Abstract
Formation of ascospores in the yeast Saccharomyces cerevisiae is driven by an unusual cell division in which daughter nuclei are encapsulated within de novo-formed plasma membranes, termed prospore membranes. Generation of viable spores requires that cytoplasmic organelles also be captured along with nuclei. In mitotic cells segregation of mitochondria into the bud requires a polarized actin cytoskeleton. In contrast, genes involved in actin-mediated transport are not essential for sporulation. Instead, efficient segregation of mitochondria into spores requires Ady3p, a component of a protein coat found at the leading edge of the prospore membrane. Other organelles whose mitotic segregation is promoted by actin, such as the vacuole and the cortical endoplasmic reticulum, are not actively segregated during sporulation but are regenerated within spores. These results reveal that organellar segregation into spores is achieved by mechanisms distinct from those in mitotic cells.
In response to nitrogen starvation in the presence of a nonfermentable carbon source, diploid cells of the yeast Saccharomyces cerevisiae exit mitotic growth and undergo meiosis and sporulation to form haploid spores (11, 37). The spores are generated by a distinct form of cell division that takes place within the cytoplasm of the mother cell. During meiosis II, the four sets of haploid chromosomes separate into distinct lobes of the nucleus, which subsequently pinch off to form discrete nuclei (29). As the nucleus is dividing, each lobe is encapsulated within a double membrane, termed the prospore membrane (29, 38). After nuclear division, the ends of the prospore membranes fuse to generate four distinct prospores. These prospores then mature by the deposition of a spore wall (26).
The closure of the prospore membranes is a cytokinetic event generating four discrete daughter cells within the mother cell cytoplasm, now called the ascal cytoplasm. Each prospore must therefore have its own complement of cytoplasmic organelles. Electron microscopy studies suggest that only about 30% of the mother cell cytoplasmic contents are distributed into spores. Those organelles that do enter the prospore do so just prior to prospore membrane closure (4, 6).
In mitotic yeast cells, the actin cytoskeleton provides the basis for the segregation of many organelles, including vacuolar precursors, mitochondria, and the cortical endoplasmic reticulum (ER), into the bud (12, 14, 18, 49). The Golgi may segregate in an actin-dependent manner or regenerate from cortical-ER precursors present in the bud (43, 47). Simple vectorial transport of organelles is likely to be insufficient for sporulation, however, as organelles need to be distributed to four locations simultaneously. Indeed, though genes involved in actin-mediated endocytosis are important for spore wall assembly (32), mutation of genes required for actin-based transport of material into the bud does not effect sporulation (10, 53). These results hint that organellar segregation may be achieved differently during sporulation, though the mechanisms underlying this process have been largely unexplored.
Mitochondrial segregation during sporulation has been most extensively examined (17, 28, 50). Immediately upon transfer of cells to sporulation medium, mitochondria disperse to the cell periphery. Subsequently, they fuse to form an extensive tubular network that associates with the nuclear envelope during the meiotic divisions. After segregation into the spore, the mitochondria fragment again as the spore matures (17). Mutation of genes involved in mitochondrial fission, or both fission and fusion, alter the dynamics of mitochondrial behavior and lead to an increase in the incidence of amitochondrial and petite spores (9, 17).
Though the machinery mediating mitochondrial segregation into spores is not known, these studies make clear that incorporation of mitochondria into spores is highly efficient. By contrast, a study of vacuolar inheritance during sporulation indicated that little or none of the vacuole enters the prospore (45). Rather, the vacuolar membrane and contents are left behind in the ascal cytoplasm. Ultimately, mature spores do display vacuoles, suggesting that they may be regenerated from precursors in the spore.
In vegetative cells, a ring of septin proteins is located at the bud neck and acts as a barrier to diffusion that allows differentiation between the bud and the mother cell cytoplasms (1, 52). During spore formation, a protein coat termed the leading-edge complex forms a ring structure at the lip of the prospore membrane. This is the interface between the mother and daughter cell cytoplasms, a position analogous to that of the septin ring at the bud neck. The leading-edge complex consists of at least three subunits, Ssp1p, Ady3p, and Don1p (23, 31, 40). Though all three proteins colocalize during sporulation, deletion of each gene produces a distinct phenotype. Deletion of DON1 causes no obvious sporulation defect (23). Deletion of ADY3 leads to the accumulation of asci with fewer than four spores due to the failure of a fraction of the prospores to develop spore walls (31, 40). Also, Don1p localization to the leading edge is lost in ady3 mutants. Loss of SSP1 has the most severe phenotype, resulting in abnormalities of prospore membrane growth, a failure of any spores to form, and the delocalization of the other two components of the complex (31).
Because of its position at the junction of the prospore and mother cell cytoplasms, the possibility that the leading-edge complex might play a role in organellar segregation into the prospore was investigated. ADY3 is required for efficient mitochondrial inheritance. Moreover, genes important for the actin-based segregation of mitochondria into buds are not necessary for segregation during sporulation. The segregation of other organelles that require the actin cytoskeleton for efficient segregation during mitotic growth was also examined. The results indicate that during sporulation, rather than preexisting organelles being actively segregated, many organelles are regenerated from precursors after or during cellular division.
MATERIALS AND METHODS
Yeast strains and media.
Standard S. cerevisiae genetic methods and media were used (46). The strains used in this study are listed in Table 1. All strains used except YS87 were in the fast-sporulating SK-1 strain background. Gene insertions and replacements were performed using cassettes amplified by PCR (25) and verified by PCR. Diploids MND11, NY551, and YS60 were made by disrupting DON1, SSP1, and YPT11, respectively, in AN117-4B and AN117-16D (39) and mating of the resulting haploids. To construct YS63 (ady3Δ/ady3 ypt11Δ/ypt11Δ), a ypt11Δ haploid was crossed to an ady3Δ haploid, followed by mating of two of the ady3Δ ypt11Δ segregants. To construct MND10 (ady3Δ/ady3 gip1Δ/gip1Δ), GIP1 was disrupted using PvuII-digested pJT26-HIS3 (54) in ady3Δ haploids, and the resulting haploids were mated to generate homozygous diploids. Strain YS87 (TFP1-GFP) was generated by mating of segregants from a cross of AN117-4B to a TFP1-GFP strain from the green fluorescent protein (GFP)-tagged strain collection (20).
TABLE 1.
S. cerevisiae strains used in this study
| Name | Genotype | Source |
|---|---|---|
| AN117-4B | MATα arg4-NspI his3ΔSK hoΔ::LYS2 leu2 lys2 rme1Δ::LEU2 trp1::hisG ura3 | 39 |
| AN117-16D | MATahis3Δ/SK hoΔ::LYS2 leu2 lys2 trp1::hisG ura3 | 39 |
| AN120 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 | 39 | |
| MND11 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 don1Δ:: HIS3MX6/don1Δ::HIS3MX6 | This study | |
| AN246 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 ady3Δ::kanRMX6/ady3Δ::kanRMX6 | 40 | |
| NY501 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 gip1Δ::HIS3/gip1Δ::HIS3 | 51 | |
| MND10 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 ady3Δ::kanRMX6/ady3Δ::kanRMX6 | ||
| gip1Δ::HIS3/gip1Δ::HIS | This study | |
| YS60 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 ypt11Δ::his5+/ypt11Δ::his5+ | This study | |
| YS63 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 ady3Δ::kanRMX6/ady3Δ::kanRMX6 | ||
| ypt11Δ::his5+/ypt11Δ::his5+ | This study | |
| YS87 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 TFP1-GFP::HIS3MX6//TFP1-GFP::HIS3MX6 | This study | |
| HI29 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 vps13Δ::his5+/vps13Δ::his5+ | 35 | |
| HI3 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 sso1Δ::his5+/sso1Δ::his5+ | 34 | |
| HI32 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 mso1Δ::his5+/mso1Δ::his5+ | 35 | |
| HI41 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 sma1Δ::his5+/sma1Δ::his5+ | 35 | |
| MND57 | MATa/MATα ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ::LYS2/hoΔ::LYS2 leu2/leu2 lys2/lys2 | |
| RME1/rme1Δ::LEU2 trp1::hisG/trp1::hisG ura3/ura3 ady4Δ::HIS3MX6/ady4Δ::HIS3MX6 | 41 |
Plasmids.
The plasmids used in this study are listed in Table 2. pRS424-R20 was constructed by replacing the GFP coding region in pRS424-G20 (34) with the gene for monomeric red fluorescent protein (RFP) (7) using EcoRI and XbaI sites. The RFP gene was amplified by PCR using pTiKmRFP (16) as a template and HNO941 and HNO942 as primers (primer sequences are available upon request). To generate pRS314-RTN2-GFP, RTN2-GFP was amplified by PCR using YSO121 and MNO170 as primers and genomic DNA purified from an RTN2-GFP strain (20) as a template and cloned into SmaI-digested pRS314 (48). To generate pRS304-MPC54-RFP, a SpeI-XhoI fragment carrying the MPC54-RFP fusion from pRS306-MPC54-RFP (34) was cloned into similarly digested pRS304.
TABLE 2.
Plasmids used in this study
| Name | Relevant yeast genotype | Source |
|---|---|---|
| pJT26-HIS3 | gip1Δ::HIS3 | 54 |
| pRS424-G20 | GFP fusion to SPO20 nucleotides 151-273 under the TEF2 promoter | 33 |
| pVT100-dsRED | Fusion of a mitochondrial targeting sequence to dsRED1-T1 under the ADH1 promoter | J. Nunnari |
| pRS424-R20 | RFP fusion to SPO20 nucleotides 151-273 under the TEF2 promoter | This study |
| pDN366 | KAR2-GFP-HDEL fusion under the TDH3 promoter | D. Ng |
| pRS314-RTN2-GFP | GFP fusion to RTN2 under the RTN2 promoter | This study |
| pSSEC7-EGFPx3 | SEC7-EGFPx3 | 47 |
| pRS304-MPC54-RFP | MPC54-RFP | This study |
Sporulation assays.
Cells were induced to sporulate in liquid medium essentially as described previously (38). Cells were cultured to saturation in either yeast extract-peptone-dextrose or synthetic complete medium lacking the appropriate nutrient to select for plasmids, cultured overnight to mid-log phase in yeast extract, peptone, acetate medium, and transferred to 2% potassium acetate at a concentration of 3 × 107 cells/ml.
Fluorescence microscopy.
For direct detection of fluorescent proteins, cells were fixed with 3.7% formaldehyde for 10 min and mounted with mounting medium containing 4,6-diamidino-2-phenylindole (DAPI) (Vectashield; Vector Laboratories, United Kingdom). Images were acquired using a Zeiss Axioplan2 microscope (Carl Zeiss, Thornwood, NY) with a Zeiss mRM Axiocam and deconvolved using Zeiss Axiovision 4.6 software.
Transmission electron microscopy.
Sporulating cells were fixed for 1 h in 3% glutaraldehyde in cacodylate buffer (100 mM sodium cacodylate, 5 mM CaCl2, pH 6.8), washed once in cacodylate buffer, left overnight at 23°C, resuspended in 4% KMnO4 in distilled water, and incubated for 30 min at 23°C. The cells were then washed with distilled water until the supernatant was clear, resuspended in saturated uranyl acetate for 2 h, and then dehydrated by 10-minute incubations in a graded acetone series: two incubations each in 30%, 50%, 70%, and 95% acetone and four incubations in 100% acetone. For embedding, the cells were first incubated twice in acetonitrile for 10 min and then transferred to a 1:1 solution of acetonitrile-Epon Mix (50% Epon 812, 15% dodecenyl succinic anhydride, 35% nadic methyl anhydride) for 4 h. The cells were then incubated in Epon Mix for 12 h with changes every 4 h, and finally, the catalyst 2,4,6-Tris(dimethylaminomethyl) phenol (DMP-30) was added and samples were placed into a vacuum oven at 60°C for 2 days to harden. After the samples were sectioned, images were collected on an FEI BioTwin G2 microscope at 80 kV using an AMT XR-60 camera.
RESULTS
Mitochondrial segregation into spores is defective in ady3 cells.
Previous studies demonstrated that mitochondria are efficiently segregated into developing spores (17, 28, 50). During late meiosis II, the mitochondria cluster along the nuclear envelope near the center of the spindle. In electron micrographs of forming spores, mitochondria can frequently be seen in close juxtaposition to the leading edge of the prospore membrane (Fig. 1), and often it appears that the mitochondrial membranes are distorted to maintain a constant distance from the leading edge (Fig. 1D), suggesting that this space may be filled by material that is not preserved during preparation for the electron microscope. To examine if the leading-edge complex might play a role in facilitating the segregation of mitochondria into spores, a fusion of RFP to a mitochondrial signal sequence, which localizes RFP to the mitochondrial matrix (36), was coexpressed in wild-type and ady3 cells with a GFP-tagged prospore membrane marker (33), and segregation of the mitochondria was examined (Fig. 2).
FIG. 1.
Mitochondrial segregation in wild-type cells. (A and C) Transmission electron micrographs of wild-type (AN120) meiosis II cells, showing mitochondrial segregation into the spore cytoplasm. N, nucleus; m, mitochondria. The white arrowheads indicate prospore membranes. (B and D) Higher-magnification views of the boxed regions in panels A and C, respectively. N, nucleus; m, mitochondria. The black arrowheads indicate the leading edge of the prospore membrane. Scale bar = 500 nm.
FIG. 2.
Mitochondrial-segregation defect during sporulation in ady3Δ cells. AN120 (wild type [wt]) and AN246 (ady3Δ) were transformed with plasmids expressing markers for the prospore membrane (pRS424-G20) and the mitochondrial matrix (pVT100-dsRED), and the inheritance of mitochondria during sporulation was examined. The white arrowheads indicate prospores in which mitochondria are absent. Scale bar = 1 μm. Nuclei were visualized with DAPI. Mitochondrial nucleoids are visible as DAPI-stained material between the segregating nuclear chromosomes in the meiosis II cells.
Consistent with earlier reports, individual mitochondrial tubes entered the spore during meiosis II and then fragmented after prospore membrane closure. In wild-type cells, mitochondria segregated with high efficiency into all four spores of the tetrad (Fig. 2, top; Table 3). In ady3 cells, mitochondria still cluster at the spindle midzone; however, they frequently fail to enter one or more of the prospores (Fig. 2, bottom; Table 3). Comparison of differential interference contrast images to the fluorescence images revealed a strong correlation between segregation of mitochondria into prospores and the subsequent elaboration of spore walls (Fig. 3). Ninety-two percent of ady3 spores that formed visible spore walls also displayed mitochondrial RFP signals, and conversely, 98% of the prospores that failed to form spore walls lacked mitochondria. These results suggest that failure to inherit mitochondria may cause the observed spore wall defect in ady3 cells.
TABLE 3.
Efficiency of mitochondrial segregationa
| Strain | Observed frequency of prospores including mitochondria (% ± SD) |
|---|---|
| AN120 (wild type) | 100 ± 0 |
| AN246 (ady3Δ) | 48 ± 15 |
| MND11 (don1Δ) | 99 ± 1 |
| YS60 (ypt11Δ) | 100 ± 0 |
| YS63 (ady3Δ ypt11Δ) | 42 ± 9 |
| NY501 (gip1Δ) | 92 ± 10 |
| MND10 (ady3Δ gip1Δ) | 18 ± 6 |
Cells containing pRS424-G20 and pVT100-dsRED were examined in the fluorescence microscope; 100 prospores were counted for mitochondrial segregation in each clone, and the data shown are the averages of three independent experiments.
FIG. 3.
Failure of mitochondrial inheritance correlates with the spore wall defect in ady3Δ cells. AN246 (ady3Δ) was transformed with plasmids expressing markers for the prospore membrane (pRS424-G20) and the mitochondrial matrix (pVT100-dsRED), and mitochondrial morphology was examined during sporulation. Only spores with spore walls are visible in differential interference contrast (DIC). The white arrowheads indicate spores in which mitochondria are absent. Scale bar = 1 μm. Nuclei were visualized with DAPI. No nuclei are visible in the mature spores due to impermeability of the spore wall.
Mutation of ADY3 also causes the delocalization of Don1p from the leading-edge complex (31, 40), raising the possibility that the mitochondrial defect in ady3 cells is an indirect effect of the loss of Don1p. However, mitochondrial segregation was highly efficient in don1 cells (Table 3). Thus, the mitochondrial-segregation defect seems to result from the loss of Ady3p at the leading edge.
YPT11 does not contribute to mitochondrial segregation during sporulation.
ADY3 is important but not essential for mitochondrial segregation into spores, suggesting that there may be other mechanisms at work, as well. In vegetative cells, mitochondrial segregation into the bud requires the Ypt11 protein to link the mitochondria to the actin-based segregation machinery (3, 21). To determine if this vegetative system might also contribute to mitochondrial inheritance during sporulation, mitochondrial segregation in ypt11 and ypt11 ady3 diploids was examined. The absence of YPT11 resulted in no obvious sporulation or segregation defect (Table 3), indicating that it does not play a major role in mitochondrial segregation during sporulation. Moreover, deletion of YPT11 does not exacerbate the ady3 phenotype, indicating that Ypt11p-based segregation is not active as an alternate pathway during sporulation. These results are consistent with earlier studies that found no sporulation defect in ypt11 or mutants in other genes important for vegetative mitochondrial segregation, such as MMR1, JSN1, TPM1, or MYO2 (10, 53).
Septins are not required for mitochondrial segregation into spores.
During spore formation, septins form a pair of parallel sheets on the prospore membrane located just behind the leading-edge complex (13, 31). Deletion of GIP1 blocks the formation of these septin sheets (51). Whether attachment to the septins might play a role in leading the mitochondria into the spore was tested by examining mitochondrial inheritance in a gip1 mutant. Mitochondria segregated efficiently in gip1 cells (Table 3). However, in a gip1 ady3 double-mutant strain, the percentage of prospores inheriting mitochondria was about 2.5-fold lower than in the ady3 single mutant (Table 3). These data indicate that septins do not play an essential role in mitochondrial segregation during sporulation but that they may cooperate with the leading-edge complex in ensuring efficient mitochondrial inheritance.
ADY3 does not influence the segregation of the vacuole.
The importance of ADY3 in mitochondrial segregation led us to examine whether the leading-edge complex plays a role in the segregation of other organelles. Unlike mitochondria, vacuoles remain behind in mother cells during spore formation (45). In this case, it is possible that the leading-edge complex might play a role in excluding the vacuole from the prospore. The vacuolar-membrane marker Tfp1-GFP was examined in sporulating cells (Fig. 4A). Consistent with previous studies of other vacuolar markers (45), TFP1-GFP remained in the ascal cytoplasm. The behavior of Tfp1-GFP was identical in ady3 cells, indicating that ady3 plays no role in excluding the vacuole from the spore (Y. Suda, unpublished observations).
FIG. 4.
The vacuole is regenerated in the spore during maturation. (A) Strain YS87, which expresses the vacuolar-membrane marker Tfp1-GFP, was transformed with a plasmid expressing a prospore membrane marker (pRS424-R20), and vacuole segregation during sporulation was examined. Scale bar = 1 μm. wt, wild type. (B) At the indicated time points, cells were fixed and analyzed to determine the percentages of the spores with visible Tfp1p-GFP fluorescence. At least 100 spores were counted at each time point, and the values are the averages of two independent experiments. The vertical lines indicate the range of values at each time point. The arrow indicates the time at which mature spores were first observed in the population.
Though Tfp1-GFP was absent from newly formed spores, fluorescence was clearly visible in mature spores, presumably representing the regeneration of the vacuolar compartment within the spore. We performed a time course to examine the timing of the reappearance of the Tfp1-GFP signal within the spores (Fig. 4B). At 20 h after the shift to sporulation medium, roughly 50% of the spores exhibited Tfp1-GFP fluorescence. This represents an ∼10-h lag from the time at which mature spores began to appear in the population. Though some of this lag may reflect time required for spores to synthesize and accumulate Tfp1-GFP, these data suggest that several hours are required for the generation of new vacuoles within the spore.
The cortical ER disappears during meiosis and reappears within the spore.
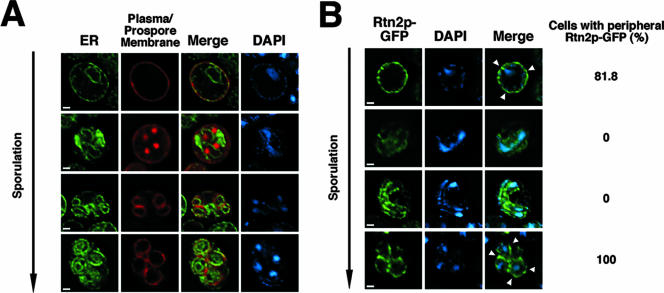
Like the vacuole, the cortical ER requires actin for delivery to the bud in mitotic cells (12, 14). The behavior of the ER during sporulation has not been previously described. To examine it, a soluble ER marker, GFP-HDEL, and a prospore membrane marker, RFP fused to a lipid binding domain of the Spo20 protein (33), were coexpressed in sporulating cells. In vegetative cells, the ER is found predominantly as the nuclear envelope (perinuclear ER) and in a network underneath the plasma membrane (cortical ER) (42). This same distribution of ER is seen in cells early in sporulation (Fig. 5A, top row). As cells enter meiosis, perinuclear ER fluorescence persists, but the signal from the cortical ER is progressively lost, so that by the time cells enter meiosis II, the cortical ER appears to be absent (Fig. 5A, second row from top). During meiosis II, GFP-HDEL was found in rings surrounding the DAPI-stained chromatin of the forming haploid nuclei, as well as in a reticular structure near the middle of the cell (Fig. 5A, third row from top). This staining presumably represented the envelope of the remnant nucleoid that was left behind in the ascus (15). Importantly, during meiosis II, the perinuclear ER is distinguishable from the prospore membrane both by the greater distance at which the prospore membrane surrounds the chromatin and because the perinuclear ER staining is connected by this reticular structure. In postmeiotic cells, the cortical ER rapidly reappears beneath the plasma membrane of the prospore (Fig. 5A, bottom row). These observations suggest that the cortical ER is depleted during the course of meiosis and regenerated within the newly formed spore.
FIG. 5.
The cortical ER disappears during meiosis and is regenerated during spore maturation. (A) AN120 (wild type) was transformed with plasmids expressing prospore membrane (pRS424-R20) and luminal-ER markers (pDN366), and ER morphology was visualized across a time course of sporulation. (B) AN120 (wild type) was transformed with a plasmid (pRS314-RTN2-GFP) expressing the cortical-ER marker Rtn2p-GFP and examined during sporulation. The white arrowheads indicate peripheral localization of Rtn2-GFP in premeiotic cells and in spores. The percentage of cells at each stage showing peripheral Rtn2-GFP localization is listed. At least 100 cells at each stage were scored. Scale bar = 1 μm. Nuclei were visualized with DAPI. The DAPI-stained materials evident between the segregating nuclear chromosomes in the meiosis II cells are mitochondrial nucleoids.
To look more specifically at the behavior of the cortical ER, we examined the localization of the reticulon protein Rtn2p (Fig. 5B). As reported for its paralog, Rtn1p, Rtn2-GFP localized exclusively to the cortical ER in vegetative cells (8) (Suda, unpublished). Similarly, early in sporulation, Rtn2-GFP was seen only in the peripheral ER (Fig. 5B, top row). As cells progressed through meiosis, peripheral localization progressively decreased, so that by meiosis II Rtn2-GFP was found predominantly in the nuclear envelope (Fig. 5B, third row from top). In postmeiotic cells, Rtn2-GFP rapidly appeared at the periphery of the spore and disappeared from the nuclear envelope (Fig. 5B, bottom row). These results mirror those seen with the general ER marker and suggest that cortical-ER material is pulled back into the nuclear envelope during meiosis. As with the vacuole, mutation of ADY3 did not alter the efficiency of cortical-ER inheritance (Suda, unpublished).
Golgi apparatus segregation occurs early in prospore membrane growth.
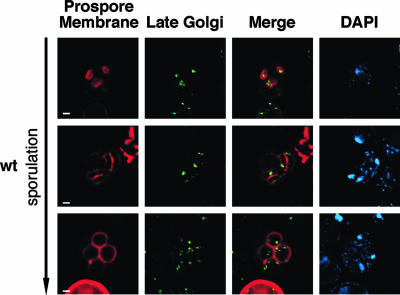
To examine the segregation of Golgi apparatus and trans-Golgi network elements into spores, a GFP fusion to the Sec7 protein was used (43). In wild-type cells, late Golgi structures were present as scattered foci throughout the cytoplasm in early meiotic cells. These Golgi puncta remain throughout meiosis. However, early in meiosis II, a bright perinuclear Golgi apparatus signal appeared within the prospore membrane, often close to the spindle pole body (SPB). This signal was present throughout the remainder of meiosis II, and multiple Sec7-GFP-containing structures were present within the prospores in postmeiotic cells (Fig. 6).
FIG. 6.
Golgi elements segregate early during sporulation. AN120 (wild type [wt]) was transformed with plasmids expressing prospore membrane (pRS424-R20) and late Golgi apparatus (pSSEC7-EGFPx3) markers, and the segregation of late Golgi elements was examined in sporulating cells. Scale bar = 1 μm. Nuclei were visualized with DAPI. DAPI staining between the segregating nuclear chromosomes evident in the meiosis II cells indicates mitochondrial nucleoids.
The localization of the Golgi apparatus inside the prospore membrane from very early in meiosis II suggested that an active recruitment of the Golgi apparatus to the spindle pole region, where prospore membrane formation starts, might be important for segregation. To test this possibility, Golgi apparatus localization was examined in an sso1 mutant that failed to form prospore membranes. However, in the absence of sso1, bright Golgi elements were less common and were found distributed in the cytoplasm rather than in perinuclear positions (Fig. 7A). This result suggests an alternative possibility, that capture of Golgi elements between the nuclear envelope and the prospore membrane during prospore membrane growth might be the basis for Golgi apparatus segregation.
FIG. 7.
Perinuclear Golgi elements correlate with the presence of a prospore membrane. (A) AN120 (wild type [wt]) and HI3 (sso1Δ) were transformed with plasmids expressing markers for the late Golgi apparatus (pSSEC7-EGFPx3) and the meiotic SPB (MPC54-RFP), and the distribution of late Golgi elements with respect to the SPB was analyzed in meiosis II cells. Scale bar = 1 μm. Nuclei were visualized with DAPI. DAPI staining between the segregating nuclear chromosomes evident in the meiosis II cells indicates mitochondrial nucleoids. (B) AN120 (wild type), AN246 (ady3Δ), HI29 (vps13Δ), MND57 (ady4Δ), HI3 (sso1Δ), HI41 (sma1Δ), and HI32 (mso1Δ) were transformed with plasmids expressing markers for the late Golgi apparatus (pSSEC7-EGFPx3) and the meiotic SPB (MPC54RFP), and the distance from each SPB in meiosis II cells to the nearest Golgi element was measured. The values shown are the averages for at least 50 SPBs in each strain. The error bars indicate the standard error of the mean for each set of measurements. (C) AN246 (ady3Δ), HI29 (vps13Δ), and MND57 (ady4Δ) were transformed with plasmids expressing prospore membrane marker (pRS424-R20) and late Golgi apparatus markers (pSSEC7-EGFPx3), and Golgi apparatus distribution was examined in sporulating cells. Scale bar = 1 μm. Nuclei were visualized with DAPI. DAPI staining between the segregating nuclear chromosomes indicates mitochondrial nucleoids.
The correlation between prospore membrane growth and Golgi apparatus inheritance was further explored by examining Golgi apparatus localization in additional mutants with prospore membrane defects (Fig. 7B). To quantify the effects on the Golgi apparatus position, the distance between the SPB, marked by Mpc54-RFP, and the nearest late Golgi element, marked by Sec7-GFP, was measured for >50 SPBs in each strain. In wild-type cells, vps13 mutants that had small prospore membranes (35), ady4 mutants that had abnormal prospore membranes (41), and ady3 mutants, the average SPB-Golgi apparatus distances were essentially the same. By contrast, in three different mutants that formed very small or absent prospore membranes, sso1, mso1, or sma1 (22, 34, 44), the distance from the SPB to the Golgi apparatus was much greater. Therefore, a shorter SPB-Golgi apparatus distance correlates with the presence of a prospore membrane. Moreover, as in wild-type cells, Golgi elements in vps13, ady3, and ady4 mutants were found within the prospore membrane (Fig. 7C). The simplest interpretation of these results is that the shorter SPB-Golgi apparatus distances are a consequence of the capture of the Golgi apparatus within the prospore membrane.
DISCUSSION
A unique mechanism for mitochondrial segregation in sporulating cells.
During mitotic growth of S. cerevisiae, mitochondria are actively segregated into the forming bud along actin cables in a process that involves the small GTP binding protein Ypt11p, as well as the myosin motor Myo2p (3, 21). While mitochondrial segregation into spores also appears to be an active process, Ypt11p is not required. In vegetative cells, several other proteins appear to contribute to mitochondrial inheritance independently of YPT11 (2), so the absence of a role for YPT11 in sporulation does not eliminate the possibility that some of the vegetative machinery might function during sporulation. However, unlike budding cells, in which the polarized actin cables run from the mother cell into the bud, the few actin cables present during sporulation are not directed toward or into the prospore (53). Combined with our results, these observations suggest that actin-based movement is not the basis for mitochondrial inheritance following meiosis.
We report here that a sporulation-specific protein, Ady3p, is important for efficient segregation of the mitochondria into spores. Ady3p is a component of the leading-edge complex, which forms a ring structure at the lip of the prospore membrane. The position of the complex at the mouth of the prospore membrane places it at the junction of the mother (ascal) and daughter (spore) cell cytoplasms and therefore in a position to function in the delivery of cytoplasmic components into the spore. Previous studies have demonstrated a role for this structure in the proper formation of the prospore membrane and in closure of the membrane (27, 31). However, these functions require only the Ssp1p component of the complex. Thus, different subunits of the complex play distinct roles in spore morphogenesis. We found a strong correlation between the failure of prospores to inherit mitochondria and failure to elaborate spore walls. As sporulation occurs under conditions that require aerobic respiration, the prospores that lack mitochondria should be unable to generate ATP. It seems likely, therefore, that the mitochondrial-inheritance defect accounts for the spore wall phenotype of ady3 cells. However, an earlier study reported that certain MDM mutants generate amitochondrial spores with visible spore walls but which fail to germinate (9). It may be, therefore, that the mitochondrial defect in ady3 mutants is linked to, but not a direct cause of, the spore wall defect.
ADY3 is important but not essential for distribution of mitochondria to prospores, indicating that alternative mechanisms of segregation must exist. In the absence of ADY3, septins appear to play a role in mitochondrial inheritance. Perhaps interaction with septin sheets helps to anchor the mitochondria inside the prospore membrane. However, even in cells lacking both Ady3p and septins ∼20% of the spores still inherit mitochondria, suggesting that other means of segregation are present. One possibility is that the association of mitochondria with the nuclear envelope may be the underlying means of mitochondrial distribution.
In wild-type cells, mitochondria associate with the nuclear envelope and cluster near the spindle midzone in meiosis II, placing them near the mouth of the prospore membrane (28, 50). As meiosis II proceeds and nuclear lobes containing haploid chromosome sets are engulfed by prospore membranes, the mitochondria would also be encapsulated. However, as these lobes pinch off the nucleus to generate the four haploid nuclei, a remnant nucleoid body that contains nucleolar material is formed which remains behind in the ascus (15, 30). Because much of the nucleus is left behind, association of the mitochondria with the nucleus may not be sufficient to ensure proper segregation. Moreover, there may be limited room for an individual mitochondrion to fit between the prospore membrane and the nuclear envelope. We propose that Ady3p helps to facilitate mitochondrial movement through this opening, thereby ensuring proper segregation. Consistent with the idea that mitochondrial entry into the prospore is limiting, even in wild-type cells, much of the mitochondrial mass is left behind in the ascus (4, 28).
Many organelles are regenerated within the spore.
Similar to mitochondria, the vacuole and the cortical ER also rely on actin-mediated transport to promote inheritance during budding. In meiotic cells, however, the vacuole and the cortical ER are not actively segregated, like mitochondria, but are regenerated after prospore membrane closure. This has been reported previously for the vacuole (45), and we show here that about 10 hours is required for a vacuolar-membrane marker to appear in spores. The cortical ER is progressively lost from the cytoplasm as cells progress through meiosis and then reappears relatively rapidly after prospore membrane closure. The localization of the cortical-ER marker Rtn2-GFP suggests that the cortical ER membrane might be “pulled back” into the nuclear envelope during meiosis. The reasons for this behavior are not clear, but during meiosis II the nuclear envelope is greatly distorted by the movement of chromatin within it (30). It may be that the additional membrane is required to allow the nucleus to accommodate two anaphase spindles at once.
It should be noted that regeneration within the spore is not limited to cytoplasmic organelles. The portion of the nucleus containing the nucleolus is left behind in the ascus after prospore membrane closure (5, 15). The spore nuclei then rapidly regenerate visible nucleoli (15).
Finally, the segregation of the Golgi apparatus during budding has been proposed to be based on generation of a new Golgi apparatus from cortical-ER elements within the bud (43). We found that the Golgi apparatus was the first organelle to segregate into the forming prospore and that this “segregation” correlates with the appearance of prospore membranes large enough to cover some of the nuclear envelope surface. This early segregation of the Golgi apparatus could be due to active transport of existing Golgi elements into the prospore membrane or to the trapping of Golgi apparatus newly generated from the nuclear envelope/ER between the nuclear envelope and the prospore membrane. Though we cannot distinguish between these two possibilities at present, in the second case, Golgi apparatus segregation would be based on de novo generation of the organelle during both budding and sporulation. During budding, the Golgi apparatus would be generated from the cortical ER, and during sporulation, directly from the nuclear envelope.
Taken as a whole, our data suggest that during sporulation the mitochondria and the nuclear envelope are actively segregated into prospores, whereas most, or all, other organelles are reformed from precursors within the spore. This stands in contrast to mitotic cells, in which efficient inheritance requires active transport of most organelles into the bud.
Developmental-stage-specific modes of organellar inheritance.
If segregation of the nuclear envelope and mitochondria is sufficient to allow the regeneration of a full complement of cellular organelles, why do mitotically dividing cells maintain extensive systems for the active delivery and segregation of organelles? One likely explanation is that the rapid division of mitotically dividing cells may not leave time for organelles to regenerate. For instance, the reappearance of vacuolar signals within the spores takes ∼8 h, whereas a mitotic cell cycle is only 90 min. Thus, delivery of fully formed and functioning organelles to the daughter may be necessary to maintain a rapid growth rate. Additionally, the fact that organelles are largely left behind during sporulation may be important for development of the ascus. In the case of the vacuole, the vacuolar proteases left behind in the ascal cytoplasm are important for the subsequent collapse of the ascal cell into a sheath around the spores (55).
This difference may also be due in part to the distinct challenges raised by sporulation. In contrast to the vectorial delivery of organelles to the daughter cell during budding, during spore formation the mother cell must evenly distribute organelles (or organellar precursors) to four distinct daughter cells simultaneously. If segregation is based to some extent on the stochastic entry of organelles into the developing prospore, then the fewer organelles that must be delivered in this way, the more likely that every spore will receive all of the essential organelles.
In sum, the mechanisms of segregation of organelles to daughter cells vary at different stages of the S. cerevisiae life cycle. During sporulation, a distributive mechanism that relies on regeneration of many organelles from a few precursors is used. It may be that distributive delivery will also prove to be important in organellar segregation in other instances where multiple daughter cells are formed simultaneously, such as cellularization of the Drosophila blastoderm or the division of apicomplexan parasites in host cells (19, 24).
Acknowledgments
We thank Nancy Hollingsworth for comments on the manuscript; Jodi Nunnari (UC Davis) for helpful discussions; and Jodi Nunnari, Benjamin Glick (University of Chicago), and Davis Ng (Life Sciences Institute, Singapore) for providing plasmids. We are grateful to Susan Van Horn (Stony Brook University) for assistance with the electron microscopy.
This work was supported by NIH grant GM072154 to A.M.N.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841-851. [DOI] [PubMed] [Google Scholar]
- 2.Boldogh, I. R., K. L. Fehrenbacher, H. C. Yang, and L. A. Pon. 2005. Mitochondrial movement and inheritance in budding yeast. Gene 354:28-36. [DOI] [PubMed] [Google Scholar]
- 3.Boldogh, I. R., S. L. Ramcharan, H. C. Yang, and L. A. Pon. 2004. A type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol. Biol. Cell 15:3994-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewer, B. J., and W. L. Fangman. 1980. Preferential inclusion of extrachromosomal genetic elements in yeast meiotic spores. Proc. Natl. Acad. Sci. USA 77:5380-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer, B. J., V. A. Zakian, and W. L. Fangman. 1980. Replication and meiotic transmission of yeast ribosomal RNA genes. Proc. Natl. Acad. Sci. USA 77:6739-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byers, B. 1981. Cytology of the yeast life cycle, p. 59-96. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 7.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Craene, J. O., J. Coleman, P. Estrada de Martin, M. Pypaert, S. Anderson, J. R. Yates III, S. Ferro-Novick, and P. Novick. 2006. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol. Biol. Cell 17:3009-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durr, M., M. Escobar-Henriques, S. Merz, S. Geimer, T. Langer, and B. Westermann. 2006. Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast. Mol. Biol. Cell 17:3745-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enyenihi, A. H., and W. S. Saunders. 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esposito, R. E., and S. Klapholz. 1981. Meiosis and ascospore development, p. 211-287. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Estrada, P., J. Kim, J. Coleman, L. Walker, B. Dunn, P. Takizawa, P. Novick, and S. Ferro-Novick. 2003. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 163:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fares, H., L. Goetsch, and J. R. Pringle. 1996. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 132:399-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehrenbacher, K. L., D. Davis, M. Wu, I. Boldogh, and L. A. Pon. 2002. Endoplasmic reticulum dynamics, inheritance, and cytoskeletal interactions in budding yeast. Mol. Biol. Cell 13:854-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, J., and J. Loidl. 2004. Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 12:427-438. [DOI] [PubMed] [Google Scholar]
- 16.Gao, X. D., H. Tachikawa, T. Sato, Y. Jigami, and N. Dean. 2005. Alg14 recruits Alg13 to the cytoplasmic face of the endoplasmic reticulum to form a novel bipartite UDP-N-acetylglucosamine transferase required for the second step of N-linked glycosylation. J. Biol. Chem. 280:36254-36262. [DOI] [PubMed] [Google Scholar]
- 17.Gorsich, S. W., and J. M. Shaw. 2004. Importance of mitochondrial dynamics during meiosis and sporulation. Mol. Biol. Cell 15:4369-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill, K. L., N. L. Catlett, and L. S. Weisman. 1996. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. J. Cell Biol. 135:1535-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, K., T. Mann, B. Striepen, C. J. Beckers, D. S. Roos, and J. M. Murray. 2002. Daughter cell assembly in the protozoan parasite Toxoplasma gondii. Mol. Biol. Cell 13:593-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 21.Itoh, T., A. Watabe, E. A. Toh, and Y. Matsui. 2002. Complex formation with Ypt11p, a Rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:7744-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knop, M., K. J. Miller, M. Mazza, D. Feng, M. Weber, S. Keranen, and J. Jantti. 2005. Molecular interactions position Mso1p, a novel PTB domain homologue, in the interface of the exocyst complex and the exocytic SNARE machinery in yeast. Mol. Biol. Cell 16:4543-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knop, M., and K. Strasser. 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19:3657-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lecuit, T. 2004. Junctions and vesicular trafficking during Drosophila cellularization. J. Cell Sci. 117:3427-3433. [DOI] [PubMed] [Google Scholar]
- 25.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 26.Lynn, R. R., and P. T. Magee. 1970. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J. Cell Biol. 44:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maier, P., N. Rathfelder, M. G. Finkbeiner, C. Taxis, M. Mazza, S. Le Panse, R. Haguenauer-Tsapis, and M. Knop. 2007. Cytokinesis in yeast meiosis depends on the regulated removal of Ssp1p from the prospore membrane. EMBO J. 26:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyakawa, I., H. Aoi, N. Sando, and T. Kuroiwa. 1984. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J. Cell Sci. 66:21-38. [DOI] [PubMed] [Google Scholar]
- 29.Moens, P. B. 1971. Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can. J. Microbiol. 17:507-510. [DOI] [PubMed] [Google Scholar]
- 30.Moens, P. B., and E. Rapport. 1971. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J. Cell Biol. 50:344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno-Borchart, A. C., K. Strasser, M. G. Finkbeiner, A. Shevchenko, A. Shevchenko, and M. Knop. 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20:6946-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morishita, M., and J. Engebrecht. 2005. End3p-mediated endocytosis is required for spore wall formation in Saccharomyces cerevisiae. Genetics 170:1561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakanishi, H., P. de los Santos, and A. M. Neiman. 2004. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15:1802-11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakanishi, H., M. Morishita, C. L. Schwartz, A. Coluccio, J. Engebrecht, and A. M. Neiman. 2006. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 119:1406-1415. [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi, H., Y. Suda, and A. M. Neiman. 2007. Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 120:908-916. [DOI] [PubMed] [Google Scholar]
- 36.Naylor, K., E. Ingerman, V. Okreglak, M. Marino, J. E. Hinshaw, and J. Nunnari. 2006. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 281:2177-2183. [DOI] [PubMed] [Google Scholar]
- 37.Neiman, A. M. 2005. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:565-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neiman, A. M. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neiman, A. M., L. Katz, and P. J. Brennwald. 2000. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 155:1643-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nickas, M. E., and A. M. Neiman. 2002. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics 160:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nickas, M. E., C. Schwartz, and A. M. Neiman. 2003. Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot. Cell 2:431-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preuss, D., J. Mulholland, C. A. Kaiser, P. Orlean, C. Albright, M. D. Rose, P. W. Robbins, and D. Botstein. 1991. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast 7:891-911. [DOI] [PubMed] [Google Scholar]
- 43.Reinke, C. A., P. Kozik, and B. S. Glick. 2004. Golgi inheritance in small buds of Saccharomyces cerevisiae is linked to endoplasmic reticulum inheritance. Proc. Natl. Acad. Sci. USA 101:18018-18023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riedel, C. G., M. Mazza, P. Maier, R. Korner, and M. Knop. 2005. Differential requirement for phospholipase D/Spo14 and its novel interactor Sma1 for regulation of exocytotic vesicle fusion in yeast meiosis. J. Biol. Chem. 280:37846-37852. [DOI] [PubMed] [Google Scholar]
- 45.Roeder, A. D., and J. M. Shaw. 1996. Vacuole partitioning during meiotic division in yeast. Genetics 144:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose, M. D., and G. R. Fink. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Rossanese, O. W., C. A. Reinke, B. J. Bevis, A. T. Hammond, I. B. Sears, J. O'Connor, and B. S. Glick. 2001. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 153:47-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon, V. R., S. L. Karmon, and L. A. Pon. 1997. Mitochondrial inheritance: cell cycle and actin cable dependence of polarized mitochondrial movements in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton 37:199-210. [DOI] [PubMed] [Google Scholar]
- 50.Stevens, B. 1981. Mitochondrial structure, p. 471-504. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Tachikawa, H., A. Bloecher, K. Tatchell, and A. M. Neiman. 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155:797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takizawa, P. A., J. L. DeRisi, J. E. Wilhelm, and R. D. Vale. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290:341-344. [DOI] [PubMed] [Google Scholar]
- 53.Taxis, C., C. Maeder, S. Reber, N. Rathfelder, K. Miura, K. Greger, E. H. Stelzer, and M. Knop. 2006. Dynamic organization of the actin cytoskeleton during meiosis and spore formation in budding yeast. Traffic 7:1628-1642. [DOI] [PubMed] [Google Scholar]
- 54.Tu, J., W. Song, and M. Carlson. 1996. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4199-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zubenko, G. S., and E. W. Jones. 1981. Protein degradation, meiosis and sporulation in proteinase-deficient mutants of Saccharomyces cerevisiae. Genetics 97:45-64. [DOI] [PMC free article] [PubMed] [Google Scholar]