Abstract
Entamoeba histolytica is a leading cause of parasitic death globally. However, the molecular framework regulating pathogenesis is poorly understood. We have previously used expression profiling to identify Entamoeba genes whose expressions were strictly associated with virulent strains (R. C. MacFarlane and U. Singh, Infect. Immun. 74:340-351, 2006). One gene, which we have named EhSTIRP (Entamoeba histolytica serine-, threonine-, and isoleucine-rich protein), was exclusively expressed in virulent but not in nonvirulent Entamoeba strains. EhSTIRP is predicted to be a transmembrane protein and is encoded by a multigene family. In order to characterize its function in amebic biology, we used a double-stranded RNA-based approach and were able to selectively down-regulate expression of this gene family. Upon EhSTIRP down-regulation, we were able to ascribe cytotoxic and adhesive properties to the protein family using lactate dehydrogenase release and Chinese hamster ovary cell adhesion assays. EhSTIRP thus likely represents a novel determinant of virulence in Entamoeba histolytica. This work validates the fact that genes expressed exclusively in virulent strains may represent virulence determinants and highlights the need for further functional analyses of other genes with similar expression profiles.
Entamoeba histolytica is a protozoan parasite that causes significant morbidity and mortality in the developing world (48). The life cycle is composed of two stages: the transmissible cyst that is released in the feces of infected humans, and the motile trophozoite, which resides in the large bowel. The infection begins when cysts are ingested, usually through contaminated food or water. Excystation occurs in the small bowel, and the motile trophozoites eventually colonize the large bowel.
The majority of individuals with E. histolytica infection remain asymptomatic; however, in some cases the parasites penetrate the colon wall causing dysentery and colitis and occasionally disseminate hematogenously to cause liver abscesses. The potential reasons for why so many individuals remain asymptomatic include differences in host immunity and gut microflora as well as genetic differences between Entamoeba isolates. The molecular processes that underlie the progression from gut commensal to pathogen are not completely understood (46). Invasion begins when the parasite attaches to host cells via the galactose/N-acetylgalactosamine (Gal/GalNAc)-inhibitable lectin. This binding event is followed by rapid cell death, which has been characterized as having both necrotic and apoptotic features (9, 44). Additional molecules important in amebic pathogenesis include the cysteine proteinases, papain family proteases that have been proven to be important in virulence. Parasites genetically engineered to produce low levels of cysteine proteinase activity showed significantly less gut inflammation and were unable to form liver abscesses in animal models of amebic disease (4, 5, 51). The amoebapores, pore-forming peptides, insert into the membranes of bacteria or eukaryotic cells and form pores, resulting in cell lysis. E. histolytica trophozoites expressing decreased levels of amoebapore A showed defects in cytolytic capability and were less virulent in the hamster model of amebic liver abscess (10), but these proteins appear not to be involved in causing amebic colitis (50). Important differences between the virulent E. histolytica HM-1:IMSS and the morphologically identical but nonpathogenic species Entamoeba dispar (20) have been described previously. The differences include lack of functional cysteine proteinase genes EhCP1 and EhCP5 in E. dispar (12, 49) as well as reduced expression of EhCP8 (13). Moreover, the pore-forming peptide amoebapore A is 25-fold less active in E. dispar than in E. histolytica (36). Finally, KERP1, a lysine- and glutamic acid-rich protein with roles in host cell attachment (42) and liver abscess formation (41) is divergent in E. dispar.
Most strains of E. histolytica are believed to have virulence potential or the ability to cause disease. However, E. histolytica strain Rahman is an exception. This strain, originally isolated from an asymptomatically infected person (21, 34) has long been described as having attenuated virulence (14). E. histolytica Rahman was recently shown to have decreased levels of cytotoxicity, decreased ability to phagocytose erythrocytes, decreased ability to elicit colitis in human colonic xenografts, and decreased resistance to oxidative stress (19). Interestingly, relatively few genetic differences were noted between E. histolytica HM-1:IMSS and E. histolytica Rahman based on comparative genomic hybridization analysis (45). However, a number of genes solely expressed in virulent strains and species of Entamoeba have recently been identified (18, 24, 33). Using a genomic DNA microarray, we demonstrated that out of 2,110 genes analyzed, 29 genes had low expression in both E. histolytica Rahman and E. dispar compared to that in E. histolytica HM-1:IMSS (33). Davis et al. used a microarray containing 70-mer oligonucleotides and reported 152 genes with significantly higher expression in E. histolytica HM-1:IMSS than in E. histolytica Rahman (18). Using an E. histolytica microarray based on the Affymetrix platform, Ehrenkaufer et al. identified 141 genes with significantly higher expression in E. histolytica HM-1:IMSS than in E. histolytica Rahman (24). Of these 141 genes, approximately 52 were not expressed in E. histolytica Rahman whereas the remainder were expressed but at lower levels than in E. histolytica HM-1:IMSS. The expression profile of E. dispar cannot be reliably determined using the oligonucleotide microarrays.
Some genes involved in amebic pathogenesis are expressed at lower levels in E. histolytica Rahman, including cysteine proteases (EhCP6, EhCP4, EhCP7), gene products important in bacterial killing (AIG1, lysozyme) and stress response (70-kDa heat shock protein, peroxiredoxin) (18, 19), and the light subunit of the Gal/GalNAc lectin (3). Additionally, the proteophosphoglycans coating the surface of E. histolytica Rahman have truncated glucan side chains compared to those of the virulent E. histolytica HM-1:IMSS (35). The authors hypothesized that the glycans may protect parasite adhesion molecules from proteolysis or that the proteophosphoglycans may regulate the ability of parasite surface molecules to interact with host cell receptors. Two genes with lower expression in E. histolytica Rahman have been genetically linked to virulence: overexpression of the peroxiredoxin gene in E. histolytica Rahman imparted increased resistance to oxidative stress (19), and antisense inhibition of the light lectin subunit in E. histolytica HM-1:IMSS resulted in decreased cytotoxicity and fewer amebic liver abscesses in hamsters (3).
We previously identified a protein family rich in serine, threonine, and isoleucine residues which we have named EhSTIRP (Entamoeba histolytica serine-, threonine-, and isoleucine-rich protein) (33). We originally identified EhSTIRP as a putative virulence determinant based on its expression profile. EhSTIRP is highly expressed in two virulent strains (E. histolytica HM-1:IMSS and 200:NIH) as well as in parasites causing disease in a mouse model of colitis (24, 27). Conversely, EhSTIRP was not expressed in the nonvirulent E. histolytica Rahman or E. dispar (33). Additionally, EhSTIRP had low expression in the cyst stage of the parasite, which is noninvasive (24).
In order to determine if EhSTIRP is involved in parasite virulence, we down-regulated EhSTIRP expression in the virulent E. histolytica HM-1:IMSS via episomal production of double-stranded RNA (dsRNA). Upon EhSTIRP down-regulation, parasites exhibited decreased levels of cytotoxicity and reduced ability to adhere Chinese hamster ovary (CHO) cells, implicating a role in amebic pathogenesis. This work validates our hypothesis that genes expressed exclusively in virulent strains may represent novel virulence pathways and increases the repertoire of genes that have been genetically linked to amebic virulence.
MATERIALS AND METHODS
Parasite strains and culture.
E. histolytica strains HM-1:IMSS and Rahman were obtained from the American Type Culture Collection (ATCC) (http://www.atcc.org/). The parasites were grown under axenic conditions in Trypticase-yeast extract-iron-serum medium (TYI-S-33) with 15% adult bovine serum (Sigma), supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml) (Gibco BRL), and 1× Diamond's vitamins (Biosource International) in 15-ml glass culture tubes at 37°C as previously described (21). Parasite genotypes were tested using restriction fragment length polymorphism analysis of the EhSREHP gene using previously described methods (6, 15). The SREHP PCR products were digested with AluI, and results for all PCR products were compared to banding patterns from previously published studies (15). Growth rates were determined for parasites transfected with the control or dsRNA construct. Parasites (50,000) were inoculated into glass tubes at time point zero and grown under standard culture conditions, and cell counts were determined using a hemacytometer at 12- to 18-h intervals.
Sequence analysis.
Genome sequences were downloaded from The Institute for Genomic Research (TIGR) (http://www.tigr.org/tdb/e2k1/eha1/) and the Sanger Institute Entamoeba histolytica Genedb (http://www.genedb.org/genedb/ehistolytica/) websites. A ClustalW alignment was performed using nucleotide and amino acid sequences for all gene family members. Signal peptides were predicted using the SignalP algorithm (http://www.cbs.dtu.dk/services/SignalP/); transmembrane sites were annotated on the TIGR website.
Constructs for expressing EhSTIRP dsRNA.
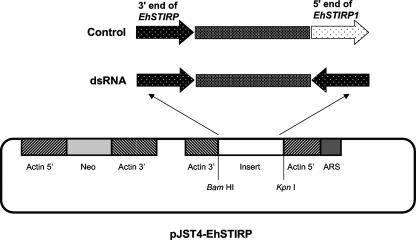
To down-regulate EhSTIRP, we utilized a methodology previously described in reference 30. Briefly, a 395-bp segment from the 3′ end of the EhSTIRP1 gene (99% nucleotide similarity to EhSTIRP2 and EhSTIRP3) was amplified by PCR and subcloned into the TOPO-TA 2.1 vector (Invitrogen). The gene fragment was subcloned in opposing orientations separated by a 500-bp nonspecific stuffer region in pBluescript SK(−) and then subcloned into the E. histolytica expression construct pJST4 at the KpnI and BamHI sites (26). This construct would be expected to generate dsRNA specific to the EhSTIRP gene family. A control construct was created using a similar methodology, except that 395 bp from the 3′ end of the gene and 350 bp from the 5′ end of EhSTIRP1 were cloned downstream and upstream of the 500-bp stuffer region. This construct would not be expected to generate dsRNA.
Transfection of E. histolytica.
Entamoeba histolytica HM-1:IMSS trophozoites (2 × 105 to 4 × 105) were transformed using 6 to 20 μg of plasmid DNA and 15 to 30 μl of Superfect (QIAGEN) in either 35-mm petri dishes or 1.5-ml tubes according to the manufacturer's instructions and as published previously (40). Stable transfectants were initially selected with G418 at 3 to 6 μg/ml, and cultures were maintained at 48 μg/ml G418.
Northern blot analysis.
Northern blots were performed as previously described (32). Briefly, total parasite RNA was isolated using TRIZOL reagent (Invitrogen) according to the manufacturer's instructions. RNA (10 to 20 μg) was separated by denaturing 1.2% agarose gel electrophoresis, transferred to Hybond-N membrane filters (Amersham-Pharmacia), and hybridized with radioactive probes using the ExpressHyb (Clontech) hybridization buffer. PCR product probes were sequenced and labeled with [α-32P]dATP with a random primed DNA labeling kit (Roche). Oligonucleotide probes were labeled with [γ-32P]dATP and T4 polynucleotide kinase (New England Biolabs). Blots were exposed to film or a phosphorimaging screen, scanned, and prepared for publication using Adobe Photoshop (version 7; Adobe, San Jose, CA). Blots were stripped using 0.5% sodium dodecyl sulfate and reused for subsequent hybridizations per the manufacturer's suggestions. For loading controls, a probe for EhActin (locus 8.m00351) was PCR amplified from E. histolytica HM-1:IMSS genomic DNA and labeled as described previously, or rRNA bands were stained with methylene blue, destained with dH2O, and scanned. The following primers were used in this study: EhActin forward, 5′-GCTGGTATGGGTCAAAAGGA-3′; EhActin reverse, 5′-TTTCTGTGGACAATAGCTGGTC-3′; EhSTIRP1 5′ forward, 5′-TGCAACTTCTTCAACTGTTTCA-3′; EhSTIRP1 5′ reverse, 5′-ATTACCATTACCAAGTGAACATG-3′; EhSTIRP1 3′ forward, 5′-GCCATTGGTTGAAGTAGAAGG-3′; EhSTIRP1 3′ reverse, 5′-GAACATTTGCTTCAACTCCTTTT-3′; EhSTIRP1 specific, 5′-CTGGATTTTCTTTTGTTTGACCTGTTA-3′; EhSTIRP2 specific, 5′-TTAATTCTGGATTTTCTTTTGTTTGACC-3′; EhSTIRP3 specific, 5′-TTAATTCATTTCCATCTTCTGTTTGACC-3′; 5′ RACE outer primer, 5′-GCTGATGGCGATGAATGAACACTG-3′; 5′ RACE inner primer, 5′-CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG-3′; EhSTIRP1 5′ RACE outer primer, 5′-TGTGAATCAATAATCAGTGGACAA-3′; EhSTIRP2 5′ RACE outer primer, 5′-TTGTGGGTTGAACAGTTTTCC-3′; EhSTIRP3 5′ RACE outer primer, 5′-TGTGATTTGAGAGATCTGAAGGA-3′; EhSTIRP1 5′ RACE inner primer, 5′-GTATCAATATCAATACTTTCATTGTTCT-3′; EhSTIRP2 5′ RACE inner primer, 5′-AATATTTGTATTTTCATTGTTCTCTTCAA-3′; EhSTIRP3 5′ RACE inner primer, 5′-AATCTGGTTATTTTCTTTGTTTTCTCC-3′; EhSTIRP1 promoter forward, 5′-GCTAAACCAATTTGTATTGAACCA-3′; EhSTIRP2 promoter forward, 5′-GTTGCAAATAAACGTGTTGAAA-3′; and EhSTIRP3 promoter forward, 5′-GAATTGATAAAAGAAGGTCCCAAT-3′.
5′ RACE and sequencing.
5′ Rapid amplification of cDNA ends (RACE) was carried out using a First Choice RLM 5′ RACE kit (Ambion) according to manufacturer's instructions. PCR products were cloned using the TOPO-TA cloning kit and sequenced with M13F or M13R primers. Promoter analysis for E. histolytica Rahman was performed by PCR amplifying ∼500 bp upstream of the predicted start codon from each of the three full-length EhSTIRPs from E. histolytica Rahman genomic DNA and sequencing the PCR products using an ABI PRISM 310 genetic analyzer (PE Applied Biosystems).
Cytotoxicity assay.
Host cell cytotoxicity was measured using a lactate dehydrogenase (LDH) release assay (Biovision) as previously published (32). Colonic epithelial (Caco-2) cells were grown on 24-well culture dishes in Dulbecco's modified Eagle's media, supplemented with 10% serum and penicillin/streptomycin until polarized. On the day of the experiment, Caco-2 cells were washed and the media changed to TYI-S-33 media. Amebae were added at a 1:15 (ameba:Caco-2 cell) ratio and the cultures were incubated at 37°C. Samples of culture supernatant were taken at 3, 6, and 9 h, diluted 1:10 in 150 mM phosphate-buffered saline, mixed 1:1 with reaction mixture, and transferred to a 96-well plate. The absorbance at 490 nm was read using a Bio-Tek Instruments EL311SX plate reader. Caco-2 cells incubated in TYI-S-33 media alone or syringe lysed were used as controls.
Adhesion assay.
Adhesion experiments were conducted as published previously (39). Briefly, E. histolytica cells were mixed with CHO cells in M199 media at a 1:20 ratio and incubated on ice for 90 min. The amebae were washed twice and samples counted on a hemacytometer. Amebae with three or more CHO cells attached were considered positive for adhesion in this assay.
RESULTS AND DISCUSSION
Characterization of the EhSTIRP gene family.
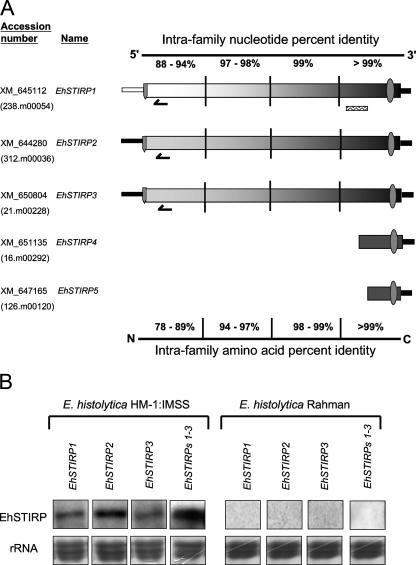
In the current E. histolytica genome assembly and annotation, the EhSTIRP gene family consists of five members: EhSTIRP1 (GenBank accession no. XM_645112), EhSTIRP2 (XM_644280), EhSTIRP3 (XM_650804), EhSTIRP4 (XM_651135), and EhSTRIP5 (XM_ 647165). Three members (EhSTIRP1, EhSTIRP2, and EhSTIRP3) are approximately 8 kb in length, making them some of the largest annotated genes in the E. histolytica genome. The other two, EhSTIRP4 and EhSTIRP5, appear to be truncated forms (Fig. 1A). In the TIGR database, only EhSTIRP1 is annotated as having a signal peptide. In order to determine if the absence of a signal peptide in EhSTIRP2 and EhSTIRP3 may be a result of an annotation error, we determined the transcription start site by performing 5′ RACE for each full-length gene family member. Utilizing the first in-frame start codon downstream of the transcription initiation site, both EhSTIRP2 and EhSTIRP3 are predicted to have signal peptides with 86% and 93% probability, respectively, according to the SignalP 3.0 program (7) (data not shown). All gene family members are predicted to contain a single transmembrane domain. These data together with additional topology information (WoLF PSORTII [28]) indicate that the gene family members are likely transmembrane proteins with the vast majority of the protein facing the luminal side and only a short (34-amino-acid) cytoplasmic tail and are likely to be present on the plasma or vesicular membranes (data not shown). Future studies with antisera to EhSTIRPs will be important to definitively prove the localization of the protein products.
FIG. 1.
EhSTIRP gene family structure and expression. (A) The EhSTIRP gene family is made up of five members. The GenBank accession number is listed for each gene. EhSTIRP4 and EhSTIRP5 may represent truncated versions of the gene. The thinner bars at either end of the gene represent the 5′ promoter region and presumptive 3′ untranslated region. EhSTIRP2 and EhSTIRP3 have very similar 5′ promoter regions (98% nucleotide identity), whereas the 5′ promoter region from EhSTIRP1 was only 45% and 47% identical to EhSTIRP3 and EhSTIRP2, respectively. The gene family members are highly similar at both the nucleotide and amino acid levels towards the 3′ end/C terminus (∼99%) and less similar to one another at the 5′ end/N terminus. The overall similarity is higher between EhSTIRP2 and EhSTIRP3 than between either gene and EhSTIRP1. A signal peptide 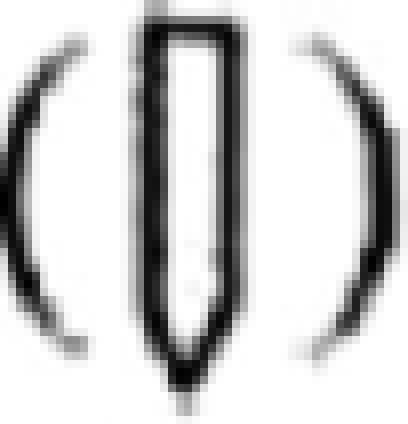 is shown for the three full-length gene family members and a transmembrane domain
is shown for the three full-length gene family members and a transmembrane domain 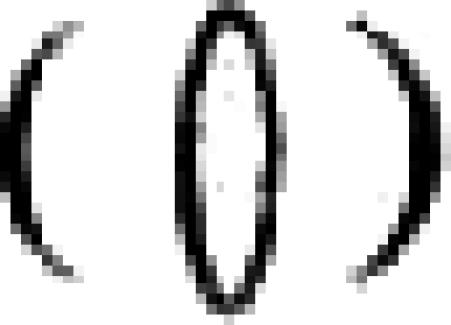 is shown for each gene. Gene-specific oligonucleotide probes are denoted as follows:
is shown for each gene. Gene-specific oligonucleotide probes are denoted as follows:  . A PCR-generated probe capable of detecting all full-length EhSTIRPs and used in the dsRNA constructs is denoted as follows:
. A PCR-generated probe capable of detecting all full-length EhSTIRPs and used in the dsRNA constructs is denoted as follows:  . (B) E. histolytica HM-1:IMSS expresses each of the EhSTIRP full-length members, whereas E. histolytica Rahman does not express any of the full-length gene family members. Separate blots were probed for EhSTIRP expression in E. histolytica HM-1:IMSS, whereas since no signal was observed in E. histolytica Rahman, the same blot was sequentially probed. rRNA stained with methylene blue is shown as a loading control.
. (B) E. histolytica HM-1:IMSS expresses each of the EhSTIRP full-length members, whereas E. histolytica Rahman does not express any of the full-length gene family members. Separate blots were probed for EhSTIRP expression in E. histolytica HM-1:IMSS, whereas since no signal was observed in E. histolytica Rahman, the same blot was sequentially probed. rRNA stained with methylene blue is shown as a loading control.
Comparisons of the intrafamily nucleotide and amino acid percent identities revealed nearly complete conservation at the 3′ end between EhSTIRP1, EhSTIRP2, and EhSTIRP3 and much less conservation at the 5′ end of the EhSTIRP gene family. We found that the promoter for EhSTIRP1 (defined as the region 500 bp upstream of the predicted start codon) was significantly different from EhSTIRP2 and EhSTIRP3 (47% and 45% identity, respectively) while those two gene promoters (EhSTIRP3 and EhSTIRP2) were quite similar to each other (98% identity).
EhSTIRP expression is restricted to Entamoeba strains and stages with virulence potential.
Using gene-specific oligonucleotide probes from a region of low intrafamily nucleotide similarity (63% to 77% identity), Northern blot analysis was performed and indicated that each of the three full-length family members were expressed under standard axenic culture conditions in E. histolytica HM-1:IMSS; we did not detect any transcript from any of the gene family members in the nonvirulent E. histolytica Rahman (Fig. 1B). EhSTIRP has an interesting expression profile: highly expressed in all strains with virulence potential (E. histolytica HM-1:IMSS, E. histolytica 200:NIH, and trophozoites during colonic invasion) and low or absent expression under all conditions with low virulence potential (E. histolytica Rahman, E. dispar, and E. histolytica cysts). Furthermore, although a number of genes have higher expression in virulent strains than in nonvirulent strains, the completely absent expression in E. histolytica Rahman and the high expression in E. histolytica HM-1:IMSS is an unusual expression profile for the E. histolytica transcriptome.
We previously reported that Entamoeba dispar does not contain a full-length intact copy of EhSTIRP according to the available genome sequence (33). We are currently investigating the mechanism by which EhSTIRP is silenced in E. histolytica Rahman. We have sequenced the majority of the EhSTIRP1 coding region in E. histolytica Rahman and found it to be nearly identical to that in E. histolytica HM-1:IMSS, indicating that E. histolytica Rahman most likely contains at least one full-length copy of the gene (33). The average promoter length in E. histolytica is ∼350 bp (Jason Hackney, personal communication). We sequenced approximately 500 bp of the promoter for each of the three full-length gene EhSTIRP family members in E. histolytica Rahman and found them to be highly similar (≥98%) to the promoter sequences for each gene in E. histolytica HM-1:IMSS. DNA methylation is known to silence expression of virulence genes in E. histolytica (1). We treated E. histolytica Rahman with 23 μM 5-azacytidine for 72 h and found no change in expression of EhSTIRPs, indicating that silencing via DNA methyltransferase is an unlikely mechanism of silencing in the Rahman strain (I. K. M. Ali and U. Singh, unpublished data). We also treated E. histolytica Rahman with trichostatin A (TSA) at 75 nM for 48 h and 150 nM and 300 nM for 24 h. TSA has been shown to be a potent inhibitor of histone deacetylase activity in E. histolytica (8, 38). TSA treatment modulates amebic gene expression (23) and has been shown to affect the expression level of other genes involved in amebic virulence (31). After treatment with TSA, we noted no change in EhSTIRP expression in E. histolytica Rahman by Northern blot analysis (data not shown), making it unlikely that deacetylation is responsible for the lack of EhSTIRP expression in E. histolytica Rahman. Transcriptional gene silencing has been previously documented in E. histolytica. Bracha et al. (11) showed that upon transfection with a plasmid containing the amoebapore gene and a portion of the short interspersed nuclear element EhSINE1, both chromosomal and episomal copies of the amoebapore gene were permanently silenced, and this silencing was stable even after removal of the plasmid (2). This permanent and stable epigenetic silencing mechanism is not completely understood, and whether a similar mechanism is functional in E. histolytica Rahman is not currently known. We previously reported that the mobile elements EhLINE1 (long interspersed nuclear element) and EhLINE3 had lower expression in E. histolytica Rahman (33). We are currently investigating whether the silencing of EhSTIRP in E. histolytica Rahman is related to the LINE or SINE elements or small RNAs.
Down-regulation of EhSTIRP expression.
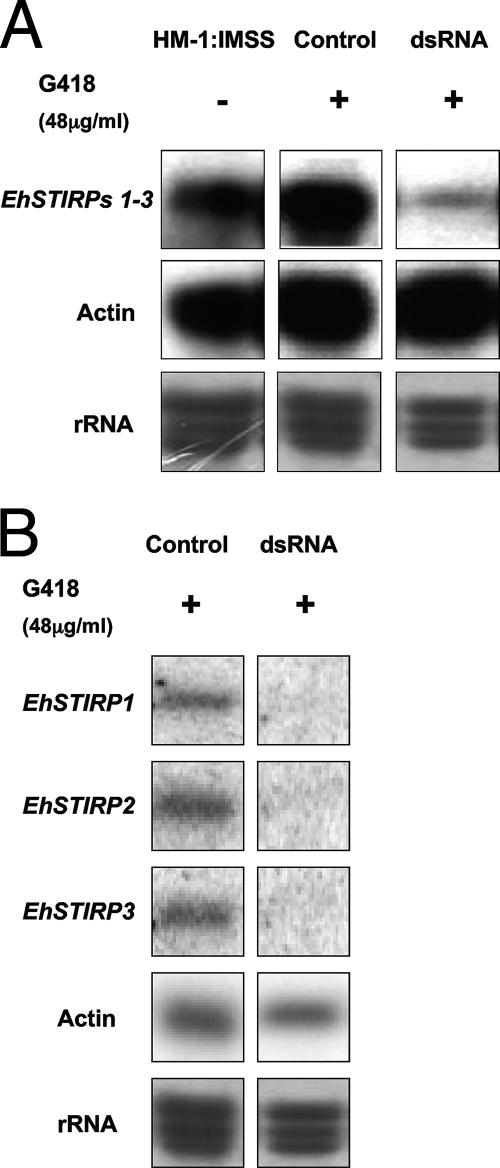
In order to down-regulate the EhSTIRP gene family, we transfected E. histolytica HM-1:IMSS parasites with a construct coding for the production of dsRNA specific to the 3′ region of the gene family, which should target all family members for silencing. As a control, we constructed a plasmid containing nonannealing ends (Fig. 2). This methodology was previously used to down-regulate expression of the formin homology gene EhDia1, as well as the kinesin family gene EhKlp5 (17, 30). After selection with G418 at 48 μg/ml, the expression levels of all EhSTIRP genes were drastically reduced as shown by reverse transcription-PCR (data not shown) and Northern blot analysis (Fig. 3). The PCR-amplified gene segment used as a probe in the Northern blot analysis (shown in Fig. 1A) would be predicted to hybridize with all gene family members. We also looked individually at each full-length EhSTIRP gene family member and found that each transcript was significantly reduced in parasites producing dsRNA specific to the EhSTIRP gene family (Fig. 3B). In order to determine whether the EhSTIRP dsRNA parasites had altered growth, we compared their growth rates to the growth of parasites transfected with a control plasmid maintained at the same drug selection. We found no significant difference in growth compared to amebae transfected with control plasmid during the first 20 h. At time points beyond 20 h and up to 72 h, the EhSTIRP dsRNA parasites grew slightly slower than the control strains (data not shown). All parasite strains were routinely passed every 48 h and all experiments for cell killing and adhesion were performed with equal numbers of parasites for short time points (≤9 h), time points at which there were no detectable growth differences between EhSTIRP dsRNA and control parasite strains.
FIG. 2.
Constructs used in analysis of EhSTIRP. Two different versions of the pJST4-EhSTIRP vector were created. The dsRNA construct contained two identical 395-bp segments in opposing orientations capable of producing dsRNA upon transcription. The 395-bp segment was amplified from the 3′ end of EhSTIRP in a region of high similarity among all family members and would be predicted to down-regulate the entire EhSTIRP gene family. The control construct contained two different regions of EhSTIRP1 and was incapable of forming dsRNA. Both the insert and the selectable marker Neo were placed under the control of the EhActin1 5′ and 3′ regulatory regions.
FIG. 3.
Northern blotting indicates a significant decrease in EhSTIRP gene expression when exposed to dsRNA targeting the 3′ end of the EhSTIRP gene family. (A) Northern blot analysis shows substantial down-regulation of EhSTIRP expression in the presence of dsRNA targeted to the 3′ end of the EhSTIRP gene family (parasites selected at 48 μg/ml G418). A PCR product from the 3′ end of the gene family that should detect EhSTIRP1-3 was used as a probe. RNA probed with an actin probe and methylene blue-stained rRNA (grayscale) are shown as loading controls. (B) Gene-specific oligonucleotide probes were also used and indicated that the expression level of each full-length EhSTIRP was decreased in the dsRNA-expressing parasites. RNA probed with actin and methylene blue-stained rRNA (grayscale) are shown as loading controls.
Interestingly, after nearly 1 year in cell culture, the EhSTIRP dsRNA-based down-regulation spontaneously reverted and EhSTIRP expression reverted to wild-type levels. The spontaneous loss of dsRNA-based gene down-regulation has not previously been reported in E. histolytica, and the underlying cause of this phenomenon is currently being investigated (R. C. MacFarlane and U. Singh, unpublished data). However, all phenotypic assays described in this work were performed at a time when the gene was clearly down-regulated by either reverse transcription-PCR or Northern blot analysis.
EhSTIRP contributes to amebic cytotoxicity.
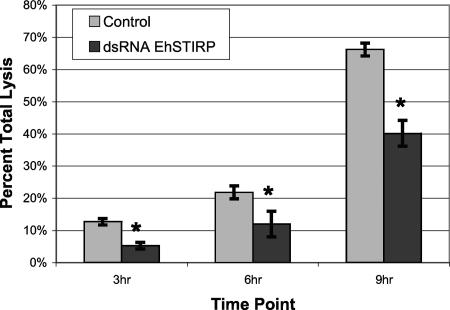
We first investigated whether the EhSTIRP gene family was involved in cytotoxicity. In order to do this we compared the ability of control and EhSTIRP-down-regulated parasites to damage Caco-2 colonic epithelial cell monolayers. In this assay, we used LDH release as the readout for damaged cells. For these experiments we combined E. histolytica HM-1:IMSS parasites with differentiated Caco-2 cells at a 1:15 ratio. Monolayer damage was assessed at 3, 6, and 9 h and compared to syringe-lysed monolayers. We found a significant difference between control and EhSTIRP-down-regulated parasites at all time points (P < 0.05) (Fig. 4). Overall, the EhSTIRP-down-regulated parasites showed a ∼45% decrease in cytotoxicity across all time points examined compared to that of the control strains exposed to similar amounts of G418 drug selection. Importantly, EhSTIRP-down-regulated parasites had a decreased cytotoxic ability at both the early (3 h) and late time points (9 h) after host cell interaction. The cytotoxicity data are a significant finding, as the relative difficulty in manipulating gene expression in E. histolytica has limited the ability to associate particular genes with pathogenicity. To date, only a few amebic genes have been genetically shown to be involved in cytotoxicity (3, 11, 16, 25, 29, 43, 47).
FIG. 4.
Down-regulation of EhSTIRP results in reduced cytotoxicity. An LDH release assay was used to measure the cytotoxicity incurred by control and EhSTIRP dsRNA parasites. The data are shown as a percent of total lysed Caco-2 cells (y axis). The averages of three independent experiments are shown with standard errors (*, denotes P of <0.05).
EhSTIRP has a role in amebic adhesion to host cells.
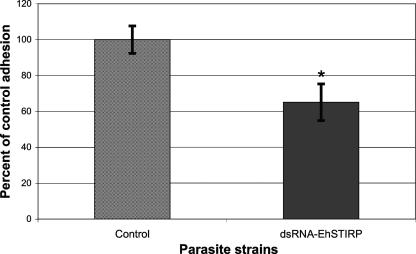
Adhesion is intimately linked to cell killing and is one of the first steps in pathogenesis on a cellular scale and a prerequisite for host cell destruction. Once a role in cytotoxicity had been established, we set out to determine whether the EhSTIRP-down-regulated parasites had a defect in adhesion to host cells. To test the adhesive ability of the parasites, we combined the control and EhSTIRP dsRNA parasites with CHO cells at a 1:20 ratio and incubated them on ice for 90 min as previously described (39). Amebae with three or more CHO cells attached after washing scored positive for the assay. The average of five experiments is shown in Fig. 5. In these experiments we noticed a significant difference (∼35% decrease in adhesion; P = 0.025) between control and EhSTIRP-down-regulated parasites in their ability to adhere CHO cells.
FIG. 5.
Down-regulation of EhSTIRP reduces the ability of E. histolytica trophozoites to adhere to CHO cells. E. histolytica trophozoites (control and EhSTIRP dsRNA) were combined with CHO cells at a 1:20 ratio and incubated on ice for 90 min. The amebae were washed twice in 1× phosphate-buffered saline, and parasites with ≥3 CHO cells attached were considered positive for adhesion. The data are shown as a percent of control adhesion (y axis). The averages of five independent experiments with standard errors are shown (*, denotes P of <0.05).
Importantly, we did not observe complete abrogation of parasite adhesion to host cells. The primary adhesive molecule on the surface of E. histolytica is the Gal/GalNAc lectin. Inhibition of the amebic lectin with galactose or N-acetylgalactosamine inhibits cell adhesion and killing (22, 37). However, complete inhibition of amebic adhesion to host cells is not seen even at very high concentrations of galactose (100 to 500 mM) (22), indicating that additional adhesins may be involved. In addition to the Gal/GalNAc lectin, a 112-kDa protein complex (25) and KERP1 (42), a lysine- and glutamic-acid rich protein, have also been implicated in adhesion, although their roles are currently less well understood. Based on the observed phenotype when EhSTIRP is down-regulated, we believe that EhSTIRP facilitates adhesion and subsequent host cell destruction and functions in the molecular program of E. histolytica pathogenesis. Studies to elucidate the role of the EhSTIRP gene family in in vivo models of amebic colitis and liver abscess formation will be crucial future experiments.
Conclusions.
As with most protozoan parasites, many of the genes in the Entamoeba histolytica genome have no homologues in other systems and are simply annotated as hypothetical proteins. These genes present a particular challenge as nothing is known regarding any potential function a priori. However, determining the function of these genes is likely to provide important details on ameba-specific biology and pathogenesis. Additionally, since these genes do not have well-conserved homologues in humans, they may represent valid drug targets. We believe that the EhSTIRP gene family likely represents a novel determinant of amebic virulence, as down-regulation of these genes decreases amebic cytotoxicity and adhesion. This work validates our approach for identification of novel virulence genes in this organism and indicates that genes differentially expressed in virulent and nonvirulent strains are good candidates for further functional studies.
Acknowledgments
We gratefully acknowledge Paul Bryson for help in cloning the dsRNA and control constructs, James Nelson for kindly providing the Caco-2 cells, and all members of the Singh lab for helpful discussions.
This work is supported by a Cellular and Molecular Biology Training Program Grant (NIH 5 T32 GM007276) for R.C.M. and a grant from the NIAID (AI-053724) to U.S.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Ali, I. K., G. M. Ehrenkaufer, J. A. Hackney, and U. Singh. 2007. Growth of the protozoan parasite Entamoeba histolytica in 5-azacytidine has limited effects on parasite gene expression. BMC Genomics 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anbar, M., R. Bracha, Y. Nuchamowitz, Y. Li, A. Florentin, and D. Mirelman. 2005. Involvement of a short interspersed element in epigenetic transcriptional silencing of the amoebapore gene in Entamoeba histolytica. Eukaryot. Cell 4:1775-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ankri, S., F. Padilla-Vaca, T. Stolarsky, L. Koole, U. Katz, and D. Mirelman. 1999. Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNac lectin complex inhibits Entamoeba histolytica virulence. Mol. Microbiol. 33:327-337. [DOI] [PubMed] [Google Scholar]
- 4.Ankri, S., T. Stolarsky, R. Bracha, F. Padilla-Vaca, and D. Mirelman. 1999. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect. Immun. 67:421-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ankri, S., T. Stolarsky, and D. Mirelman. 1998. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol. Microbiol. 28:777-785. [DOI] [PubMed] [Google Scholar]
- 6.Ayeh-Kumi, P. F., I. M. Ali, L. A. Lockhart, C. A. Gilchrist, W. A. Petri, Jr., and R. Haque. 2001. Entamoeba histolytica: genetic diversity of clinical isolates from Bangladesh as demonstrated by polymorphisms in the serine-rich gene. Exp. Parasitol. 99:80-88. [DOI] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 8.Bernes, S., R. Siman-Tov, and S. Ankri. 2005. Epigenetic and classical activation of Entamoeba histolytica heat shock protein 100 (EHsp100) expression. FEBS Lett. 579:6395-6402. [DOI] [PubMed] [Google Scholar]
- 9.Berninghausen, O., and M. Leippe. 1997. Necrosis versus apoptosis as the mechanism of target cell death induced by Entamoeba histolytica. Infect. Immun. 65:3615-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bracha, R., Y. Nuchamowitz, M. Leippe, and D. Mirelman. 1999. Antisense inhibition of amoebapore expression in Entamoeba histolytica causes a decrease in amoebic virulence. Mol. Microbiol. 34:463-472. [DOI] [PubMed] [Google Scholar]
- 11.Bracha, R., Y. Nuchamowitz, and D. Mirelman. 2003. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot. Cell 2:295-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruchhaus, I., T. Jacobs, M. Leippe, and E. Tannich. 1996. Entamoeba histolytica and Entamoeba dispar: differences in numbers and expression of cysteine proteinase genes. Mol. Microbiol. 22:255-263. [DOI] [PubMed] [Google Scholar]
- 13.Bruchhaus, I., B. J. Loftus, N. Hall, and E. Tannich. 2003. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2:501-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bujanover, S., U. Katz, R. Bracha, and D. Mirelman. 2003. A virulence attenuated amoebapore-less mutant of Entamoeba histolytica and its interaction with host cells. Int. J. Parasitol. 33:1655-1663. [DOI] [PubMed] [Google Scholar]
- 15.Clark, C. G., and L. S. Diamond. 1993. Entamoeba histolytica: a method for isolate identification. Exp. Parasitol. 77:450-455. [DOI] [PubMed] [Google Scholar]
- 16.Coudrier, E., F. Amblard, C. Zimmer, P. Roux, J. C. Olivo-Marin, M. C. Rigothier, and N. Guillen. 2005. Myosin II and the Gal-GalNAc lectin play a crucial role in tissue invasion by Entamoeba histolytica. Cell. Microbiol. 7:19-27. [DOI] [PubMed] [Google Scholar]
- 17.Dastidar, P. G., S. Majumder, and A. Lohia. 2007. Eh Klp5 is a divergent member of the kinesin 5 family that regulates genome content and microtubular assembly in Entamoeba histolytica. Cell. Microbiol. 9:316-328. [DOI] [PubMed] [Google Scholar]
- 18.Davis, P. H., J. Schulze, and S. L. Stanley, Jr. 2007. Transcriptomic comparison of two Entamoeba histolytica strains with defined virulence phenotypes identifies new virulence factor candidates and key differences in the expression patterns of cysteine proteases, lectin light chains, and calmodulin. Mol. Biochem. Parasitol. 151:118-128. [DOI] [PubMed] [Google Scholar]
- 19.Davis, P. H., X. Zhang, J. Guo, R. R. Townsend, and S. L. Stanley, Jr. 2006. Comparative proteomic analysis of two Entamoeba histolytica strains with different virulence phenotypes identifies peroxiredoxin as an important component of amoebic virulence. Mol. Microbiol. 61:1523-1532. [DOI] [PubMed] [Google Scholar]
- 20.Diamond, L. S., and C. G. Clark. 1993. A redescription of Entamoeba histolytica Schaudinn, 1903 (Emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J. Eukaryot. Microbiol. 40:340-344. [DOI] [PubMed] [Google Scholar]
- 21.Diamond, L. S., D. R. Harlow, and C. C. Cunnick. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431-432. [DOI] [PubMed] [Google Scholar]
- 22.Dodson, J. M., P. W. Lenkowski, Jr., A. C. Eubanks, T. F. Jackson, J. Napodano, D. M. Lyerly, L. A. Lockhart, B. J. Mann, and W. A. Petri, Jr. 1999. Infection and immunity mediated by the carbohydrate recognition domain of the Entamoeba histolytica Gal/GalNAc lectin. J. Infect. Dis. 179:460-466. [DOI] [PubMed] [Google Scholar]
- 23.Ehrenkaufer, G. M., D. J. Eichinger, and U. Singh. 2007. Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica. BMC Genomics 8:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehrenkaufer, G. M., R. Haque, J. A. Hackney, D. J. Eichinger, and U. Singh. 2007. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell. Microbiol. 9:1426-1444. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Rivera, G., M. A. Rodriguez, R. Ocadiz, M. C. Martinez-Lopez, R. Arroyo, A. Gonzalez-Robles, and E. Orozco. 1999. Entamoeba histolytica: a novel cysteine protease and an adhesin form the 112 kDa surface protein. Mol. Microbiol. 33:556-568. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh, S. K., A. Lohia, A. Kumar, and J. Samuelson. 1996. Overexpression of P-glycoprotein gene 1 by transfected Entamoeba histolytica confers emetine-resistance. Mol. Biochem. Parasitol. 82:257-260. [DOI] [PubMed] [Google Scholar]
- 27.Gilchrist, C. A., E. Houpt, N. Trapaidze, Z. Fei, O. Crasta, A. Asgharpour, C. Evans, S. Martino-Catt, D. J. Baba, S. Stroup, S. Hamano, G. Ehrenkaufer, M. Okada, U. Singh, T. Nozaki, B. J. Mann, and W. A. Petri, Jr. 2006. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol. Biochem. Parasitol. 147:163-176. [DOI] [PubMed] [Google Scholar]
- 28.Horton, P., K.-J. Park, T. Obayashi, and K. Nakai. 2006. Protein subcellular localization prediction with WoLF PSORT, p. 39-48. Proceedings of the 4th Annual Asia Pacific Bioinformatics Conference, Taipei, Taiwan.
- 29.Katz, U., S. Ankri, T. Stolarsky, Y. Nuchamowitz, and D. Mirelman. 2002. Entamoeba histolytica expressing a dominant negative N-truncated light subunit of its gal-lectin are less virulent. Mol. Biol. Cell 13:4256-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaur, G., and A. Lohia. 2004. Inhibition of gene expression with double strand RNA interference in Entamoeba histolytica. Biochem. Biophys. Res. Commun. 320:1118-1122. [DOI] [PubMed] [Google Scholar]
- 31.Lavi, T., E. Isakov, H. Harony, R. Siman-Tov, C. Weber, N. Guillen, and S. Ankri. 2007. Characterization of an unusual epigenetic machinery in the protozoan parasite Entamoeba histolytica. Fifth European Congress on Tropical Medicine and International Health, Amsterdam, Netherlands.
- 32.MacFarlane, R. C., P. H. Shah, and U. Singh. 2005. Transcriptional profiling of Entamoeba histolytica trophozoites. Int. J. Parasitol. 35:533-542. [DOI] [PubMed] [Google Scholar]
- 33.MacFarlane, R. C., and U. Singh. 2006. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect. Immun. 74:340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattern, C. F., D. B. Keister, and P. A. Caspar. 1978. Experimental amebiasis. III. A rapid in vitro assay for virulence of Entamoeba histolytica. Am. J. Trop. Med. Hyg. 27:882-887. [PubMed] [Google Scholar]
- 35.Moody-Haupt, S., J. H. Patterson, D. Mirelman, and M. J. McConville. 2000. The major surface antigens of Entamoeba histolytica trophozoites are GPI-anchored proteophosphoglycans. J. Mol. Biol. 297:409-420. [DOI] [PubMed] [Google Scholar]
- 36.Nickel, R., C. Ott, T. Dandekar, and M. Leippe. 1999. Pore-forming peptides of Entamoeba dispar. Similarity and divergence to amoebapores in structure, expression and activity. Eur. J. Biochem. 265:1002-1007. [DOI] [PubMed] [Google Scholar]
- 37.Petri, W. A., Jr., R. D. Smith, P. H. Schlesinger, C. F. Murphy, and J. I. Ravdin. 1987. Isolation of the galactose-binding lectin that mediates the in vitro adherence of Entamoeba histolytica. J. Clin. Investig. 80:1238-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramakrishnan, G., C. A. Gilchrist, H. Musa, M. S. Torok, P. A. Grant, B. J. Mann, and W. A. Petri, Jr. 2004. Histone acetyltransferases and deacetylase in Entamoeba histolytica. Mol. Biochem. Parasitol. 138:205-216. [DOI] [PubMed] [Google Scholar]
- 39.Ravdin, J. I., and R. L. Guerrant. 1981. Role of adherence in cytopathogenic mechanisms of Entamoeba histolytica. Study with mammalian tissue culture cells and human erythrocytes. J. Clin. Investig. 68:1305-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito-Nakano, Y., B. N. Mitra, K. Nakada-Tsukui, D. Sato, and T. Nozaki. 2007. Two Rab7 isotypes, EhRab7A and EhRab7B, play distinct roles in biogenesis of lysosomes and phagosomes in the enteric protozoan parasite Entamoeba histolytica. Cell. Microbiol. 9:1796-1808. [DOI] [PubMed] [Google Scholar]
- 41.Santi-Rocca, J., C. Weber, G. Guigon, O. Sismeiro, J. Y. Coppee, and N. Guillen. 17 August 2007, posting date. The lysine- and glutamic acid-rich protein KERP1 plays a role in Entamoeba histolytica liver abscess pathogenesis. Cell. Microbiol. doi: 10.1111/j.1462-5822.2007.01030. [DOI] [PubMed]
- 42.Seigneur, M., J. Mounier, M. C. Prevost, and N. Guillen. 2005. A lysine- and glutamic acid-rich protein, KERP1, from Entamoeba histolytica binds to human enterocytes. Cell. Microbiol. 7:569-579. [DOI] [PubMed] [Google Scholar]
- 43.Sen, A., N. S. Chatterjee, M. A. Akbar, N. Nandi, and P. Das. 2007. The 29-kilodalton thiol-dependent peroxidase of Entamoeba histolytica is a factor involved in pathogenesis and survival of the parasite during oxidative stress. Eukaryot. Cell 6:664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seydel, K. B., and S. L. Stanley, Jr. 1998. Entamoeba histolytica induces host cell death in amebic liver abscess by a non-Fas-dependent, non-tumor necrosis factor alpha-dependent pathway of apoptosis. Infect. Immun. 66:2980-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah, P. H., R. C. MacFarlane, D. Bhattacharya, J. C. Matese, J. Demeter, S. E. Stroup, and U. Singh. 2005. Comparative genomic hybridizations of Entamoeba strains reveal unique genetic fingerprints that correlate with virulence. Eukaryot. Cell 4:504-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanley, S. L., Jr. 2003. Amoebiasis. Lancet 361:1025-1034. [DOI] [PubMed] [Google Scholar]
- 47.Tillack, M., N. Nowak, H. Lotter, R. Bracha, D. Mirelman, E. Tannich, and I. Bruchhaus. 2006. Increased expression of the major cysteine proteinases by stable episomal transfection underlines the important role of EhCP5 for the pathogenicity of Entamoeba histolytica. Mol. Biochem. Parasitol. 149: 58-64. [DOI] [PubMed] [Google Scholar]
- 48.WHO/PAHO/UNESCO. 1997. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28-29 January, 1997. Epidemiol. Bull. 18:13-14. [PubMed] [Google Scholar]
- 49.Willhoeft, U., L. Hamann, and E. Tannich. 1999. A DNA sequence corresponding to the gene encoding cysteine proteinase 5 in Entamoeba histolytica is present and positionally conserved but highly degenerated in Entamoeba dispar. Infect. Immun. 67:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, X., Z. Zhang, D. Alexander, R. Bracha, D. Mirelman, and S. L. Stanley, Jr. 2004. Expression of amoebapores is required for full expression of Entamoeba histolytica virulence in amebic liver abscess but is not necessary for the induction of inflammation or tissue damage in amebic colitis. Infect. Immun. 72:678-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, Z., L. Wang, K. B. Seydel, E. Li, S. Ankri, D. Mirelman, and S. L. Stanley, Jr. 2000. Entamoeba histolytica cysteine proteinases with interleukin-1 beta converting enzyme (ICE) activity cause intestinal inflammation and tissue damage in amoebiasis. Mol. Microbiol. 37:542-548. [DOI] [PubMed] [Google Scholar]