoclonal mouse anti-HA antibody (Santa Cruz Biotech) coupled to magnetic beads (Dynabeads pan-mouse immunoglobulin G; Dynal Biotech, Brown Deer, WI) at 4°C. Beads were then washed twice with lysis buffer, twice with lysis buffer supplemented with 360 mM NaCl, twice with wash buffer (10 mM Tris-HCl, pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Na deoxycholate, 1 mM EDTA), and once with TE buffer (10 mM Tris, pH 8.0, 1 mM EDTA). The cross-links were reversed by incubating the washed beads overnight in 50 μl of TE-1% SDS at 65°C. The eluted material was then treated with proteinase K and RNase A and extracted twice with phenol:chloroform:isoamyl alcohol (25:24:1), and the DNA was precipitated with 100% ethanol and resuspended in 50 μl of TE. Forty microliters of the immunoprecipitated (IP) DNA fragments was used for amplification and Cy5/Cy3 dye labeling prior to hybridization to C. albicans intergenic DNA microarrays.
DNA labeling, hybridization to intergenic microarrays, and data analysis.
DNA labeling was conducted as described by Drouin and Robert (23). Briefly, the IP DNA fragments were blunted with T4 DNA polymerase and ligated to unidirectional linkers. The DNA was then amplified by ligation-mediated PCR in the presence of aminoallyl-modified dUTP. The labeling was carried out post-PCR using monoreactive Cy dye N-hydroxysuccinimide esters (Cy5/Cy3 monoreactive dye packs; Amersham Biosciences) that react specifically with the aminoallyl-modified dUTP [5-(3-aminoallyl)-2′ deoxyuridine-5′-triphosphate; Sigma-Aldrich]. Both pools of labeled IP DNA from the tagged SZY63 strain (Cy5) and the untagged parental CAI4 strain (Cy3) were mixed and hybridized to a C. albicans intergenic DNA microarray described elsewhere (H. Hogues, H. Lavoie, A. Sellam, M. Mangos, T. Roemer, E. Purisima, A. Nantel, and M. Whiteway, submitted for publication). Images of Cy5 and Cy3 fluorescence intensities were generated by scanning arrays using an Axon 4000b scanner and analyzed with GenePix Pro 4.1 software (Molecular Devices, Downington, PA). Normalization of the data was conducted using ArrayPipe 1.7 (32), and replicate slides (n = 3) were combined using a weighted-average method as described by Ren et al. (60).
Q-PCR confirmation of the genome-wide location data.
Q-PCR was performed with three independent CAI4 and SZY63 ChIP samples prepared as described above, except that the cell cultures were carried out in the presence of dimethyl sulfoxide (0.08%). Quantification of the DNA recovered from the CAI4 and SZY63 ChIPs was performed using a Quant-iT PicoGreen double-stranded DNA assay kit (Molecular Probes/Invitrogen). A standard curve was prepared using C. albicans SC5314 genomic DNA quantified by fluorometry and serially diluted in TE buffer (0, 0.1, 0.01, and 0.001 ng/μl). The CAI4 and SZY63 ChIP samples (1 μl) were resuspended in 49 μl of TE buffer. The samples (50 μl) were transferred, in duplicate, in a black 96-well plate (Costar 3694; Corning, Inc., Corning, NY), and 50 μl of the PicoGreen reagent was added. The fluorescence was measured using an Envision luminometer (Perkin-Elmer, Waltham, MA) at excitation and emission wavelengths of 485 and 535 nm, respectively. The DNA concentration of the ChIP samples was an average 500 pg/μl, yielding approximately 25 ng of total IP DNA per ChIP sample (50 μl).
Q-PCR assays were designed using the Universal ProbeLibrary (Roche Applied Science, Indianapolis, IN) (formerly the Exiqon ProbeLibrary) and TaqMan (Integrated DNA Technologies [IDT], Coralville, IA) methodologies. The different primer and probe combinations used for Q-PCR are listed in Table 3. The CDR1, PDR16, TAC1, and FUR1 promoter sequences were submitted to the web-based ProbeFinder software (version 2.34; Roche Applied Science) available on the Roche Applied Science website. The software assigned optimal PCR-specific primer sequences to be combined with the corresponding Universal ProbeLibrary probe for each promoter sequence (Universal ProbeLibrary probes; Roche Diagnostics Corp., Basel, Switzerland). We also used the PrimerQuest tool from the IDT website (http://www.idtdna.com/Scitools/Applications/Primerquest/) to design a custom TaqMan probe for the CDR1 promoter (with its corresponding forward and reverse primers) that binds closer to the DRE motif than the Universal ProbeLibrary probe. The C. albicans homologue of S. cerevisiae SPS4 (orf19.7568) was used as the reference promoter and FUR1 was used as a control to perform statistical analyses. The SPS4 and FUR1 genes were selected because (i) they were not modulated in our microarray expression study and (ii) their promoters were not enriched in the ChIP-chip experiments. A set of probe and primers was designed for each of the SPS4 and FUR1 promoters by use of ProbeFinder software (Table 3).
TABLE 3.
Primers used for quantitative real-time PCR binding assays
| Promoter | Primer/probe sequence (5′-3′)a | Amplicon locationb |
|---|---|---|
| CDR1 | F, GGTGCACACACACACAAACACACA | −435 → −341 |
| R, TTGAGCTCCCACTATCCGATCCCTA | ||
| P, TaqMan probe CCGCCCTCACTCTGTTCCATACAAAT | ||
| PDR16 | F, GAAAAGAAAAAGAAATGGCAACAT | −424 → −501 |
| R, TCGACACGTCTTTCCATCAC | ||
| P, Universal ProbeLibrary probe no. 77 (catalog no. 04689003001) GGTGGTGG | ||
| TAC1 | F, CACGAAGATAAAAATTGTGGTAGC | −1024 → −948 |
| R, AATTGTGCTGATATTTAATTGTTGGT | ||
| P, Universal ProbeLibrary probe no. 18 (catalog no. 04686918001) CAGCAGGA | ||
| FUR1 | F, GGTGCTTTTGGGAGAATGAA | −987 → −913 |
| R, CTTCCTCAAAACAAAACTGCAA | ||
| P, Universal ProbeLibrary probe no. 27 (catalog no. 04687582001) GCTGCCTG | ||
| SPS4c | F, TACAGTTGCCCCAGTCAACA | −636 → −574 |
| R, TGTCTTGGAACGGAAACTCA | ||
| P, Universal ProbeLibrary probe no. 15 (catalog no. 04685148001) TCCTGCTC |
F, forward; R, reverse; P, probe. The TaqMan probe was from IDT, and the Universal ProbeLibrary probes were from Roche.
Position according to the ATG start codon.
orf19.7568.
Q-PCR mixtures were prepared using TaqMan universal PCR master mix (Applied Biosystems, Inc.) according to the manufacturer's instructions. For reactions using probes from the Universal ProbeLibrary, 200 pg of ChIP DNA, 250 nM of each forward and reverse primer, 100 nM of the probe, 5 μl of TaqMan universal PCR master mix, and water were combined in a final volume of 10 μl. Mixtures for Q-PCRs using the TaqMan probe were prepared using the same conditions except that the probe and the primers (forward and reverse) were added to final concentrations of 100 nM and 200 nM, respectively. Reactions were performed in a MicroAmp optical 384-well reaction plate (Applied Biosystems, Inc.) by use of an ABI 7900 HT real-time PCR instrument with 1 cycle at 95°C for 10 min and 40 cycles at 95°C for 15 s and 60°C for 1 min. Each biological replicate sample (three) was processed in triplicate.
Data analysis was performed using Sequence Detection System software (SDS 2.2.2; ABI). For each sample, CT values were determined using the Sequence Detection System software. The severalfold enrichments of the targets (CDR1, PDR16, and TAC1 promoters) were calculated using relative quantification according to the 2−ΔΔCT method, where ΔCT = CT target − CT reference(SPS4 promoter) and ΔΔCT = ΔCT test(SZY63 ChIP sample) − ΔCT calibrator(CAI4 ChIP sample) (46). The FUR1 promoter was used as a negative control to confirm the severalfold enrichment obtained using the 2−ΔΔCT method. Statistical analysis was performed using R software (version 2.5.1; www.r-project.org), and the ΔΔCT values were compared using Welch's t test. The statistical significance threshold was fixed at α = 0.001.
DRE motif analysis.
The promoter sequences (1.5 kb upstream of the starting ATG) were retrieved from the Candida Genome Database (CGD) (http://www.candidagenome.org) and analyzed for the presence of potential DRE motifs by use of fuzznuc software from EMBOSS (http://emboss.bioinformatics.nl/).
Microarray data accession number.
Data files of each scanned chip were submitted to the Gene Expression Omnibus database (GEO; www.ncbi.nlm.nih.gov/geo/). The accession number for the series is GSE8727.
RESULTS
Global gene expression profile.
As a means to identify genes that are coregulated with the CDR1 and CDR2 multidrug efflux pumps and thus potentially new transcriptional targets of Tac1p, we performed gene expression profiling analyses of four matched sets of azole-susceptible and -resistant C. albicans clinical isolates in which the acquisition of azole resistance is associated with CDR1 and CDR2 overexpression (Table 1). Three of these azole-resistant isolates possess previously defined gain-of-function mutations in the C terminus of TAC1: an N977D amino acid substitution in isolate C56 (also known as DSY296) (15), an N972D amino acid substitution in isolate 5674 (85), and an A736C amino acid substitution in one allele and an L962 N969 deletion in the second allele of TAC1 in isolate 17 (14). Isolate 17 is the last in a series of 17 clinical isolates taken from a single human immunodeficiency virus-infected patient (80, 83). Relative to the parental isolate 2, isolate 3 exhibits increased expression of MDR1 whereas isolate 17 exhibits increased expression of CDR1 and CDR2 as well as MDR1 (80, 83). We therefore compared isolate 17 to isolate 3 to identify the genes coregulated with CDR1 and CDR2. The other azole-resistant isolate, Gu5, overexpresses CDR1 and CDR2 and presumably also contains gain-of-function mutations in Tac1p. The eight strains did not show major chromosomal rearrangements or aneuploidies, as determined by comparative genomic hybridization analyses (data not shown and J. Berman, personal communication).
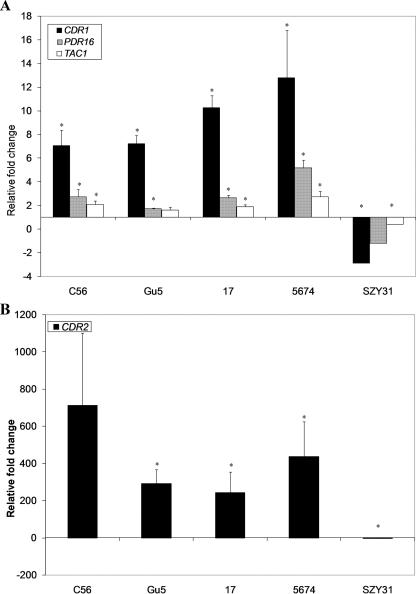
Three independent RNA samples per strain were hybridized to custom-designed Affymetrix C. albicans microarrays and the data analyzed as described in Materials and Methods. Genes were initially considered to be differentially expressed if (i) their average change (n-fold) in expression was ≥1.5 for each matched isolate, (ii) their expression changed by at least 1.5-fold in at least two of the three experiments for each matched isolate, and (iii) the average change (n-fold) was statistically significant by the t test. By use of these criteria, there were 222 genes up-regulated and 150 genes down-regulated in C56 compared to C43, 104 genes up-regulated and 63 genes down-regulated in Gu5 compared to Gu2, 126 genes up-regulated and 92 genes down-regulated in 17 compared to 3, and 327 genes up-regulated and 243 genes down-regulated in 5674 compared to 5457 (see Table S1 in the supplemental material). Genes which met these criteria for all four matched isolates and whose average change (n-fold) was statistically significant by the t test in at least two of the four matched isolates were included in a final data set (Tables 4 and 5) . We found 31 genes to be up-regulated in all four matched clinical isolate sets (Table 4), including the known Tac1p targets CDR1, CDR2, IFU5, HSP12, and RTA3 (16). Other up-regulated genes included GPX1 (putative glutathione peroxidase), CHK1 (histidine kinase), LCB4 (sphingoid long-chain base kinase), NDH2 (NADH dehydrogenase), SOU1 (sorbose dehydrogenase), orf19.3047 (transcription cofactor), and orf19.4531 (ABC transporter), as well as TAC1 itself, consistent with our previous proposition that TAC1 transcription is autoactivated, either directly or indirectly, as a consequence of gain-of-function mutations in Tac1p (85). We also found 12 genes to be down-regulated in all four matched clinical isolate sets. These included FTR1 (iron transporter), IHD1 (putative glycosylphosphatidylinositol-anchored protein of unknown function), and OPT6 (oligopeptide transporter), all of which encode integral membrane proteins, and SOD5 (superoxide dismutase), which encodes a cell wall protein (Table 5). As judged from the expression data, clinical isolate 5674, which carries the Tac1p N972D amino acid substitution, results in the highest impact on gene expression among the isolates tested, as 38 out of the 43 genes modulated in a TAC1-dependent manner had the highest modulation (n-fold) in this strain (Tables 4 and 5).
TABLE 4.
Genes up-regulated in all four matched clinical isolate sets
| Systematic namea | CGD nameb | CandidaDB namec | GO annotation (molecular function)d | Fold change in expressione in isolates:
|
||||
|---|---|---|---|---|---|---|---|---|
| C56 vs C43 | Gu5 vs Gu2 | 17 vs 3 | 5674 vs 5457 | SZY31 vs 5457 | ||||
| orf19.6000 | CDR1 | CDR1 | Multidrug transporter activity | 1.7 | 3.0 | 2.4 | 4.4 | −1.0 |
| orf19.5958 | CDR2 | CDR2 | Transporter activity | 50.6 | 14.1 | 17.1 | 50.5 | −1.3 |
| orf19.86 | GPX1 | Glutathione peroxidase activity | 2.3 | 3.0 | 2.3 | 4.0 | −1.2 | |
| orf19.896 | CHK1 | HK1 | Protein histidine kinase activity | 2.5 | 1.9 | 1.8 | 4.5 | 1.3 |
| orf19.3160 | HSP12 | HSP12 | 5.3 | 2.9 | 3.1 | 16.7 | −1.7 | |
| orf19.157 | IFA24.3 | 1.5 | 1.7 | 1.6 | 2.1 | 1.1 | ||
| orf19.2568 | IFU5 | IFU5 | 3.3 | 3.0 | 3.7 | 5.4 | 1.5 | |
| orf19.5257 | LCB4 | d-Erythro-sphingosine kinase activity | 2.2 | 1.7 | 2.2 | 4.0 | 1.1 | |
| orf19.278 | MTR | 3.1 | 1.8 | 1.9 | 5.2 | −1.1 | ||
| orf19.5713 | YMX6 | NDH2 | NADH dehydrogenase activity | 5.3 | 3.1 | 2.9 | 15.3 | 1.2 |
| orf19.7218 | RBE1 | PRY2 | 2.1 | 1.8 | 1.7 | 1.7 | 1.2 | |
| orf19.23 | RTA3 | RTA3 | Phospholipid-translocating ATPase activity | 40.4 | 15.9 | 22.0 | 41.0 | 1.0 |
| orf19.2896 | SOU1 | SOU1 | Sorbose dehydrogenase activity | 2.0 | 1.5 | 1.5 | 7.0 | −2.0 |
| orf19.3104 | YDC1 | 3.3 | 1.6 | 2.1 | 5.3 | 1.4 | ||
| orf19.2726 | IPF10262 | 2.5 | 2.8 | 1.9 | 6.2 | −2.9 | ||
| orf19.4459 | IPF11849 | 2.0 | 4.1 | 4.3 | 5.0 | −1.5 | ||
| orf19.2244 | IPF12897 | 1.8 | 1.7 | 1.5 | 5.4 | −1.7 | ||
| orf19.344 | IPF1514 | 15.5 | 4.7 | 4.0 | 28.9 | 1.9 | ||
| orf19.4907 | IPF17283 | 3.2 | 1.6 | 2.3 | 2.0 | 1.1 | ||
| orf19.5313 | IPF19810 | 36.0 | 29.4 | 18.1 | 143.1 | 1.7 | ||
| orf19.5777 | IPF19961 | 2.0 | 1.7 | 2.2 | 4.2 | −1.3 | ||
| orf19.6840 | IPF3535 | 1.8 | 2.3 | 1.9 | 2.3 | −1.4 | ||
| orf19.3047 | SIP3 | IPF3598 | Transcription cofactor activity | 1.5 | 1.5 | 1.7 | 3.0 | 1.3 |
| orf19.2531 | CSP37 | IPF4991 | ERf-to-Golgi vesicle-mediated transport | 2.5 | 1.8 | 5.5 | 6.8 | −1.3 |
| orf19.7310 | IPF5981 | 4.9 | 2.7 | 2.0 | 16.8 | 1.2 | ||
| orf19.1887 | IPF6464 | 2.3 | 1.8 | 1.7 | 4.5 | 1.2 | ||
| orf19.2790 | IPF7260 | Histone-lysine N-methyltransferase activity | 1.8 | 1.6 | 1.6 | 2.8 | −1.0 | |
| orf19.4531 | IPF7530 | ATPase activity | 2.7 | 3.8 | 3.5 | 4.0 | −1.6 | |
| orf19.4898 | IPF8950 | 2.4 | 1.6 | 1.8 | 3.4 | 1.1 | ||
| orf19.3188 | TAC1 | IPF9191.3f | 2.7 | 2.8 | 2.8 | 4.2 | −11.7 | |
| orf19.3644 | IPF9683 | 2.5 | 1.5 | 2.1 | 3.4 | 1.1 | ||
orf19 nomenclature according to the assembly 19 version. Systematic names in bold designate overlap with ChIP-chip experiments.
Gene name at CGD (http://www.candidagenome.org/).
Gene name at CandidaDB (http://genolist.pasteur.fr/CandidaDB/).
GO annotation found at CGD (http://www.candidagenome.org/).
Gene expression values in bold designate statistical significance (P < 0.05) in all three experiments.
ER, endoplasmic reticulum.
TABLE 5.
Genes down-regulated in all four matched clinical isolate sets
| Systematic namea | CGD nameb | CandidaDB namec | GO annotation (molecular function)d | Fold change in expressione in isolates:
|
||||
|---|---|---|---|---|---|---|---|---|
| C56 vs C43 | Gu5 vs Gu2 | 17 vs 3 | 5674 vs 5457 | SZY31 vs 5457 | ||||
| orf19.7219 | FTR1 | FTR1 | Ferrous iron transporter activity | −2.7 | −3.0 | −2.1 | −4.0 | −1.6 |
| orf19.5025 | MET3 | MET3 | Sulfate adenylyltransferase (ATP) activity | −2.0 | −7.9 | −1.8 | −2.5 | −1.2 |
| orf19.2947 | SNZ1 | SNZ1 | −3.7 | −1.8 | −1.7 | −2.2 | 1.3 | |
| orf19.5760 | IHD1 | IPF10662 | −1.9 | −2.2 | −3.1 | −9.0 | 1.7 | |
| orf19.3475 | IPF11725 | −3.9 | −2.1 | −2.7 | −5.8 | −1.2 | ||
| orf19.4655 | OPT6 | IPF12193 | Oligopeptide transporter activity | −1.8 | −2.0 | −2.4 | −3.1 | −1.4 |
| orf19.2060 | SOD5 | IPF1222 | Copper, zinc superoxide dismutase activity | −2.3 | −8.6 | −17.8 | −26.3 | −1.4 |
| orf19.2059 | IPF1228 | −2.0 | −1.7 | −1.7 | −2.9 | 1.2 | ||
| orf19.4749 | IPF13921 | −2.3 | −1.8 | −2.4 | −3.0 | −1.4 | ||
| orf19.670.2 | IPF20159 | −2.3 | −2.0 | −1.7 | −4.9 | −1.9 | ||
| orf19.6679 | IPF2314 | −1.5 | −2.1 | −1.8 | −2.2 | 1.1 | ||
| orf19.7561 | DEF1 | IPF946 | −3.4 | −1.5 | −1.6 | −2.8 | −1.4 | |
orf19 nomenclature according to the assembly 19 version.
Gene name at CGD (http://www.candidagenome.org/).
Gene name at CandidaDB (http://genolist.pasteur.fr/CandidaDB/).
GO annotation found at CGD (http://www.candidagenome.org/).
Gene expression values in bold designate statistical significance (P < 0.05) in all three experiments.
In order to determine which of these genes require Tac1p for their constitutive differential expression in these isolates, the gene expression profile of strain SZY31, a tac1Δ/tac1Δ derivative of the azole-resistant strain 5674 (Table 1), was compared with that of the azole-susceptible parental strain 5457 (85). Genes whose expression was modulated in strain 5674 compared to strain 5457 and whose expression in strain SZY31 was returned to levels similar to those observed for strain 5457 were determined to be dependent upon Tac1p for their up- or down-regulation. As shown in Tables 4 and 5, the majority of commonly differentially expressed genes were found to be Tac1p dependent.
We selected four genes of interest for confirmation of differential expression by real-time RT-PCR: CDR1, CDR2, TAC1, and PDR16. PDR16 was selected as it has been shown to be coregulated with CDR1 and CDR2 in association with azole resistance; yet, this is one of a limited number of genes not represented on the microarray used in this study (19, 66). As expected, all four genes were confirmed to be up-regulated in all four isolate sets (Fig. 1). Likewise, up-regulation of all four of these genes in isolate 5674 was ablated in strain SZY31 (Fig. 1).
FIG. 1.
Quantitative real-time RT-PCR analysis of selected genes differentially expressed in the microarray experiments. (A) Genes differentially expressed in each of the azole-resistant clinical isolates compared to their parent isolates. (B) CDR2 gene expression in each of the azole-resistant clinical isolates. Asterisks denote statistical significance by the t test (P ≤ 0.05). Error bars denote standard deviations.
Identification of Tac1p-binding sites in vivo.
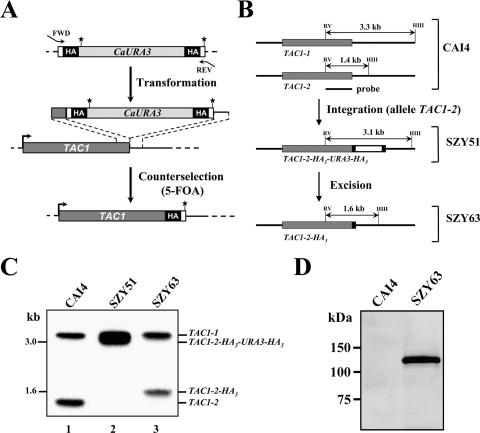
To determine if the TAC1-dependent genes that were differentially expressed in the clinical isolates have their promoters bound in vivo by Tac1p and to identify additional direct targets of Tac1p, we conducted genome-wide location analyses (ChIP-chip), a procedure combining ChIP with hybridization to DNA intergenic microarrays (60). For this purpose, Tac1p was tagged at its C terminus with a triple HA (HA3) epitope by use of homologous recombination at the TAC1 chromosomal locus (Fig. 2). First, we adapted the S. cerevisiae PCR epitope-tagging vector pMPY-3×HA (72) for its use in C. albicans by substituting the S. cerevisiae URA3 marker with the C. albicans URA3 gene (see Materials and Methods for details). The resulting vector (pCaMPY-3×HA) contains the C. albicans URA3 marker flanked by direct repeats of the HA3 epitope. The TAC1-tagging cassette was amplified with 100-bp primers (16 bp of vector sequences and 84 bp from the gene to be tagged), allowing tagging of Tac1p at its C terminus (Fig. 2A). After transformation of the CAI4 strain and integration of the cassette by homologous recombination, the marker was excised via recombination between the two directly repeated HA3 tags and the excision mutants were selected on 5-FOA medium (Fig. 2A). The resulting preexcision (strain SZY51) and postexcision (strain SZY63) strains were characterized by Southern hybridization using HindIII and EcoRV double-digested genomic DNA and the TAC1 probe (Fig. 2B). As predicted (Fig. 2B), the probe detected two fragments, one of 3.3 kb and one of 1.4 kb, in the CAI4 strain, corresponding to the wild-type TAC1-1 (orf19.3188) and TAC1-2 (orf19.10700) alleles, respectively (Fig. 2C, lane 1). Integration of the HA3-tagging cassette occurred at the TAC1-2 allele in the preexcision strain SZY51, as the 1.4-kb band was shifted to the 3.1-kb band (Fig. 2C, lane 2). Excision of the C. albicans URA3 marker in strain SZY63 was confirmed by the presence of the 1.6-kb band (Fig. 2C, lane 3). To detect the Tac1p-HA3 fusion protein, Western blotting was performed with strains CAI4 and SZY63 by use of an anti-HA monoclonal antibody (Fig. 2D). A signal corresponding to the Tac1p-HA3 fusion protein was detected in the HA-tagged SZY63 strain but not in strain CAI4 (control) (Fig. 2D). The size of this band was approximately 120 kDa, consistent with the predicted molecular size of the Tac1p-HA3 fusion protein (118 kDa).
FIG. 2.
Chromosomal tagging of Tac1p. (A) PCR epitope-tagging strategy for C-terminally tagging transcription factor Tac1p with the triple HA epitope. (Top) Primers (100 nucleotides) (see Materials and Methods) were designed such that the 5′ 84 bases of the forward (FWD) and reverse (REV) primers are homologous to sequences of the TAC1 gene and the 3′ 16 bases are complementary and in-frame to unique sequences (open boxes) in the tagging cassette which contains the C. albicans URA3 marker (CaURA3, light-gray box) flanked by direct repeats of the HA3-encoding sequences (HA, black boxes). The Tac1p stop codon is indicated by the asterisk. (Middle) PCR amplification results in a fragment whose ends include the primer sequences, allowing integration by homologous recombination of the tagging cassette upstream of the TAC1 3′-untranslated region (thick horizontal line). The orientation of the TAC1 ORF (dark-gray box) is indicated by the arrow. (Bottom) C. albicans URA3 marker excision results in the final product, TAC1-HA3. (B) Schematic representations of the TAC1-1 (orf19.3188) and TAC1-2 (orf19.10700) alleles (gray boxes) in strain CAI4. Sizes of the HindIII (HIII)/EcoRV (RV) double-digested fragments detected by the TAC1 probe (top, thick line) used for the Southern blot experiment are given for the TAC1-2 allele following integration of the HA3-tagging cassette (open box) and excision of the C. albicans URA3 marker through HA3 recombination. (C) Southern blot analysis of genomic DNA from the CAI4 strain and its URA3 preexcision (SZY51) and postexcision (SZY63) derivatives, digested with EcoRV and HindIII and hybridized with the TAC1 probe shown in panel B. Marker sizes are indicated on the left. (D) Western blot analysis of protein extracts from strains CAI4 and the Tac1p-HA3 integrant SZY63 with an anti-HA monoclonal antibody. Molecular size markers are indicated on the left.
Because the SZY63 integrant is heterozygous for the HA3 tag and carries a wild-type copy of the TAC1 gene (TAC1-1), it was not possible to test the functionality of the Tac1p-HA3 fusion protein in this background. Using growth on sorbose-containing medium, which induces the loss of one copy of chromosome 5 in which TAC1 is located (64), we constructed strains carrying only the HA3 epitope-tagged allele of Tac1p (see Fig. S1 in the supplemental material). Functional analysis of these strains by Northern blotting (see Fig. S1C in the supplemental material) and fluconazole (FLC) susceptibility testing (see Fig. S1D in the supplemental material) demonstrated that introduction of the HA3 tag at the C terminus of Tac1p did not alter Tac1p function (i.e., did not result in a loss- or gain-of-function mutation).
ChIP-chip experiments were conducted using the CAI4 and SZY63 strains and intergenic DNA microarrays containing 70-mer oligonucleotides covering the promoter regions of the C. albicans genome (see Materials and Methods). Using a P value of <0.001 (enriched binding ratio of ≥1.5), we identified 37 promoters bound by Tac1p (Table 6). These included the promoters of genes known to be regulated by Tac1p, CDR1 (10.5), IFU5 (3.0), PDR16 (2.1), RTA3 (1.8), and CDR2 (1.7) (16, 85), thus validating our ChIP-chip procedure and showing that Tac1p binds in vivo to the promoters of these genes to regulate their expression. HSP12, also known to be regulated by Tac1p, had a binding ratio of 1.2 (P = 0.09) and thus was not bound significantly by Tac1p. These experiments also identified several new targets, including genes involved in lipid metabolism and oxidative stress response as well as genes of unknown function (see Discussion). On two occurrences, Tac1p binding was found at promoters of two genes located on opposite strands (thus sharing the same probe on the chip): UGA33 (orf19.7317) and SUC1 (orf19.7319) as well as LPE10 (orf19.3455) and KIC2 (orf19.3456) (Table 6). While the expression data did not allow us to discriminate which of the LPE10 or KIC2 genes is regulated by Tac1p, we found that SUC1 was up-regulated in a TAC1-dependent manner in three out of the four clinical isolates tested by expression microarrays (see Table S1 in the supplemental material) while UGA33 expression was unchanged in three out of the four clinical isolates, suggesting that SUC1, a sucrose uptake zinc cluster regulator (37), is a target of Tac1p. The identification of several promoters bound in vivo by Tac1p under nonactivating conditions (i.e., in the absence of drugs or gain-of-function mutations) demonstrates that Tac1p is at least constitutively bound to its targets and suggests the possible involvement of postbinding mechanisms for Tac1p-mediated transcriptional activation (see Discussion).
TABLE 6.
List of Tac1p-enriched promoters (P < 0.001)
| Systematic namea | CGD/NRC nameb | CandidaDB namec | S. cerevisiae orthologd | GO terminologye | BRf | P valueg | E 5674/5457h | E SZY31/5457i |
|---|---|---|---|---|---|---|---|---|
| orf19.6000 | CDR1 | CDR1 | PDR5 | Transporter activity | 10.5 | 0.0000 | 4.4* | −1.0 |
| orf19.6869 | AST2 | IPF8041 | AST2 | Peptidase activity | 4.1 | 0.0000 | 1.6 | 1.9 |
| orf19.769 | IFE1 | IFE1 | BDH2 | 3.3 | 0.0000 | |||
| orf19.2568 | IFU5 | IFU5 | WWM1 | 3.0 | 0.0000 | 5.4* | 1.5 | |
| orf19.86 | GPX2 | GPX1 | HYR1 | Glutathione peroxidase activity | 2.8 | 0.0000 | 4.0* | −1.2 |
| orf19.3406 | IPF9898 | YHL008C | 2.5 | 0.0000 | −2.7* | −1.5 | ||
| orf19.7166 | IPF2186 | YGR110W | 2.8 | 0.0000 | 5.0 | 1.2 | ||
| orf19.7042 | IPF3080 | 2.1 | 0.0000 | 5.5* | 1.3 | |||
| orf19.93 | IPF14895 | MIC17 | 2.1 | 0.0000 | 1.5 | −1.1 | ||
| orf19.1027 | PDR16 | PDR17 | PDR16 | Phosphatidylinositol transporter activity | 2.1 | 0.0000 | 20.0j | 1.2j |
| orf19.3395 | IPF9483 | YHR048W | 2.0 | 0.0000 | ||||
| orf19.5877 | ATF1 | IPF1837 | ATF1 | Alcohol O-acetyltransferase activity | 2.0 | 0.0000 | 2.8* | 1.2 |
| orf19.6627 | IPF2557 | 1.9 | 0.0000 | 2.6* | −1.0 | |||
| orf19.23 | RTA3 | RTA3 | RSB1 | Phospholipid-translocating ATPase activity | 1.8 | 0.0000 | 41.0* | 1.0 |
| orf19.6501 | IPF3931 | YJU3 | 1.8 | 0.0000 | 4.6* | 1.1 | ||
| orf19.5037 | IPF17054 | 1.8 | 0.0000 | 7.7 | 2.1 | |||
| orf19.4898 | IPF8950 | FMP52 | 1.8 | 0.0001 | 3.4 | 1.1 | ||
| orf19.1267 | CAJ1 | IPF10278 | CAJ1 | Chaperone regulator activity | 1.7 | 0.0001 | 2.5 | −1.2 |
| orf19.1887 | IPF6464 | YEH1 | 1.7 | 0.0001 | 4.5 | 1.2 | ||
| orf19.691 | GPD2 | GPD2 | GPD1 | GAPDH (NAD+) activity | 1.7 | 0.0001 | 1.1 | 1.1 |
| orf19.2175 | IPF19998 | AIF1 | Oxioreductase activity | 1.7 | 0.0001 | 3.3 | 1.3 | |
| orf19.1444 | ENT2 | IPF17555.3 | ENT2 | Cytoskeletal adaptor activity | 1.7 | 0.0002 | 1.5 | 1.1 |
| orf19.5958 | CDR2 | CDR2 | PDR5 | Transporter activity | 1.7 | 0.0003 | 50.5* | −1.3 |
| orf19.3455k | LPE10k | IPF9782k | LPE10 | Magnesium ion transporter activity | 1.6 | 0.0003 | 1.7 | −1.7 |
| orf19.1089 | PEX11 | PEX11 | PEX11 | Peroxisome organization and biogenesis | 1.6 | 0.0003 | 1.8* | −2.4 |
| orf19.1665 | MNT1 | MNT1 | KTR1 | Alpha-1,2-mannosyltransferase activity | 1.6 | 0.0003 | ||
| orf19.7306 | IPF5987 | YPR127W | 1.7 | 0.0003 | 2.1 | 1.1 | ||
| orf19.7319k | SUC1k | SUC1k | MAL13 | Transcription factor activity | 1.6 | 0.0003 | 5.1* | 1.2 |
| orf19.1718 | ZCF8 | IPF19769 | 1.7 | 0.0004 | −1.6 | −1.8 | ||
| orf19.406 | ERG1 | ERG1 | ERG1 | Squalene monooxygenase activity | 1.7 | 0.0005 | −1.6 | 1.2 |
| orf19.7603 | IPF660 | YMR244C-A | 1.6 | 0.0005 | ||||
| orf19.5005 | OSM2 | OSM2 | OSM1 | Fumarate reductase (NADH) activity | 1.6 | 0.0005 | 2.0* | 1.2 |
| orf19.577 | IPF7353 | YDL057C | 1.6 | 0.0006 | 2.3 | 1.5 | ||
| orf19.5257 | LCB4 | LCB4 | LCB4 | d-Erythro-sphingosine kinase activity | 1.6 | 0.0007 | 4.0* | 1.1 |
| orf19.5525 | IPF4328 | YMR315W | 1.5 | 0.0008 | 2.3* | 1.1 | ||
| orf19.4476 | IFD6 | IFD6 | YPL088W | Aryl-alcohol dehydrogenase activity | 1.6 | 0.0009 | 2.2 | 1.2 |
| orf19.951 | IPF1548 | 1.6 | 0.0009 | −1.5 | 1.1 |
orf19 nomenclature according to the assembly 19 version. Systematic names in bold are modulated in the four sets of isolates, whereas those underlined are modulated at least in strain 5674.
Gene name according to CGD (http://www.candidagenome.org/) or the NRC Candida albicans database (http://candida.bri.nrc.ca/candida/index.cfm?page=CaGeneSearch).
Gene name according to CandidaDB (http://genolist.pasteur.fr/CandidaDB/).
S. cerevisiae ortholog or best hit according to CGD (http://www.candidagenome.org/).
GO terminology according to CGD.
BR, binding ratio. Probes were spotted in duplicate on the ChIP-chip arrays (H. Hogues, H. Lavoie, A. Sellam, M. Mangos, T. Roemer, E. Purisima, A. Nantel, and M. Whiteway, submitted for publication). The binding ratio with the most significant P value is shown.
P value of the corresponding binding ratio.
E 5674/5457, expression (n-fold) of the gene in strain 5674 relative to that in strain 5457. An asterisk (*) indicates that expression (n-fold) is statistically significant (see Table S1 in the supplemental material).
E SZY31/5457, expression (n-fold) of the gene in strain SZY31 relative to that in strain 5457.
Value obtained by Q-PCR.
The intergenic oligonucleotide probe corresponds to a common promoter of two adjacent genes. The orf19.3455 probe is shared by LPE10 and KIC2; the orf19.3719 probe is shared by SUC1 and UGA33.
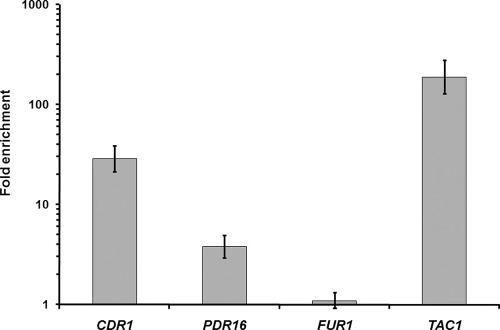
Using quantitative real-time PCR, we confirmed the binding of Tac1p to the CDR1 and PDR16 promoters, with enrichment ratios of 29.1 ± 4.0 and 4.1 ± 0.2, respectively (Fig. 3). As a control, we investigated binding of Tac1p to the promoter of the FUR1 gene, which was neither enriched in the ChIP-chip experiments nor modulated in the azole-resistant clinical isolates, and found no significant enrichment of that promoter by Q-PCR (1.1 ± 0.2), confirming the validity of the data obtained in the ChIP-chip experiments (Fig. 3).
FIG. 3.
In vivo enrichment of Tac1p binding at the CDR1, PDR16, and TAC1 promoters, determined using Q-PCR. The CAI4 and SZY63 strains were submitted to ChIP (three biological replicates), and the recovered DNA samples were analyzed by Q-PCR using Universal ProbeLibrary probes (Roche) for the PDR16, TAC1, SPS4, and FUR1 promoters or a TaqMan probe (IDT) for the CDR1 promoter. Enrichments (n-fold) are presented in log scale: 3.8 for the PDR16 promoter (95% confidence interval of 4.0, 5.0), 28.8 for the CDR1 promoter (95% confidence interval of 21.4, 38.9), 189.3 for the TAC1 promoter (95% confidence interval of 128.6, 278.6), and 1.1 for the FUR1 promoter (95% confidence interval of 0.9, 1.3), which was used as a negative control. Error bars denote standard deviations.
Although our results suggested that the expression of the TAC1 gene is controlled by Tac1p in an autoregulatory loop, the ChIP-chip data showed no enrichment for Tac1p at its own promoter (see Table S2 in the supplemental material), supporting an indirect autoregulation mechanism. However, the fact that the TAC1 promoter contains a DRE-like motif located between positions −1051 and −1060 relative to the ATG translation start codon (Table 7) (see below) and the TAC1 oligonucleotide printed on the intergenic chips is located at position −480 may have prevented us from detecting Tac1p binding at the TAC1 promoter. Therefore, we used for Q-PCR a set of TaqMan probe and primers hybridizing close to this DRE motif (Table 2). Under these conditions, we observed a strong enrichment of the TAC1 promoter (179.6 ± 25.2) (Fig. 3). These results strongly support a direct autoregulatory loop controlling TAC1 expression (see Discussion).
TABLE 7.
DRE motifs found in the Tac1p target promoters
| Motif definition | Systematic namea | CGD nameb | Motifc | Strandd | Positione |
|---|---|---|---|---|---|
| CGGN4CGG | orf19.6000 | CDR1 | CGGATATCGG | S | −457 |
| orf19.6869 | AST2 | CGGCTAACGG | A | −262 | |
| orf19.2568 | IFU5 | CGGAAATCGG | A | −235 | |
| orf19.7166 | CGGGTAACGG | S | −292 | ||
| CGGAGAACGG | S | −285 | |||
| orf19.7042 | CGGATAACGG | S | −731 | ||
| CGGTTACCGG | A | −632 | |||
| CGGATTACGG | A | −639 | |||
| orf19.5877 | ATF1 | CGGATATCGG | A | −206 | |
| orf19.6627 | CGGATATCGG | A | −176 | ||
| orf19.23 | RTA3 | CGGAACTCGG | S | −595 | |
| orf19.4898 | CGGGTGACGG | S | −232 | ||
| orf19.5958 | CDR2 | CGGAAATCGG | S | −220 | |
| orf19.1089 | PEX11 | CGGGGAACGG | A | −340 | |
| orf19.4476 | IFD6 | CGGTTGTCGG | A | −339 | |
| orf19.10700 | TAC1 | CGGAGCACGG | A | −1054f | |
| CGGN3CGG | orf19.3406 | CGGCAACGG | A | −332 | |
| orf19.1027 | PDR16 | CGGATTCGG | S | −558 | |
| orf19.5037 | CGGTTCCGG | S | −358 | ||
| CGGAACCGG | A | −351 | |||
| CGGGGGCGG | A | −250 | |||
| orf19.2175 | CGGAAGCGG | A | −167 | ||
| orf19.3455 | LPE10 | CGGCTGCGG | S | −246 | |
| orf19.1718 | ZCF8 | CGGAATCGG | S | −797 | |
| orf19.6869 | AST2 | CGGGCGCGG | A | −1161 | |
| orf19.10700g | TAC1 | CGGAAACGG | A | −1040 |
orf19 nomenclature according to the assembly 19 version.
Gene name at CGD (http://www.candidagenome.org/).
The underlined motifs have been described previously (16). Consensus sequences were obtained using the AlignX program (component of Vector NTI Advance 10.1.1; Invitrogen) and the WebLogo application (http://weblogo.berkeley.edu/) (17). The consensus sequence for CGGN4CGG motifs is CGGATAACGG, with nucleotide frequencies of 1.0, 1.0, 1.0, 0.6, 0.6, 0.7, 0.5, 1.0, 1.0, and 1.0, respectively. The consensus sequence for CGGN3CGG motifs is CGGAAGCGG, with nucleotide frequencies of 1.0, 1.0, 1.0, 0.5, 0.5, 0.4, 1.0, 1.0, and 1.0, respectively.
S, sense strand; A, antisense strand.
Nucleotide position from the ATG translation start site.
Position in contig19.20170, upstream of orf19.10700; position in contig.10170, upstream of orf19.3188, is −1051 due to sequence polymorphisms.
Present only in contig.20170 due to a sequence polymorphism (underlined) (CCGAAACGG).
Identification of potential Tac1p-binding motifs.
Tac1p belongs to the fungal-specific family of zinc cluster transcription factors that contain a DNA-binding domain formed by six cysteines coordinating two zinc atoms (48). Zinc cluster factors can bind as homodimers to CGG triplets with various orientations (everted, inverted, or direct repeats) and spacings (48). The CDR1 and CDR2 DRE consists of a direct CGG repeat with 4 intervening nucleotides (CGGAA/TATCGG), a configuration also found in the promoters of the other Tac1p-regulated genes RTA3 and IFU5 (16, 20). We thus looked for a DRE motif in the promoters of the 37 genes identified by the ChIP-chip experiments, using 1.5 kb of promoter sequence and the motif definition CGGN4CGG, which allows for complete degeneracy for the 4 nucleotides between the CGG repeats. In addition to CDR1, CDR2, RTA3, and IFU5, this analysis identified eight new genes containing the CGGN4CGG sequence, AST2, ATF1, PEX11, IFD6, and four ORFs of unknown function (orf19.7166, orf19.7042, orf19.6627, and orf19.4898), for a total of 15 matches in 12 genes (Table 7). As a control, we searched the CGGN4CGG motif in 1.5 kb of promoter sequence from 6,068 ORFs and found an average of 1.5 matches per 37 promoters, yielding a 10-fold enrichment for the presence of the CGGN4CGG sequence in the Tac1p-bound promoters. We also searched the 37 enriched promoters for the motif CGGN3CGG, since our unpublished data indicated that a CGGATTCGG sequence in the PDR16 promoter is involved in its transcriptional activation by Tac1p. This analysis identified seven genes (including PDR16), one of them with three CGGN3CGG motifs (Table 7). Searching the 6,068 promoter sequences yielded an average of 2.2 genes per 37 promoters, resulting in a 3.2-fold enrichment for that sequence in the Tac1p-bound promoters.
Finally, we examined whether the CGGN3CGG and CGGN4CGG motifs appear in the promoter regions of the genes whose expression was modulated in all four resistant isolates (31 up-regulated and 12 down-regulated) compared to their matched parental strains. We found seven up-regulated genes containing the CGGN4CGG motif, five of which were also identified by the ChIP-chip experiments (CDR1, CDR2, IFU5, RTA3, and orf19.4898) (the two others being TAC1 and orf19.3047), as well as two up-regulated genes with the CGGN3CGG motif (TAC1 and orf19.344) (Table 7). Finally, a search of the down-regulated genes identified one gene with the CGGN4CGG motif (SOD5) and one gene with the CGGN3CGG motif (OPT6).
Expression and location data mining.
When merging the expression and location data, we identified eight genes whose promoters were bound by Tac1p in vivo and which were up-regulated in the four azole-resistant clinical isolates in a TAC1-dependent manner, thus qualifying these genes as bona fide Tac1p targets. These genes were CDR1 and CDR2, GPX1 (putative glutathione peroxidase), LCB4 (putative sphingosine kinase), and RTA3 (putative phospholipid flippase), as well as three genes of unknown function: IFU5, orf19.1887, and orf19.4898 (Tables 4 and 6). We also identified nine genes whose promoters were bound by Tac1p in vivo and which were significantly modulated in a TAC1-dependent manner in at least strain 5674, the isolate displaying the strongest severalfold change in gene expression. These genes were orf19.5877 (ATF1, alcohol acetyltransferase), orf19.1089 (PEX11, fatty acid oxidation), orf19.5005 (OSM2, fumarate reductase), and orf19.7319 (SUC1, sucrose metabolism), as well as five ORFs of unknown function (orf19.6627, orf19.7042, orf19.5525, orf19.3406, and orf19.6501) (Table 6; see also Table S1 in the supplemental material). These results suggest that, in addition to its function in azole drug resistance, Tac1p regulates other cellular functions, such as lipid metabolism and oxidative stress response (see Discussion).
DISCUSSION
To identify the Tac1p regulon, we combined genome-wide expression and location analyses, two technologies with complementary strengths and limitations. Expression profiling is extremely valuable since it can identify all of the changes in transcript abundance associated with the perturbation of a specific transcriptional regulator; however, it cannot distinguish between direct and indirect effects at individual target promoters. Location profiling is a very powerful tool to identify all of the direct targets of a specific transcription factor; however, it does not allow the determination of whether the bound factor acts as an activator or a repressor of these targets. Each method also gives rise to false positives and false negatives. Merging the results from the two approaches thus generates data that are most complete and cross-validated.
While merging the expression and location data sets for Tac1p, we found that, out of 37 genes whose promoters were bound by Tac1p, 20 (64%) did not have their expression modulated (Table 6). Possible technical explanations for this limited overlap are that some of these genes did not pass the stringent criteria applied to the expression microarray data or that they represent false positives. However, a biological explanation could be that the expression of the bound promoters is not modulated in response to gain-of-function mutations in Tac1p but rather in response to other potential Tac1p-activating signals. We also found that, out of 43 modulated genes, 35 (81%) were not identified in the location profiling experiments, suggesting that these genes are indirect targets of Tac1p (i.e., controlled by Tac1p-regulated transcription factors and/or other transcriptional regulators) or that they are not bound by Tac1p under nonactivating conditions (see below). It is also possible that the detection of Tac1p binding to some of these targets may have been prevented by the fact that their Tac1p-binding site is too far from the oligonucleotide sequence printed on the intergenic chip, as we found for TAC1. Last, some of these genes could have been missed by use of a P value of <0.001 as the cutoff in the location array experiments. This seems to be the case for the SIP3 gene, whose expression was up-regulated in the four azole-resistant isolates in a TAC1-dependent manner and which had a binding ratio of 1.6 but a P value of 0.0014. Taken together, combining the expression and location profiling data allowed us to identify many new genes which unambiguously belong to the Tac1p regulon and thereby gain new insights into the biological functions of Tac1p.
Previously, we examined the gene expression profiles of matched isolates 2, 3, 15, and 17 by using an earlier-generation microarray (61). In that analysis, we identified five genes as being coregulated with CDR1 and CDR2. Two of these, GPX1 and RTA3, were found in the present study to be up-regulated in all four matched isolate sets studied, whereas a third, ERG2, was found to be up-regulated in three of these isolate sets. Likewise, of the 14 genes we previously found to be down-regulated in association with azole resistance in isolate 17, orf19.3475, orf19.2060, and orf19.5760 were found to be down-regulated in all four isolate sets, and FET34 was found to be down-regulated in three of the four in this study. Using the same early-generation arrays, Coste et al. examined the gene expression profiles of isolate C56 and strain DSY2926 (isolate C43 expressing a TAC1 allele carrying a gain-of-function mutation) compared to that of their parent isolate, C43 (16). They identified seven genes to be up-regulated in common between these two experimental conditions. In addition to CDR1 and CDR2, these included RTA3 and HSP12, both of which were found to be up-regulated in all four matched isolate sets examined in the present study. In a separate study by the same group, Karababa et al. examined the gene expression profile of isolate C56 compared to that of parent isolate C43 as well as the profile of strain CAF2-1 exposed to the CDR1- and CDR2-inducing agent fluphenazine compared to the profile of this strain grown in the absence of drug (36). In addition to CDR1 and CDR2, they observed seven genes to be commonly up-regulated between these two conditions. These were HSP12, GPX1, RTA3, orf19.344, IFU5, orf19.1862, and orf19.7284, all of which were found in the present study to be up-regulated in either three or all four of the clinical isolates examined. In this report, we used a more-recent-generation microarray together with well-characterized sets of azole-resistant clinical isolates and a tac1Δ/tac1Δ mutant, which allowed us to identify 36 additional genes whose expression is modulated in azole-resistant strains, dependent upon Tac1p.
Known Tac1p targets, such as CDR1, CDR2, IFU5, and RTA3, are induced in a TAC1-dependent manner upon exposure of the cells to fluphenazine or estradiol or upon expression of a gain-of-function TAC1 allele in a tac1Δ/tac1Δ background (16, 20). This indicates that the activation of Tac1p by such inducers or by gain-of-function mutations is required for Tac1p-mediated transcriptional regulation. Our ChIP-chip experiments were done with cells grown in rich media under uninduced conditions (45). We found that, under these conditions, Tac1p binds to its target promoters, indicating that this binding is constitutive or at least partially constitutive since it cannot be excluded that Tac1p binding increases in the presence of an inducer or an activating mutation. Our functional characterization of the epitope-tagged Tac1p strain showed that Tac1p binding to its targets is not due to an activating effect of introducing the HA tag at the C terminus of Tac1p (see Fig. S1 in the supplemental material). It was reported recently that the S. cerevisiae zinc cluster regulator Pdr1p, which controls the expression of the multidrug transporters PDR5, SNQ2, and YOR1 (6), also binds constitutively to its target promoters in vivo (24). Likewise, the S. cerevisiae zinc cluster transcription factor War1p, which controls the expression of the ABC transporter Pdr12p in response to weak acid stress, has been shown to constitutively bind to the PDR12 promoter in vivo (43). Thus, the mechanisms by which Tac1p activates transcription in response to drugs or to gain-of-function mutations appear to be similar to those already documented for zinc cluster factors in S. cerevisiae and most likely involve postbinding mechanisms, such as loss of interaction with a repressor protein, as proposed for Upc2p and Ecm22p (18), or recruitment of coactivator complexes (SAGA, Mediator, SWI/SNF), as shown for Pdr1p (30).
Our analyses of the Tac1p-bound promoters for the presence of the DRE-like motif CGGN4CGG allowed us to identify eight new Tac1p targets, in addition to CDR1, CDR2, RTA3, and IFU5, containing this sequence (Table 7). The fact that 25 of the Tac1p-bound promoters do not contain this motif suggests that Tac1p may recognize additional configurations of the CGG triplets, including monomeric CGG triplets or CGG triplets with different spacings and/or orientations (48). The latest proposition is supported by our unpublished data that a CGGATTCGG sequence in the PDR16 promoter is involved in its transcriptional activation by Tac1p and the enrichment of the CGGN3CGG motif in the Tac1p-bound promoters (Table 7). Alternatively, Tac1p may bind upstream or downstream of the DNA sequences analyzed (1.5 kb of upstream sequences). Finally, it is also possible that Tac1p binds to some of its targets indirectly through its association with other DNA-binding proteins. For instance, the S. cerevisiae zinc cluster proteins Rsc30p and Rsc3p, which are part of the chromatin remodelling complex, have been shown to bind indirectly to DNA (2). Whether or not the DRE motifs identified in the Tac1p-bound promoters are functional as well as the sequences/factors mediating Tac1p binding to its target promoters in the absence of a classical DRE remains to be determined experimentally.
We showed previously that strain 5674 overexpresses the TAC1 gene and that introduction of a TAC1 allele carrying the N972D gain-of-function mutation in a tac1Δ/tac1Δ strain leads to the constitutive upregulation of the TAC1 transcript, which suggested that Tac1p is positively autoregulated, directly or indirectly (85). We show in the present study that (i) TAC1 is upregulated in three additional azole-resistant isolates in which the Tac1p pathway is activated (Table 4), (ii) the TAC1 promoter contains a DRE motif, and (iii) Tac1p binds in vivo to its own promoter (Fig. 3). Taken together, our findings support the proposition that TAC1 expression is controlled by a direct positive autoregulatory loop. This situation is similar to that of S. cerevisiae Pdr3p, which binds in vivo to two pleiotropic DREs located in the PDR3 promoter to transactivate its own expression (27). It appears that direct self-regulatory loops are a common feature among zinc cluster transcription factors in yeast, including Pdr3p and Yrr1p (pleiotropic drug resistance), Hap1p (respiration), and Stb5p (pentose phosphate pathway) (48). Studies of budding yeast have shown that transcription factor autoregulation is necessary to respond to environmental stresses. As examples, autoregulation of PDR3 in a pdr1Δ background is crucial for growth on a medium containing cycloheximide (27) and autoregulation of the basic leucine zipper transcription factor Hac1p, which controls the unfolded protein response, is required to protect the cells from prolonged endoplasmic reticulum stress (55). Similarly, autoregulation of Candida glabrata AMT1, a copper-sensing transcription factor, is necessary to protect the cells upon exposure to high environmental copper levels (84). Thus, one possible outcome of Tac1p autoregulation would be an amplifiable production of the Tac1p protein necessary for rapid and sustained response to drugs and yet-unknown activating signals (45).
Three previously identified Tac1p targets (CDR1, CDR2, and PDR16) have been shown to play a role in azole drug resistance (59, 66, 68). Thus, Tac1p confers azole resistance by activating different effectors, each contributing to some extent to the overall azole resistance of the cells. Additional Tac1p targets identified by our studies and coregulated with CDR1, CDR2, and PDR16 may also play a protective role against toxic injuries. For instance, orf19.4531 is found among the group of genes differentially expressed under the gene ontology (GO) molecular function term “ATPase activity coupled to movement of substances,” including CDR1, CDR2, and RTA3 (Table 4). orf19.4531 encodes a putative ABC transporter of the PDR subfamily (to which CDR1 and CDR2 belong). Whether this transporter impacts azole resistance, alone or in conjunction with Cdr1p and Cdr2p, remains to be determined. This efflux pump may also protect the cell from other toxic compounds. As another example, orf19.86 (GPX1), which encodes a putative glutathione peroxidase, is an integral component of the glutathione and glutathione-dependent enzyme system, which has been implicated in the resistance of tumor cells to anticancer agents (7, 22). Increased activity of this enzyme system is often observed in conjunction with increased activity of the ABC transporter P glycoprotein in drug-resistant human cancer cells (9).
Although not found among the 37 promoters observed to be bound by Tac1p, CHK1 was among the 31 genes that were consistently coregulated with CDR1 and CDR2 in all four matched isolate sets. Its up-regulation in these isolates is at least influenced by Tac1p, as deletion of TAC1 in isolate 5674 reduced its expression to normal levels. Chk1p is a histidine kinase involved in a two-component signaling pathway, along with the response regulator Ssk1p, which regulates cell wall biosynthesis (10, 11). Interestingly, it was shown recently that strains of C. albicans lacking either of these signal transduction proteins are hypersensitive to FLC (13). Up-regulation of CHK1 in association with CDR1- and CDR2-mediated azole resistance suggests that, in addition to its requirement for baseline azole tolerance in azole-susceptible cells, this protein may contribute to azole resistance in clinical isolates. In the related fungal species C. glabrata, the ATPase activity of the ABC transporter Cdr1p and the drug efflux activity of the ABC transporter Pdh1p (Cdr2p) are regulated by phosphorylation (77, 78). Likewise, phosphorylation has been shown to modulate transcriptional activity of the transcription factor Gal4p (65) and has been suggested to regulate the activity of the transcriptional regulator Pdr3p (49). It is therefore tempting to speculate that Chk1p is involved in the phosphorylation and activity of the efflux pumps Cdr1p and Cdr2p or possibly Tac1p itself. Further investigation into the contribution of this signaling pathway to azole resistance is required to address this question.
Our genome-wide location experiments revealed that Tac1p binds to the promoters of a group of genes involved (or predicted to be involved) in lipid metabolism. These genes are CDR1, CDR2, PDR16, RTA3, ATF1, ERG1, LCB4, orf19.6501, orf19.7166, and orf19.1887. With the exception of ERG1, all of these genes were significantly up-regulated in a TAC1-dependent manner at least in one out of the four clinical isolates tested by expression microarrays, strongly suggesting (i) that these genes are direct transcriptional targets of Tac1p and (ii) that there is a role for Tac1p in lipid metabolism. It has been shown that Rsb1p, the S. cerevisiae orthologue of C. albicans Rta3p, plays an essential role in the translocation of long-chain bases across the plasma membrane (39), suggesting a role for Rsb1p, and thus potentially Rta3p, in regulating the sphingolipid composition of the plasma membrane. Interestingly, S. cerevisiae LCB4 encodes a major sphingolipid long-chain base kinase required for synthesis of long-chain base phosphates and for the rapid incorporation of long-chain bases from the culture medium into sphingolipids (29). We also found that orf19.3104, the orthologue of S. cerevisiae YDC1 encoding an alkaline dihydroceramidase, is up-regulated in a TAC1-dependent manner in all four sets of clinical isolates (Table 4), although binding of Tac1p to the promoter of this gene was not detected. Ydc1p hydrolyzes dihydroceramide to free fatty acid and dihydrosphingosine, the substrate of LCB4 (50). Taken together, these findings suggest a role for Tac1p in the synthesis and translocation of sphingolipids into the plasma membrane.
Likewise, C. albicans Cdr1p and Cdr2p have been shown to function as plasma membrane energy-dependent translocators of phospholipids, mediating their in-to-out movement (floppases) (75). Thus, Cdr1p and Cdr2p would act in concert with Rta3p to establish the asymmetry of membrane lipids in the plasma membrane. Interestingly, Tac1p binds to the promoter of the S. cerevisiae AST1 homologue, orf19.6869 (AST2). S. cerevisiae Ast1p was shown to be required for the raft association and restoration of surface delivery of a mutant of the major plasma membrane proton ATPase, Pma1p, that was mistargeted to the vacuole (5). Elsewhere, it has been shown that Cdr1p is a lipid raft-associated protein (34). An attractive model would be that Tac1p contributes to the raft association and proper routing of its target gene products Cdr1p, Cdr2p, and Rta3p to the plasma membrane by regulating AST2 expression. Recent studies have demonstrated that both sphingolipids and sterols are important determinants of surface localization of Cdr1p, as reduced membrane localization of Cdr1p was associated with increased susceptibility of C. albicans to ketoconazole in mutants defective in ergosterol biosynthesis (53). Interestingly, we found that Tac1p binding was enriched at the ERG1 (1.7-fold, P value of 0.0005) and ERG2 (1.6-fold, P value of 0.0011) promoters. Moreover, ERG2 gene expression was significantly up-regulated in a TAC1-dependent manner in isolates C56, Gu5, and 17 (see Table S1 in the supplemental material), suggesting a role of Tac1p in ergosterol biosynthesis. PDR16, another target of Tac1p, is the functional homologue of the S. cerevisiae PDR16, encoding a lipid transfer protein of the Sec14 family (58, 66). S. cerevisiae Pdr16p was shown to localize to lipid particles and to transport phosphatidylinositol (71). There is strong evidence showing that lipid transfer proteins exchange lipid molecules between organelle membranes and plasma membrane at membrane contact sites (33). Thus, by controlling the expression of the PDR16 gene, Tac1p would act as a regulator of phosphatidylinositol transfer within membrane contact sites, further supporting the role of Tac1p in membrane lipid traffic.
Consistent with the potential role of Tac1p in membrane lipid metabolism, we found that the Tac1p targets orf19.7166, orf19.6501, and orf19.1887 encode three putative steryl ester hydrolases or triglyceride lipases, as (i) a Pfam analysis detected an α/β hydrolase fold in their primary sequences (Pfam entries PF04083.6 for orf19.7166 and PF00561 for orf19.1887 and orf19.6501), (ii) their primary sequences contain the lipase consensus sequence motif GXSXG (21), and (iii) Kyte-Doolittle hydropathy plots revealed hydrophobic regions in the primary sequences, suggesting that these proteins are potentially membrane anchored to lipid particles (data not shown). Steryl esters and triacylglycerols are neutral lipids stored in lipid particles and serve as an energy source as well as a rapid fatty acid source needed upon lipid depletion conditions (79). The orf19.1887 protein is highly homologous to S. cerevisiae Yeh1p, a steryl ester hydrolase localized to lipid particles (41, 42), whereas the orf19.7166 and orf19.6501 proteins display moderate homology to the products of the S. cerevisiae YGR110W and YJU3 genes. Yju3p was shown to be localized in lipid particles as well (3, 54). Another Tac1p target whose product was shown to be localized in lipid particles in S. cerevisiae is ATF1, encoding an alcohol acetyltransferase (76). This gene was significantly up-regulated in a TAC1-dependent manner in strain 5674 (see Table S1 in the supplemental material). The fact that Atf1p localizes to lipid particles and functions in the esterification process suggests that it is involved in the metabolism of lipids probably by transferring acetyl groups to free hydroxyl groups of fatty acids. Thus, in addition to being a potential regulator of membrane lipid traffic, Tac1p appears to be an important regulator of lipid mobilization in C. albicans, a function needed for rapid restoration of membrane lipids upon lipid depletion conditions (79).
Our studies identified two Tac1p target genes, GPX1 and SOD5, which have been shown to be involved in the oxidative stress response. We provide strong evidence that Tac1p directly regulates the expression of GPX1, which is implicated in response to oxidative stress (44). Interestingly, although the C. albicans genome encodes two other putative glutathione peroxidases (encoded by orf19.85 and orf19.87) with high homology to Gpx1p, only Gpx1p was found to be overproduced upon treatments with diamide or hydrogen peroxide (44), suggesting that, among the three putative glutathione peroxidases, GPX1 plays an important role in the oxidative stress response. It was also shown that S. cerevisiae GPX1 encodes a phospholipid hydroperoxide glutathione peroxidase which protects the cell against phospholipid hydroperoxides during oxidative stress (4).
SOD5, a copper, zinc superoxide dismutase found in the cell wall of C. albicans, has been shown to be important in protection against osmotic and oxidative stress (51). Since SOD5 seems to play an important role in this process, it is interesting that the gene is in fact down-regulated in all four azole-resistant clinical isolates studied. Martchenko et al. showed that while deleting SOD5 does not decrease cell viability, susceptibility to hydrogen peroxide under nutrient-poor conditions is increased (51). Indeed, we have shown previously that azole-resistant isolate 5674, while resistant to diamide, is hypersusceptible to hydrogen peroxide (31). While this hypersusceptibility to hydrogen peroxide could be explained by the down-regulation of SOD5, resistance to diamide could be attributed to the up-regulation of the CDR2 gene by Tac1p since CDR2 expression was shown to confer diamide resistance in S. cerevisiae (31). Taken together, these observations indicate that TAC1 differentially protects the cell against different oxidative stresses.
Supplementary Material
Acknowledgments
We thank Osman Zin-Al Abdin, Perrine Bomme, and Qing Zhang for their excellent technical assistance. We are indebted to François Robert and Simon Drouin for their assistance with the ChIP-chip experiments and to Tracey Rigby for performing the comparative genomic hybridization experiments. We are grateful to Raphaëlle Lambert and Pierre Chagnon from the IRIC's genomic platform for their support with the Q-PCR experiments and to Sébastien Lemieux and Martin Larose for their assistance with the statistical and bioinformatic analyses. We also thank Dominique Sanglard, Spencer Redding, and Ted White for providing clinical isolates used in this study.
This work was supported by research grants to P.D.R. from the National Institutes of Health (R01 AI058145) and to M.R. from the Canadian Institutes of Health Research (MT-15679 and HOP-67260). S.Z. and S.S. are supported by doctoral studentships from the Fonds de la Recherche en Santé du Québec (FRSQ). This is NRC publication number 49522.
Footnotes
Published ahead of print on 28 September 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Albert, T. J., J. Norton, M. Ott, T. Richmond, K. Nuwaysir, E. F. Nuwaysir, K. P. Stengele, and R. D. Green. 2003. Light-directed 5′->3′ synthesis of complex oligonucleotide microarrays. Nucleic Acids Res. 31:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 3.Athenstaedt, K., D. Zweytick, A. Jandrositz, S. D. Kohlwein, and G. Daum. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181:6441-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avery, A. M., and S. V. Avery. 2001. Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J. Biol. Chem. 276:33730-33735. [DOI] [PubMed] [Google Scholar]
- 5.Bagnat, M., A. Chang, and K. Simons. 2001. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 12:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balzi, E., and A. Goffeau. 1995. Yeast multidrug resistance: the PDR network. J. Bioenerg. Biomembr. 27:71-76. [DOI] [PubMed] [Google Scholar]
- 7.Black, S. M., and C. R. Wolf. 1991. The role of glutathione-dependent enzymes in drug resistance. Pharmacol. Ther. 51:139-154. [DOI] [PubMed] [Google Scholar]
- 8.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 9.Buser, K., F. Joncourt, H. J. Altermatt, M. Bacchi, A. Oberli, and T. Cerny. 1997. Breast cancer: pretreatment drug resistance parameters (GSH-system, ATase, P-glycoprotein) in tumor tissue and their correlation with clinical and prognostic characteristics. Ann. Oncol. 8:335-341. [DOI] [PubMed] [Google Scholar]
- 10.Calera, J. A., and R. Calderone. 1999. Histidine kinase, two-component signal transduction proteins of Candida albicans and the pathogenesis of candidosis. Mycoses 42(Suppl. 2):49-53. [PubMed] [Google Scholar]
- 11.Calera, J. A., G. H. Choi, and R. A. Calderone. 1998. Identification of a putative histidine kinase two-component phosphorelay gene (CaHK1) in Candida albicans. Yeast 14:665-674. [DOI] [PubMed] [Google Scholar]
- 12.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792-798. [DOI] [PubMed] [Google Scholar]
- 13.Chauhan, N., M. Kruppa, and R. Calderone. 2007. The Ssk1p response regulator and Chk1p histidine kinase mutants of Candida albicans are hypersensitive to fluconazole and voriconazole. Antimicrob. Agents Chemother. 51:3747-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coste, A., A. Selmecki, A. Forche, D. Diogo, M.-E. Bougnoux, C. d'Enfert, J. Berman, and D. Sanglard. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell 6:1889-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coste, A., V. Turner, F. Ischer, J. Morschhauser, A. Forche, A. Selmecki, J. Berman, J. Bille, and D. Sanglard. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies, B. S., H. S. Wang, and J. Rine. 2005. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol. Cell. Biol. 25:7375-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Deken, X., and M. Raymond. 2004. Constitutive activation of the PDR16 promoter in a Candida albicans azole-resistant clinical isolate overexpressing CDR1 and CDR2. Antimicrob. Agents Chemother. 48:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 21.Derewenda, Z. S., and U. Derewenda. 1991. Relationships among serine hydrolases: evidence for a common structural motif in triacylglyceride lipases and esterases. Biochem. Cell Biol. 69:842-851. [DOI] [PubMed] [Google Scholar]
- 22.Doroshow, J. H., S. Akman, F. F. Chu, and S. Esworthy. 1990. Role of the glutathione-glutathione peroxidase cycle in the cytotoxicity of the anticancer quinones. Pharmacol. Ther. 47:359-370. [DOI] [PubMed] [Google Scholar]
- 23.Drouin, S., and F. Robert. Genome-wide location analysis of chromatin-associated proteins by ChIP on chip: controls matter. Methods, in press.
- 24.Fardeau, V., G. Lelandais, A. Oldfield, H. Salin, S. Lemoine, M. Garcia, V. Tanty, S. Le Crom, C. Jacq, and F. Devaux. 2007. The central role of PDR1 in the foundation of yeast drug resistance. J. Biol. Chem. 282:5063-5074. [DOI] [PubMed] [Google Scholar]
- 25.Feigal, D. W., M. H. Katz, D. Greenspan, J. Westenhouse, W. Winkelstein, Jr., W. Lang, M. Samuel, S. P. Buchbinder, N. A. Hessol, and A. R. Lifson. 1991. The prevalence of oral lesions in HIV-infected homosexual and bisexual men: three San Francisco epidemiological cohorts. AIDS 5:519-525. [DOI] [PubMed] [Google Scholar]
- 26.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhauser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franz, R., M. Ruhnke, and J. Morschhauser. 1999. Molecular aspects of fluconazole resistance development in Candida albicans. Mycoses 42:453-458. [DOI] [PubMed] [Google Scholar]
- 29.Funato, K., R. Lombardi, B. Vallee, and H. Riezman. 2003. Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. J. Biol. Chem. 278:7325-7334. [DOI] [PubMed] [Google Scholar]
- 30.Gao, C., L. Wang, E. Milgrom, and W. C. Shen. 2004. On the mechanism of constitutive Pdr1 activator-mediated PDR5 transcription in Saccharomyces cerevisiae: evidence for enhanced recruitment of coactivators and altered nucleosome structures. J. Biol. Chem. 279:42677-42686. [DOI] [PubMed] [Google Scholar]
- 31.Gauthier, C., S. Weber, A. M. Alarco, O. Alqawi, R. Daoud, E. Georges, and M. Raymond. 2003. Functional similarities and differences between Candida albicans Cdr1p and Cdr2p transporters. Antimicrob. Agents Chemother. 47:1543-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hokamp, K., F. M. Roche, M. Acab, M. E. Rousseau, B. Kuo, D. Goode, D. Aeschliman, J. Bryan, L. A. Babiuk, R. E. Hancock, and F. S. Brinkman. 2004. ArrayPipe: a flexible processing pipeline for microarray data. Nucleic Acids Res. 32:W457-W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holthuis, J. C., and T. P. Levine. 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6:209-220. [DOI] [PubMed] [Google Scholar]
- 34.Insenser, M., C. Nombela, G. Molero, and C. Gil. 2006. Proteomic analysis of detergent-resistant membranes from Candida albicans. Proteomics 6(Suppl. 1):S74-S81. [DOI] [PubMed] [Google Scholar]
- 35.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karababa, M., A. T. Coste, B. Rognon, J. Bille, and D. Sanglard. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 48:3064-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly, R., and K. J. Kwon-Chung. 1992. A zinc finger protein from Candida albicans is involved in sucrose utilization. J. Bacteriol. 174:222-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly, S. L., A. Arnoldi, and D. E. Kelly. 1993. Molecular genetic analysis of azole antifungal mode of action. Biochem. Soc. Trans. 21:1034-1038. [DOI] [PubMed] [Google Scholar]
- 39.Kihara, A., and Y. Igarashi. 2002. Identification and characterization of a Saccharomyces cerevisiae gene, RSB1, involved in sphingoid long-chain base release. J. Biol. Chem. 277:30048-30054. [DOI] [PubMed] [Google Scholar]
- 40.Klein, R. S., C. A. Harris, C. B. Small, B. Moll, M. Lesser, and G. H. Friedland. 1984. Oral candidiasis in high-risk patients as the initial manifestation of the acquired immunodeficiency syndrome. N. Engl. J. Med. 311:354-358. [DOI] [PubMed] [Google Scholar]
- 41.Köffel, R., and R. Schneiter. 2006. Yeh1 constitutes the major steryl ester hydrolase under heme-deficient conditions in Saccharomyces cerevisiae. Eukaryot. Cell 5:1018-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Köffel, R., R. Tiwari, L. Falquet, and R. Schneiter. 2005. The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis. Mol. Cell. Biol. 25:1655-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kren, A., Y. M. Mamnun, B. E. Bauer, C. Schuller, H. Wolfger, K. Hatzixanthis, M. Mollapour, C. Gregori, P. Piper, and K. Kuchler. 2003. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol. Cell. Biol. 23:1775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kusch, H., S. Engelmann, D. Albrecht, J. Morschhauser, and M. Hecker. 2007. Proteomic analysis of the oxidative stress response in Candida albicans. Proteomics 7:686-697. [DOI] [PubMed] [Google Scholar]
- 45.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 46.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, T. C. White, D. Sanglard, and T. F. Patterson. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42:2932-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacPherson, S., M. Larochelle, and B. Turcotte. 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70:583-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mamnun, Y. M., R. Pandjaitan, Y. Mahe, A. Delahodde, and K. Kuchler. 2002. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol. Microbiol. 46:1429-1440. [DOI] [PubMed] [Google Scholar]
- 50.Mao, C., R. Xu, A. Bielawska, Z. M. Szulc, and L. M. Obeid. 2000. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J. Biol. Chem. 275:31369-31378. [DOI] [PubMed] [Google Scholar]
- 51.Martchenko, M., A. M. Alarco, D. Harcus, and M. Whiteway. 2004. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15:456-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morschhauser, J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587:240-248. [DOI] [PubMed] [Google Scholar]
- 53.Mukhopadhyay, K., T. Prasad, P. Saini, T. J. Pucadyil, A. Chattopadhyay, and R. Prasad. 2004. Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob. Agents Chemother. 48:1778-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Natter, K., P. Leitner, A. Faschinger, H. Wolinski, S. McCraith, S. Fields, and S. D. Kohlwein. 2005. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol. Cell. Proteomics 4:662-672. [DOI] [PubMed] [Google Scholar]
- 55.Ogawa, N., and K. Mori. 2004. Autoregulation of the HAC1 gene is required for sustained activation of the yeast unfolded protein response. Genes Cells 9:95-104. [DOI] [PubMed] [Google Scholar]
- 56.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfaller, M. A., C. Grant, V. Morthland, and J. Rhine-Chalberg. 1994. Comparative evaluation of alternative methods for broth dilution susceptibility testing of fluconazole against Candida albicans. J. Clin. Microbiol. 32:506-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phillips, S. E., P. Vincent, K. E. Rizzieri, G. Schaaf, V. A. Bankaitis, and E. A. Gaucher. 2006. The diverse biological functions of phosphatidylinositol transfer proteins in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 41:21-49. [DOI] [PubMed] [Google Scholar]
- 59.Prasad, R., P. De Wergifosse, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 60.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 61.Rogers, P. D., and K. S. Barker. 2003. Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob. Agents Chemother. 47:1220-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 63.Ruhnke, M., A. Eigler, I. Tennagen, B. Geiseler, E. Engelmann, and M. Trautmann. 1994. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J. Clin. Microbiol. 32:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rustchenko, E. P., D. H. Howard, and F. Sherman. 1994. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J. Bacteriol. 176:3231-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sadowski, I., C. Costa, and R. Dhanawansa. 1996. Phosphorylation of Gal4p at a single C-terminal residue is necessary for galactose-inducible transcription. Mol. Cell. Biol. 16:4879-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saidane, S., S. Weber, X. De Deken, G. St.-Germain, and M. Raymond. 2006. PDR16-mediated azole resistance in Candida albicans. Mol. Microbiol. 60:1546-1562. [DOI] [PubMed] [Google Scholar]
- 67.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 69.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schnabl, M., O. V. Oskolkova, R. Holic, B. Brezna, H. Pichler, M. Zagorsek, S. D. Kohlwein, F. Paltauf, G. Daum, and P. Griac. 2003. Subcellular localization of yeast Sec14 homologues and their involvement in regulation of phospholipid turnover. Eur. J. Biochem. 270:3133-3145. [DOI] [PubMed] [Google Scholar]
- 72.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 73.Selmecki, A., A. Forche, and J. Berman. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 75.Smriti, S. Krishnamurthy, B. L. Dixit, C. M. Gupta, S. Milewski, and R. Prasad. 2002. ABC transporters Cdr1p, Cdr2p and Cdr3p of a human pathogen Candida albicans are general phospholipid translocators. Yeast 19:303-318. [DOI] [PubMed] [Google Scholar]
- 76.Verstrepen, K. J., S. D. Van Laere, J. Vercammen, G. Derdelinckx, J. P. Dufour, I. S. Pretorius, J. Winderickx, J. M. Thevelein, and F. R. Delvaux. 2004. The Saccharomyces cerevisiae alcohol acetyl transferase Atf1p is localized in lipid particles. Yeast 21:367-377. [DOI] [PubMed] [Google Scholar]
- 77.Wada, S., M. Niimi, K. Niimi, A. R. Holmes, B. C. Monk, R. D. Cannon, and Y. Uehara. 2002. Candida glabrata ATP-binding cassette transporters Cdr1p and Pdh1p expressed in a Saccharomyces cerevisiae strain deficient in membrane transporters show phosphorylation-dependent pumping properties. J. Biol. Chem. 277:46809-46821. [DOI] [PubMed] [Google Scholar]
- 78.Wada, S., K. Tanabe, A. Yamazaki, M. Niimi, Y. Uehara, K. Niimi, E. Lamping, R. D. Cannon, and B. C. Monk. 2005. Phosphorylation of Candida glabrata ATP-binding cassette transporter Cdr1p regulates drug efflux activity and ATPase stability. J. Biol. Chem. 280:94-103. [DOI] [PubMed] [Google Scholar]
- 79.Wagner, A., and G. Daum. 2005. Formation and mobilization of neutral lipids in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans. 33:1174-1177. [DOI] [PubMed] [Google Scholar]
- 80.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.White, T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White, T. C., M. A. Pfaller, M. G. Rinaldi, J. Smith, and S. W. Redding. 1997. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis. 3(Suppl. 1):S102-S109. [DOI] [PubMed] [Google Scholar]
- 84.Zhou, P., and D. J. Thiele. 1993. Rapid transcriptional autoregulation of a yeast metalloregulatory transcription factor is essential for high-level copper detoxification. Genes Dev. 7:1824-1835. [DOI] [PubMed] [Google Scholar]
- 85.Znaidi, S., X. De Deken, S. Weber, T. Rigby, A. Nantel, and M. Raymond. 2007. The zinc cluster transcription factor Tac1p regulates PDR16 expression in Candida albicans. Mol. Microbiol. 66:440-452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.