Abstract
In the African trypanosome Trypanosoma brucei nearly all control of gene expression is posttranscriptional; sequences in the 3′-untranslated regions of mRNAs determine the steady-state mRNA levels by regulation of RNA turnover. Here we investigate the roles of two related proteins, TbUBP1 and TbUBP2, containing a single RNA recognition motif, in trypanosome gene expression. TbUBP1 and TbUBP2 are in the cytoplasm and nucleus, comprise ca. 0.1% of the total protein, and are not associated with polysomes or RNA degradation enzymes. Overexpression of TbUBP2 upregulated the levels of several mRNAs potentially involved in cell division, including the CFB1 mRNA, which encodes a protein with a cyclin F-box domain. CFB1 regulation was mediated by the 3′-untranslated region and involved stabilization of the mRNA. Depletion of TbUBP2 and TbUBP1 inhibited growth and downregulated expression of the cyclin F box protein gene CFB2; trans splicing was unaffected. The results of pull-down assays indicated that all tested mRNAs were bound to TbUBP2 or TbUBP1, with some preference for CFB1. We suggest that TbUBP1 and TbUBP2 may be relatively nonspecific RNA-binding proteins and that specific effects of overexpression or depletion could depend on competition between various different proteins for RNA binding.
The RNA recognition motif (RRM) consists of about 90 amino acid residues, including the canonical RNP1 octapeptide: (K/R)G(F/Y)(G/A)FVX(F/Y). Examination of eukaryotic genomes—from protists to humans—reveals that between 0.6 and 1.6% of all open reading frames encode proteins with one or more RRM motifs (14). Such proteins may be involved in rRNA biogenesis, mRNA splicing, mRNA-cap binding, export of RNAs from the nucleus, tRNA processing, translation, and transport of mRNAs in the cytoplasm (14). In addition, several RRM-motif proteins play key roles in the regulation of mRNA degradation. For the vast majority of RRM proteins, however, no function has yet been defined.
The roles of RRM proteins in mammalian mRNA degradation have been studied in some detail. A large class of very short-lived mRNAs, including many involved in signal transduction and in modulating the immune response, contains AU-rich elements (AREs) in the 3′-untranslated region (3′-UTR) (2). Degradation of ARE-mRNAs is controlled by a variety of RNA-binding proteins; in mammalian cells, these include ELAV family proteins such as HuR, which have three RRMs and stabilize ARE-containing mRNAs, and the destabilizing factor AUF1, which has two RRMs (2, 8, 49). In yeast, Sbp1p, with two RRMs, influences decapping (50), and the ELAV-like protein Pub1p influences the stability of over 50 mRNAs (22).
Protists of the genus Kinetoplastida are interesting models for the study of mRNA degradation because they depend almost exclusively on posttranscriptional mechanisms for the control of gene expression (10). The protein-coding genes are arrayed in long multigene transcription units (5, 23, 31, 44). Transcription by RNA polymerase II initiates from only a few positions on each chromosome (41, 42), and individual mRNAs are generated by 5′ trans splicing and 3′ polyadenylation (37). Although mRNA processing and export are potential points for control of mRNA levels, there is thus far strong evidence mainly for regulation of mRNA degradation and translation; in nearly all cases studied the sequences responsible were found to be in the 3′-UTR (10).
The African trypanosome Trypanosoma brucei has two major replicative stages, the procyclic form which grows in the tsetse fly vector and the bloodstream form which parasitizes mammals. At least 200 genes show differential expression in the two forms (7). For example, the EP (major surface protein), PGKB (cytosolic phosphoglycerate kinase), and PPDK (pyruvate phosphate dikinase) mRNAs are abundant and stable in procyclic trypanosomes but are extremely unstable and barely detectable in bloodstream forms. The instability of these mRNAs is caused by U-rich elements in the 3′-UTR, which strongly resemble AREs (34, 47), and they can even be stabilized by expression of HuR (47). Similarly, an ARE in the 3′-UTR of the SMUG mucin mRNA of Trypanosoma cruzi is responsible for the developmental regulation of degradation of that mRNA (16).
Two small RRM-containing proteins, T. cruzi UBP1 (TcUBP1) and TcUBP2, have been implicated in controlling T. cruzi SMUG mucin mRNA levels: mild overexpression of TcUBP1 in epimastigotes accelerated mRNA decay, and binding of TcUBP1 to the mRNA was shown (18). TcUBP1 and TcUBP2 each contain a single RRM and show a preference, in RNA-binding assays, for U- and G-rich sequences (13). In the present study, however, only 10 potential target RNAs for TcUBP1 and TcUBP2 were investigated, and more general roles of the proteins in parasite mRNA metabolism were not analyzed. We have therefore turned to the more experimentally amenable T. brucei in order to further characterize the functional roles of these proteins.
MATERIALS AND METHODS
Gene cloning and plasmid construction.
The open reading frame of TcUBP1 was amplified by using pT7-TcUBP1 as a template (18), and the product was cloned between the HindIII and BamHI sites of pHD 615 and pHD 617 (6) to give pHD 1260 and pHD 1261. The 5′ end of T. brucei UBP2 mRNA was identified by PCR using a forward spliced leader primer and two nested reverse coding-region primers. For tetracycline-inducible overexpression in trypanosomes, the full open reading frame was PCR amplified as two fragments which were religated and then inserted into pHD 615 and pHD 617 (6) to yield pHD 1325 and pHD 1326, respectively.
For RNA interference the open reading frame of TbUBP2 without the initiation codon was cloned twice in opposite orientations surrounding a “stuffer” from the spliced leader array (52). The inverted repeat was cloned into pHD 1146 (a version of pHD 617 lacking the T7 promoter (24), to give pHD 1459. This plasmid, which should allow tetracycline-inducible expression of an RNA stem-loop, was used to transfect procyclic forms. A similar construct (used for bloodstream forms) was made starting with pHD 1145, yielding pHD 1458 (24). (The resulting constructs differ only in the 3′-UTR downstream of the stem-loop, which has actually been shown to have no influence on RNA interference [RNAi] efficiency [20].) Since there are no sequences in the trypanosome genome with at least 85% identity to this double-stranded RNA apart from TbUBP1, the RNAi should specifically target TbUBP1 and TbUBP2 mRNAs and cause their degradation by RNAi (21).
For CAT reporter gene regulation experiments, a complete CFB locus intergenic region (from the termination codon of Tb927.1.4540 to the start codon of Tb927.1.4560) was PCR amplified and cloned between the BamHI and SpeI sites of pHD1437 (30) to give pHD 1453. A fortuitously obtained fragment from 1 to 450 of the 3′-UTR, obtained by the same PCR, was cloned between the BamHI and XhoI sites of pHD 1437, as was the fragment from nucleotides (nt) 630 to 1010 of the 3′-UTR (pHDs 1454 and 1455, respectively). Fragment 630-1010 was also inserted into the middle of the EPI 3′-UTR, replacing the destabilizing 26mer sequence: this was done by inserting the CFB fragment into the unique BglII site of pHD 523 (34) and then transferring the hybrid 3′-UTR and downstream actin 5′-UTR into BamHI and SpeI-cut pHD 1437 (pHD 1509). Finally, a plasmid containing nt 657 onward of the intergenic region was created by digesting pHD 1453 with BamHI and BsaAI, filling the ends and religating to give pHD 1510.
To in situ tag the TbUBP1, TbUBP2, and CFB genes, we used the Bla-V5 plasmid (51). Fragments from the 5′-UTR and coding regions of the three target genes were amplified from genomic DNA. Oligonucleotides used are given in Table 1. The 5′-UTR fragment was inserted immediately upstream of the blasticidin resistance cassette, and the start of the coding region was inserted in frame 3′ to the sequence encoding the V5 tag. The plasmids were pHD 1612 (UBP1), pHD 1613 (CFB1), and pHD 1614 (UBP2).
TABLE 1.
Oligonucleotides used in this study
| DNA target | Sequencea |
|---|---|
| TbUBP2 CDS | AGCTCGCTCGAGATGTCCCAGCAACCGCAGTA |
| AGCTCGGGGCCCGCGCTCTGAAACTTCACGAA | |
| TbUBP1 CDS (in situ tagging) | AGCTCGCTCGAGATGTCTCAGGTTCCACTGGC |
| AGCTCGGGGCCCGCGACTTTGACGAGTCTC | |
| TbUBP1 upstream (in situ tagging) | AGCTCGCCGCGGCTGCGGGATACTTCCGATG |
| AGCTCGTCTAGATTGTTTTGCGACTGCACCGTC | |
| CFB coding region (in situ tagging) | AGCTCGCTCGAGATGTTTTTGAAGGAGGGAAACAGT |
| AGCTCGGGGCCCTGACAACCCTGACGGCTC | |
| CFB upstream (in situ tagging) | AGCTCGCCGCGGGCAGCGGTAGATTGGACGTG |
| AGCTCGTCTAGA CGTGTACACCGCTATTCAAAT | |
| CFB1 IGR (50°C) | CZ 2478: GAA TGC CCT TAT GTA AGT AA |
| CZ 2481: CAG CAA ATG GAG CAC CAC TG | |
| ACT (52°C) | CZ 2578: ATG TCG GAC GAG GAA CAA AC |
| CZ 2579: CGA CTC GTC GTA TTC ACT CT | |
| AAT11 (52°C) | CZ 1952: GTA AGG ATC CGG AGG GGA GGC GGC GTA GCA GGA G |
| CZ 2089: ATA TGT TAA CCC CGA AAG TAT CGC CTG | |
| 6470 (55°C) | CZ2759: AAT GCA GGC TGT AGC GAA GT |
| CZ2760: AGC TTG GGC TCT GTA GCA TC | |
| TUB Q-PCR | CZ2724: TGA CTC GCC GCA ACC TCG AT |
| CZ2581: CCT TTG GCA CAA CGT CAC CAC GG | |
| CAT Q-PCR | ATCCGGCCTTTATTCACATTC |
| TCGTGGTATTCACTCCAGAGC | |
| SRP Q-PCR | CGTTGACTTGGTGTTCTGCTT |
| CTCGGTGTGCTTCTGCAAC |
Restriction sites are underlined, and initiation codons are in boldface. Additional nucleotides to allow fragment cleavage are in italics.
To express N-terminally myc-tagged TbUBP1 and TbUBP2, the open reading frames were cloned in frame with two myc tags in the vector pHD1701 (11). C-terminally TAP-tagged versions were obtained by excising the open reading frames from the myc vector by using BamHI (Klenow) and HindIII and cloning them into pHD918 cut with ApaI, end filled (Klenow), and then cut with HindIII (25). All of these constructs are tetracycline inducible.
Expression of TbUBP1, TbUBP2, and TbDHH1 in Escherichia coli.
To make glutathione S-transferase (GST)-tagged UBPs, the open reading frames were amplified and cloned into pGEX-GST cut with EcoRI. GST-UBP1 was insoluble, but GST-UBP2 was soluble and was purified on GST-Sepharose, and the purity was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. A single dominant band with a few very minor bands was obtained; the concentration of the major band was estimated from the gel stain relative to dilutions of a bovine serum albumin standard. Binding to poly(U), poly(A), poly(C), or poly(G) coupled to agarose beads was tested as previously described (13). Binding to the beads alone was similar to binding in the presence of homopolymers.
The complete DHH1 open reading frame (Tb10.70.3290) was amplified from genomic DNA and cloned into a derivative of pET15b in-frame with an N-terminal His6 tag. After expression in E. coli BL21(DE3), the insoluble recombinant polypeptide was purified from inclusion bodies by preparative SDS-PAGE and used to raise a rabbit antiserum. The serum reacts with a single band in Western blots of trypanosome extracts.
Trypanosomes.
Bloodstream-form or procyclic-form T. brucei, strain Lister 427, expressing the Tn10 tet repressor from integrated plasmid pHD 449 or pHD 1313 was transfected with overexpression plasmids based on pHD 615 (bloodstream form) or pHD 617 (procyclic form) and stable lines selected for hygromycin resistance as described previously (6, 54). To induce expression, tetracycline was added to 100 ng ml−1. For RNAi, the TbUBP stem-loop plasmid was transfected into trypanosomes containing pHD449 with all transfection and culture methods as described previously (1). To analyze the effects of intergenic regions on CAT gene expression, the bloodstream-form cell lines were transfected with various derivatives of pHD 1437, and transformants were selected by using 0.2 μg of puromycin/ml. RNA and protein were prepared only from exponentially growing cells (less than 2 × 106/ml for bloodstream forms and 5 × 106/ml for procyclic forms).
In situ tagging constructs were restriction digested at the ends of the genomic fragments designed for homologous recombination and transfected into various cell lines as indicated in the text.
DNA arrays.
RNA preparation and analysis were essentially as previously described (7, 15). Briefly, for the genome-wide analysis of gene expression, 30 μg of total RNA was incubated for 10 min at 100°C with 2.6 μg of oligo(T)12-18 (Amersham). The mixture was put on ice before addition of 6 μl of 5× superscript buffer (Invitrogen), 0.6 μl of 50× deoxynucleoside triphosphates (dATP, dGTP, dCTP, and 7.5 mM dTTP [25 mM each]), 3 μl of 2 mM amino-allyl dUTP, 3 μl of 0.1 M DTT, and 1.9 μl of Superscript II (Invitrogen). The reaction was incubated for 2 h at 42°C. Afterward, the RNA was hydrolyzed by the addition of 10 ml of 0.1 M NaOH and an incubation at 65°C for 10 min. The solution was neutralized by addition of the same volume of 0.1 M Tris-HCl (pH 7.5), and the cDNA was purified by using a QIAquick PCR purification kit. The amino-allyl cDNA was then coupled either to Cy3 or Cy5 dye in 50 mM Na2CO3 in 50% dimethyl sulfoxide for 2 h in the dark. The cDNA was then repurified by using QIAquick PCR purification columns, ethanol precipitated, and resuspended in hybridization buffer.
Arrays of 23,000 to 24,000 (7, 15) random 2-kb genomic fragments on glass slides were preincubated in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS, and 1% BSA for 1 h at 42°C; dipped in water and then isopropanol; and allowed to dry. Hybridization with mixed Cy3- and Cy5-labeled cDNAs was performed in 50% deionized formamide, 5% dextran sulfate, 3× SSC, 1% SDS, and 5× Denhardt solution at 42°C for 16 h, using 24-to-60-mm coverslips and glass array hybridization cassettes (Ambion). Slides were then washed for 5 min each in 1× SSC, 0.2% SDS, 0.1× SSC, 0.2% SDS, and 0.1× SSC at room temperature. The results were analyzed as previously described (7, 15) by using M-CHIPS software (26, 27). To choose regulated clones for sequence analysis, we selected only those with the following parameters: fitted intensities of at least 50,000, a minimum/maximum separation of 0.2, and a TbUBP2/wild-type ratio of at least 2.0 or −2.0.
Northern and Western blots.
Northern and Western blotting was done by using standard procedures; probes for the Northern blots were labeled with 32P and quantitated by phosphorimager. The antibodies for the Western blots were rabbit polyclonal anti-TcUBP1 (18) used at 1:5,000, a rabbit anti-peptide antibody reacting with a cytosolic protein (CSM) (24, 28), a monoclonal antibody to the V5 tag (Invitrogen), anti-TbRRP6 (29), and anti-TbDHH1 (the present study). Detection was done by enhanced chemiluminescence (Amersham, Braunschweig, Germany) according to the manufacturer's instructions. Western results were quantified by densitometry.
Analysis of splicing intermediates.
To assess the effect of TbUBP1 and TbUBP2 depletion on splicing, the amounts of SLRNA and of the Y-structure splicing intermediate were assayed by primer extension (38) using the oligonucleotide CZ2711 (5′-GCAGGAACCAACAGCACAATGCG-3′), using RNAi cells without tetracycline or 24 or 48 h after tetracycline addition. Trypanosomes treated with Sinefungin (30 min) served as a control; these showed an absence of cap methylation and of the Y-intermediate. Extension of the U3 RNA (primer CZ2712 5′-TGCCGTTCATCGAAC-3′) served as a loading control.
Affinity purification and analysis of TAP-tagged TbUBP1 and TbUBP2.
To identify RNAs associated with TbUBP1 and TbUBP2 in vivo, lysates of 109 trypanosomes were bound to immunoglobulin G (IgG)-Sepharose, washed, and then eluted with Tobacco Etch Virus (TEV) protease as previously described (40). The extracts, flowthrough, and eluate were each in 1 ml. RNA was made from the entire eluate and from 30 μl of the loaded extract and flowthrough fractions (equivalent to 3 × 107 cells). The resulting RNA was converted to cDNA as described previously (40). 1/10 or 1/20 of the cDNA and fivefold serial dilutions thereof were used for PCR (maximally equivalent to 3 × 106 cells for the load and flowthrough and to 108 cells for the eluate). To identify proteins associated with TAP-tagged TbUBP1 and TbUBP2, lysates from 109 cells expressing the proteins were subjected to a complete tandem affinity procedure. For mass spectrometry total proteins from a single purification were separated by SDS-PAGE and stained with SyproRuby. For Western blots, samples representing 5 × 106 cells were loaded.
RESULTS
Arrangement and expression of the TbUBP1 genes.
A BLASTp search of the genome of T. brucei with the TcUBP1 amino acid sequence yielded two genes encoding UBP1-like proteins: Tb11.03.0620 and Tb11.03.0580. Alignment of the RNA-binding domains with those of TcUBP1 and the closely related TcUBP2 (see Fig. S8 in the supplemental material) indicated that the T. brucei proteins are more closely related to each other than to the T. cruzi proteins. The Tb11.03.0620 and Tb11.03.0580 genes are located together on chromosome 11. A map of this chromosome, as annotated in GeneDB, is shown in Fig. 1A. The TcUBP1 and TcUBP2 genes are also next to each other on a chromosome, and open reading frames upstream and downstream of the TcUBP genes have homologues in similar positions in T. brucei. We therefore used chromosome synteny information to name Tb11.03.0620 as TbUBP1 and Tb11.03.0580 as TbUBP2. Extending the homology search to the completed genome of Leishmania major and the partially sequenced genomes of Trypanosoma vivax and Trypanosoma congolense, we found two L. major homologues, also next to each other on a chromosome, Lmj25.0490 and Lmj25.0500, two T. vivax homologues (Tviv884h08.p1k_2, Tviv1247f12.q1k_2), and a T. congolense homologue (congo958c11.q1k_2). The UBP1 from L. major was most similar to the UBP2 protein from L. major, and the two proteins from T. vivax were similarly most closely related to each other (not shown). This suggests that a gene duplication occurred prior to divergence of leishmanias and the trypanosomes and that subsequently the genes co-evolved, maintaining similarity to each other through gene conversion, selective pressure, or both.
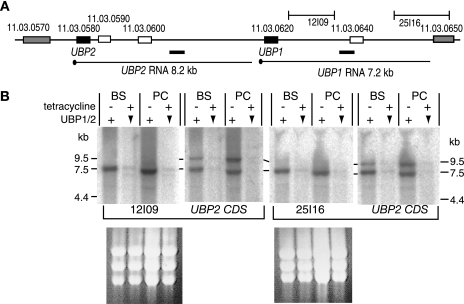
FIG. 1.
(A) Map of the UBP2-UBP1 region of chromosome 11. Distances in kilobases are shown above the map, and the gene identity numbers are above the open reading frames. The locations of two array clones, 12I09 and 25I16, are indicated, and the positions deduced for the transcripts are beneath the map with corresponding predicted sizes. The dark bars below the map and above the putative transcripts represent EST sequences from the EMBL database. Three hypothetical open reading frames shown as open boxes downstream of TbUBP1 and TbUBP2 have repetitive AT-rich sequences, are not conserved between trypanosomatids, and are unlikely to encode proteins. (B) TbUBP2 and TbUBP1 mRNA identification. Two independent Northern blots are shown. Total RNA was prepared from either bloodstream-form (BS) or procyclic-form (PC) trypanosomes containing an integrated plasmid designed for tetracycline-inducible expression of a UBP2 stem-loop. After 24 h in the presence of tetracycline (+) the level of mRNA encoding TbUBP1 and TbUBP2 is reduced through RNAi (downward arrowheads). The first blot was hybridized with clone 12I09, stripped, and then rehybridized with a TbUBP2 coding region probe, which hybridizes with both TbUBP1 and TbUBP2 mRNA. The second blot was hybridized with clone 25I16, followed by TbUBP2. Marker sizes are in kilobases. Ethidium bromide-stained rRNA is shown beneath the blots.
All of the UBP1/2 proteins contain a single RRM, connected by a more variable domain with a glycine-rich region (see Fig. S8 in the supplemental material). The short N and C termini are less well conserved but are all glutamine-rich. The RNA-binding region is very well conserved: of 83 amino acids, 72 are identical in all of the organisms examined, and of the remaining 11 positions only 5 show nonconservative substitutions. Residues that have been identified as being likely to be involved in the RNA binding by TcUBP1 (55) are conserved in all of the UBP proteins, and there is only a single G→S difference between the T. cruzi and T. brucei RRM domains.
The TbUBP1 and TbUBP2 open reading frames are 7.6 kb apart (Fig. 1A). In trypanosomes, polyadenylation of an mRNA depends on downstream trans splicing and usually occurs 100 nt upstream of the splice acceptor site of the next mRNA downstream (4, 37). Using this as a guide, we predict lengths of 8.2 and 7.2 kb, respectively, for the TbUBP2 and TbUBP1 mRNAs. Northern blot analyses (Fig. 1B) using a TbUBP2 open reading frame probe (UBP2 CDS), which carries sequence common to both genes, indeed hybridized to two RNAs of approximately the appropriate sizes (9.5 and 7.5 kb), which were expressed at similar levels in bloodstream and procyclic trypanosomes (Fig. 1B and data not shown). No smaller mRNAs were seen. Both mRNAs were degraded when targeted by RNA interference directed to the conserved coding region (Fig. 1B). To determine which of these mRNAs encoded TbUBP1 and TbUBP2, we hybridized the Northern blots with two genomic clones, 12I09 and 25I16, derived from the untranslated region downstream of the TbUBP1 gene. Neither of these clones was homologous to the predicted TbUBP2 mRNA, and both hybridized to the smaller, 7.5-kb mRNA (Fig. 1B), suggesting that this encodes TbUBP1. Apart from TbUBP1 and TbUBP2, there is no other gene in the genome with homology to the TbUBP2 coding region probe. Since both proteins are expressed (see below), the longer mRNA presumably—by default—encodes TbUBP2.
We also searched for expressed sequenced tags (ESTs) from this region. The EST libraries in the EMBL database were made by oligo(T) priming, which could initiate cDNA synthesis either from the poly(A) tail or from internal oligo(A) sequences (which abound in the TbUBP1 and TbUBP2 3′-UTRs). The cDNAs were sequenced unidirectionally from their 5′ ends; this means that the 3′ ends of the mRNA templates must be downstream of the EST ends. We found two relevant sequences, shown as dark bars below the map in Fig. 1A. A 337-nt MVAT4 bloodstream trypanosome EST (17) terminates 3.9 kb downstream of the TbUBP2 stop codon (accession no. W68918) and a 444-nt YiTat1 EST (poor quality) probably ends about 5.4 kb downstream of the TbUBP1 start codon (accession no. AA736273). This confirms that both of these regions are present in mature transcripts.
The 9.5-kb RNA appeared longer than the 8.2 kb expected for a TbUBP2 mRNA, but mRNA lengths are not always accurate when measured using formamide-formaldehyde agarose gels. The presence of a bicistronic RNA, as has been suggested for the TcUBP genes (35), is possible since the distance from the TbUBP2 start codon to the TbUBP1 stop codon is 8.76 kb. To confirm the mRNA structure, we attempted to generate probes specific for TbUBP2 (the EST clones are no longer available) and, more importantly, to map the polyadenylation sites of both mRNAs by reverse transcription-PCR (RT-PCR). Despite the use of several different primers, these experiments failed, probably because the untranslated regions of both genes are very repetitive and have many low-complexity stretches. Thus, the precise structure of this mRNA remains uncertain.
We conclude that the 7.5-kb mRNA encodes TbUBP1 and the 9.5-kb mRNA probably encodes TbUBP2.
Expression of TbUBP2 and TbUBP1.
To analyze the expression of TbUBP2 and TbUBP1, we probed Western blots with a polyclonal antibody raised using recombinant TcUBP1 as an immunogen (18). A typical result is shown in Fig. 2A. The predicted molecular weights of TbUBP2 and TbUBP1 are 19.5kDa and 24.3kDa, respectively. On blots from both procyclic and bloodstream trypanosomes (Fig. 2A, lanes 1, 4, 7, and 10, and Fig. 2B), the antibody reproducibly recognized one band corresponding to the expected size of TbUBP2, at 19 kDa, and two more at 17 and 12 kDa. In addition, in procyclic extracts the antibody recognized a band which migrated at 29 kDa, which we tentatively identified as TbUBP1. When we examined extracts from bloodstream trypanosomes which are adapted for very rapid growth in vitro, the putative TbUBP1 was not detected (Fig. 2A, lanes 7 to 10, and Fig. 2B, lanes 7 to 9). If, however, we used extracts from two trypanosome strains that are readily able to differentiate into procyclic forms, TbUBP1 was detected. We do not know if this observation is general or has functional implications. In addition to these signals there was sometimes weak, irreproducible recognition of other bands from 40 to 60 kDa (Fig. 2A).
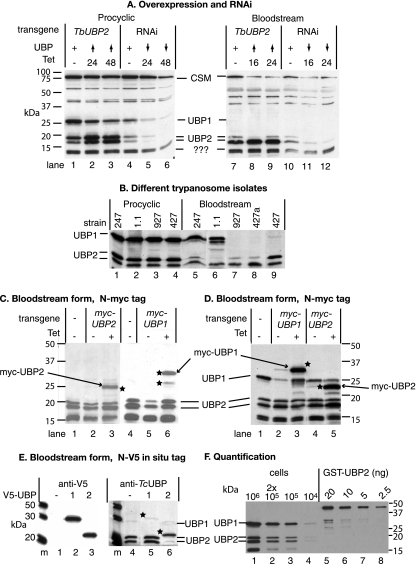
FIG. 2.
(A) TbUBP1/2 protein expression in transgenic T. brucei. Total cell lysates from bloodstream-form or procyclic-form trypanosomes were subject to denaturing SDS-gel electrophoresis and blotted; detection was done with a polyclonal antiserum to TcUBP1. The blot was also stained with antibody to the cytosolic protein CSM, which migrates at about 100 kDa. The positions of molecular mass markers are on the left. The transgenes in the different cell lines are designed for tetracycline-inducible expression as follows: TbUBP2 overexpression (lanes 1 to 3 and lanes 7 to 9) and TbUBP1/2 RNAi (lanes 4 to 6 and lanes 10 to 12). The times after tetracycline addition are indicated: time zero indicates no tetracycline (lanes 1, 4, 7, and 10); 16-h (bloodstream) and 24-h (procyclic) time points with tetracycline are lanes 2, 5, 8, and11, and the patterns after 24 h (bloodstream) or 48 h (procyclic) with tetracycline are shown in lanes 3, 6, 9, and 12. The effects on TbUBP1/2 are also shown. “+” indicates normal expression from endogenous genes; upward arrowheads denote overexpression, and downward arrowheads denote depletion. Likely identities of the protein bands are indicated in the center between the images. (B) Staining pattern of a Western blot using extracts of different trypanosome strains and anti-TcUBP1 antibody. The 247 and Antat 1.1 strains have pleomorphic bloodstream forms, whereas the 927 strain is more culture adapted but can still differentiate readily. Two different bloodstream 427 isolates were checked; the one labeled 427a can differentiate into growing procyclic trypanosomes in culture, whereas the one labeled 427 cannot. (C) Bloodstream-form trypanosomes with inducible expression of myc-tagged proteins were incubated with tetracycline to induce expression. Lanes 1 and 4 are extracts from control cells without the transgene, lanes 2 and 5 are extracts from transgenic parasites grown without tetracycline, and lanes 3 and 6 are extracts from cells grown for 24 h in 100 ng of tetracycline/ml. Proteins were separated by SDS-PAGE and detected on Western blots with anti-TcUBP1 antiserum. Bands which reacted with the anti-myc antibody (not shown) are indicated with stars. Quantitation of the bands indicated that the myc-tagged TbUBP2 was ∼2-fold overexpressed in bloodstream forms. (D) Procyclic-form trypanosomes with inducible expression of myc-tagged proteins incubated with or without tetracycline. Details are as in panel C. Lane 1 is an extract from control cells without the transgene; lanes 2 and 4 are extracts from transgenic parasites grown without tetracycline, and lanes 3 and 5 are extracts from cells grown for 24 h in 100 ng of tetracycline/ml. Quantitation of the bands indicated that the myc-tagged TbUBP2 was ∼2-fold overexpressed, relative to cells without the transgene, and myc-TbUBP1 was 3-fold overexpressed. (E) Expression of N-terminally in situ-tagged TbUBP1 and TbUBP2. A sequence encoding a V5 tag was integrated into the genome in order to give an in-frame fusion with either TbUBP1 (lane 2) or TbUBP2 (lane 3); cells without the tag are in lane 1. Details are as for panel A except that the detecting antibody was anti-V5 for the left-hand panel. Detection with anti-TcUBP1 antibody followed the V5 detection; lanes 1, 2, and 3 are here marked 4, 5, and 6, respectively, for clarity of description in the text. Three more cell lines for each tag gave indistinguishable results. The stars mark the positions of the tagged proteins. (F) Quantitation of TbUBP1 and TbUBP2 protein levels in procyclic trypanosomes. Lanes 1 to 4 contained various numbers of trypanosomes, and lanes 5 to 8 contained serial dilutions of purified GST-tagged TbUBP2.
After induction of RNAi targeting the TbUBP1 and TbUBP2 mRNAs, the bands at 29, 19, and 17 kDa diminished in intensity by 10- to 20-fold within 24 h (Fig. 2A, compare lanes 5 and 6 with lane 4 and lanes 11 and 12 with lane 10; see also the mRNA results in Fig. 1B). The other bands were unaffected by the RNAi. It was not possible to design double-stranded RNAs of adequate length to target the two RNAs individually due to the repetitive nature of the 3′-untranslated regions and the similarity of the coding regions. In cells in which TbUBP2 was overexpressed using a tetracycline-inducible RNA polymerase I promoter (lanes 2 and 3 and lanes 8 and 9 in Fig. 2A), the 17- and 19-kDa bands became more prominent after tetracycline addition, with a twofold increase in procyclic forms and a nearly sixfold increase in bloodstream forms. TbUBP2 overexpression had no reproducible effect on TbUBP1 expression. These results indicated that the 19- and 17-kDa bands were both TbUBP2 and that TbUBP1 was not present in rapidly growing bloodstream trypanosomes.
To investigate expression of the proteins further, we expressed tagged versions. We made bloodstream (Fig. 2C) and procyclic (Fig. 2D) trypanosomes in which N-terminally myc-tagged TbUBP1 or TbUBP2 was inducibly expressed from the tetracycline-inducible promoter. Myc-UBP2 (Fig. 2C and D, lane 3) and myc-UBP1 (Fig. 2C, lane 3, and Fig. 2D, lane 5) were detected as single bands, both with the anti-TcUBP1 antibody (Fig. 2C and D) and with anti-myc (not shown). Thus, myc-tagged TbUBP1 can indeed be detected in bloodstream trypanosomes if overexpressed. The overexpression had no effect on the levels of untagged protein. Although the double myc tag has a molecular mass of about 3.2 kDa, the increase as judged by migration on SDS-gels was somewhat more.
To detect the two proteins individually without overexpression, we N-terminally tagged the genes in situ by integration of a sequence encoding the V5 tag into the genome of bloodstream trypanosomes (51). Tagged TbUBP1 (Fig. 2E, lane 2) and tagged TbUBP2 (Fig. 2E, lane 3) were easily detected with the V5 antibody, since single bands migrated slightly slower than the expected sizes, as for the myc-tagged versions. To our considerable surprise, however, the anti-TcUBP1 antibody readily detected endogenous TbUBP2 (Fig. 2E, lanes 4 and 5) and tagged TbUBP2 (lane 6), but tagged TbUBP1 was barely visible (Fig. 2E, lane 5). This suggested to us that in bloodstream forms, the reactivity of TbUBP1 with the polyclonal anti-TcUBP1 antibody was impaired. Expression of V5-TbUBP2 protein from one allele is expected to reduce the total amount of untagged TbUBP2 by 50%. The signal was indeed markedly reduced (compare lane 6 with lanes 4 and 5 in Fig. 2E). Similarly, tagging of one TbUBP1 locus resulted in the disappearance of the (already faint) signal for untagged TbUBP1. Exactly the same results were obtained with three completely independent cell lines. Further attempts to determine why the untagged or V5-tagged TbUBP1 was not detected by the TcUBP1 antibody (e.g., mass spectrometry and searches for phosphorylation) were unsuccessful. Similarly, we were unable, despite exhaustive attempts, to unequivocally determine the relationship between the 17- and 19-kDa bands labeled as TbUBP2, since both V5-UBP2 and myc-UBP2 migrated as single bands. It is conceivable that one of the bands is not in fact TbUBP2, but another related protein whose presence depends on that of TbUBP2. Alternatively, N-terminal tagging may prevent a posttranslational modification. N-terminal proteolysis in vivo is one possible explanation: when we added a TAP tag to the C terminus, we did see two bands (see Fig. 7A below), but the lower band was of rather low abundance.
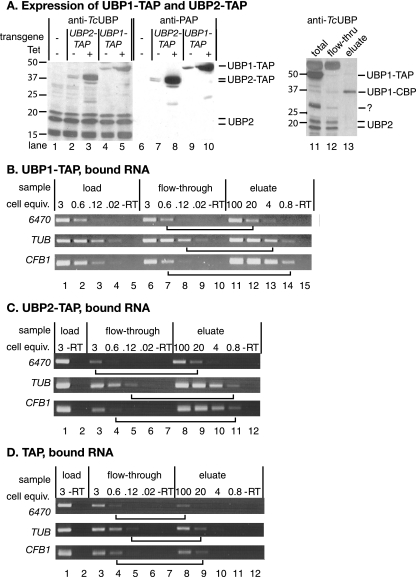
FIG. 7.
Interaction of TbUBP1 and TbUBP2 with RNA. (A) Expression of C-terminally TAP-tagged TbUBP1 and TbUBP2 in bloodstream forms. Labeling is as in Fig. 3. The IgG binding domain was detected using HRP-IgG (lanes 6 to 10), and then the blot was stripped and all UBPs were detected with the anti-UBP1 antibody (lanes 1 to 5). Lanes 11 to 13 show a Western blot (using anti-TcUBP1) of a typical first purification step (bloodstream forms). Total cell extract is in lane 11, the flowthrough from the IgG column is in lane 12, and the TEV eluate from the IgG column is in lane 13. Identical proportions of the material (equivalent cell numbers) were loaded in all three lanes. Detection is with anti-TcUBP1; using these extracts, there was cross-reaction with a thick band at 50 kDa. Note that neither TbUBP2 nor untagged TbUBP1 is found in the final eluate. The band around 30 kDa which was retained on the IgG column could be untagged TbUBP1, but this is unlikely since untagged TbUBP1 does not react with the antibody in bloodstream forms. Perhaps it was a fragment of the tagged protein. (B) Extracts from TbUBP1-TAP-expressing bloodstream forms were passed over an IgG column. The column was washed, and then bound material was eluted with TEV protease. RNA was prepared and reverse transcribed using random primers. Various dilutions of the cDNA were then subjected to PCR (30 cycles) by using gene-specific primers, and the products were detected by agarose gel electrophoresis and ethidium bromide staining. The numbers of cell equivalents used for each PCR are shown in millions above the lanes: for example, lane 1 shows the RT-PCR product from 3 × 106 cells, and lane 11 shows the RT-PCR product from 108 cells. “-RT” shows the reaction from the maximum relative number of cell equivalents, without reverse transcriptase. To assess the degree of binding, we compared signals from the eluate with signals from the flowthrough fraction. Examples of lanes with equivalent signals are joined by horizontal lines. The results of three replicate experiments were quantitated by scanning densitometry, and in each case about five times more CFB1 mRNA than control mRNAs was bound. (C) As for panel B, using cells expressing TbUBP2-TAP. (D) Same as for panel B, using cells expressing the TAP tag alone.
Both TbUBP1 and TbUBP2 were among the 770 different proteins detected in a recent analysis of the procyclic trypanosome proteome (36). To determine the abundance of the two proteins, we performed Western blots with procyclic trypanosome extracts and with various amounts of GST-tagged recombinant TbUBP2. The results (Fig. 2F) suggested that TbUBP1 and TbUBP2 are quite abundant. If the GST-tagged and endogenous proteins are reacting similarly with the antibody, the comparison suggests that there are about 1,200,000 molecules each of TbUBP1 and TbUBP2 in procyclic trypanosomes and 600,000 TbUBP2 molecules in bloodstream forms (0.1% of total cell protein).
Subcellular localization of TbUBP2 and TbUBP1.
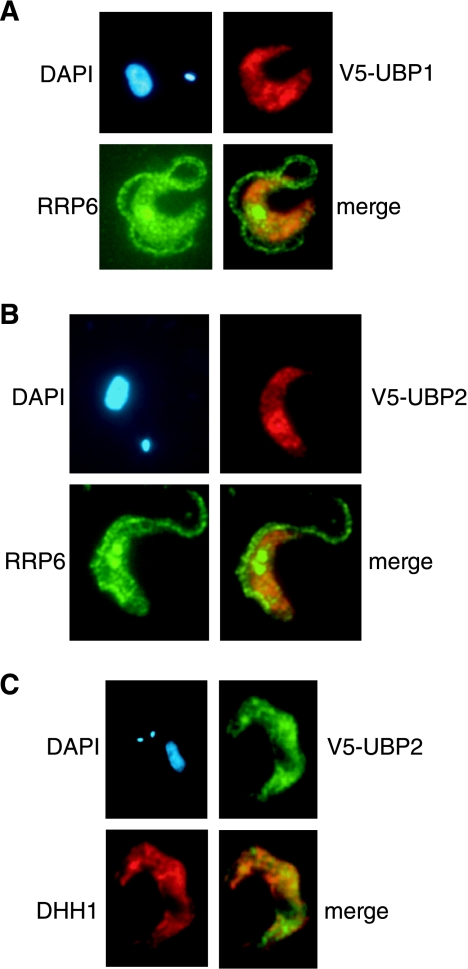
We localized V5-tagged TbUBP1 (Fig. 3A) and TbUBP2 (Fig. 3B) in bloodstream trypanosomes by immunofluorescence. The tagged proteins were mainly in the cytoplasm but were not totally excluded from the nucleus and so could play roles in both compartments. The distribution in the cytoplasm was clearly uneven. To find out whether the TbUBPs were associated with the RNA degradation machinery, we counterstained with antibody to TbRRP6, an exosome subunit (29) (Fig. 3A and B); no overlap was seen. TbUBP1 associates with polyadenylated RNA and TbDHH1 in stress granules in starved procyclic trypanosomes (9). DHH1 is also a marker for P bodies, which are subcompartments containing enzymes needed for 5′-3′ degradation of mRNAs (33, 46). In nonstressed bloodstream forms, however, only very limited overlaps between the TbUBPs and TbDHH1 were seen (Fig. 3C).
FIG. 3.
Subcellular localization of V5-tagged TbUBP1 or TbUBP2 by immunofluorescence with mouse anti-V5 antiserum. The proteins detected are indicated next to the pictures; the nucleus and kinetoplast were stained using DAPI. (A and B) colocalization with RRP6; (C) colocalization with DHH1.
Next, we looked for association of the TbUBPs with the translation machinery. After centrifugation of cycloheximide-treated procyclic cytoplasmic extracts on sucrose gradients, TbUBP1 and TbUBP2 were found partly in the pellet and partly in the S100 fraction (see Fig. S9 in the supplemental material). Further separation on sucrose gradients showed that both TbUBPs were in the lighter fractions together with soluble protein and monosomes but also smearing down into the denser fractions. Since the profile was identical in the presence of EDTA, TbUBP1 and TbUBP2 are probably not associated with polysomes. Neveretheless, the broad distribution suggested that they were associated with other macromolecules.
Effects of UBP overexpression or depletion on cell growth.
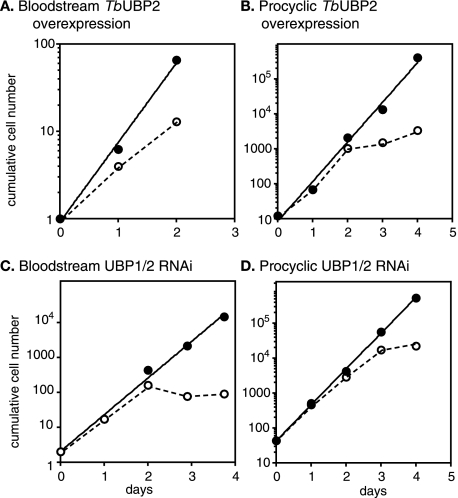
Both sixfold overexpression of TbUBP2 (Fig. 4A and B and Fig. 2A) and 10- to 20-fold depletion of TbUBP1 and TbUBP2 (Fig. 4C and D and Fig. 2A) inhibited cell growth. A similar effect was also seen upon inducible expression of TcUBP1 in both life cycle stages (not shown). The effects on bloodstream forms after RNAi were particularly dramatic, with the onset of morphological changes (rounding up) visible by 24 h and complete growth arrest within 2 days. We conducted a cell cycle analysis on bloodstream-form cells with TbUBP2 overexpression or TbUBP1/2 RNAi 24 h after tetracycline addition. We counted nuclei and kinetoplasts to determine cell cycle stages and also assayed DNA content by flow cytometry (not shown). We found that either overexpression or RNAi resulted in the appearance of some cells with abnormal numbers of nuclei and an increase (to ca. 4%) of cells with more than a 4 N DNA content. There was, however, no marked accumulation at a particular cell cycle stage, so a specific cell cycle block can be ruled out. The doses of tetracycline used here do not by themselves affect trypanosome growth or morphology or the transcriptome and proteome (40).
FIG. 4.
Effects of TbUBP1/2 up- and downregulation on cell growth. Cells were seeded at between 0.1 and 5 × 105 cells/ml and diluted as required. Cultures were bloodstream-form (A and C) or procyclic-form (B and D) trypanosomes and were grown either with (open symbols and dotted lines) or without (closed symbols) 100 ng of tetracycline/ml as indicated within each panel. Upward arrowheads indicate overexpression, and downward arrowheads indicate depletion. Expression of TcUBP1 also inhibited growth (not shown).
The expression of myc-TbUBP1 or myc-TbUBP2 (Fig. 2C and D) or TbUBP1-TAP or TbUBP2-TAP (see below) was not deleterious to growth. The degree of overexpression of myc-TbUBP2 was twofold in both bloodstream forms and procyclics; for myc-TbUBP1, overexpression in bloodstream forms could not be assessed, but it was just threefold in procyclics. This contrasts with the sixfold overexpression of untagged TbUBP2. It therefore seems likely that the effects of TbUBP2 overexpression on cell growth depend on the degree of overexpression.
Effects of TbUBP1/2 downregulation on mRNA levels.
To look for mRNAs that were affected when the amounts of TbUBP1/2 were reduced by RNAi, we compared the transcriptomes of the TbUBP1/2-depleted bloodstream forms with those of normal cells by using a T. brucei microarray (7, 15). Two independent array spots that exhibited downregulation by the TbUBP1/2 RNAi came from the TbUBP1 locus itself (see Fig. 1). The RNAi had no effect on tubulin (TUB) or EP procyclin mRNA (Northern blots, not shown). In fact, outside the TbUBP1/2 locus, only five reproducibly regulated spots, representing just two gene sequence classes, were found. Two spots contained sequences related to the genes encoding transferrin receptor subunits ESAG6 and ESAG7. Unlike the bona fide transferrin receptor mRNAs, which are bloodstream form specific, the mRNAs that reacted with the array clone probe were not stage specific. The hybridizing mRNAs (which may not be identical to the probe, since many related genes are present in the genome) were upregulated ∼2.5-fold not only by RNAi against UBP1/2 but also by overexpression (see Fig. S10 in the supplemental material). The significance of this is unknown.
The three remaining regulated spots came from five very similar genes arranged as a direct tandem repeat on chromosome 3: Tb03.26J7.120, 0.130, 0.140, 150, and 160. The RNA was twofold reduced after the TbUBP1/2 RNAi and 1.6-fold increased by TbUBP2 overexpression (not shown). These genes encode a predicted protein of unknown function, with a signal peptide and 13 to 14 transmembrane domains. The results of experiments with this locus will be presented elsewhere (31a).
Defects in trypanosome trans splicing can be detected by examining tubulin mRNAs, since splicing inhibition results in the appearance of a dicistronic transcript (see, for example, reference 32). Neither the depletion nor the upregulation of TbUBP1/2 had any effect on the pattern of tubulin transcripts (not shown). We also looked for inhibition of trans splicing by measuring the amounts of spliced leader Y-structure intermediates (see Materials and Methods). Again, no effect was seen (results not shown).
Overexpression of UBPs affects the levels of CFB1 mRNAs in bloodstream trypanosomes.
We next investigated, by using microarrays, whether the overexpression of TcUBP1 or TbUBP2 in T. brucei affected the abundances of any mRNAs.
The sequences of microarray spots that showed the greatest reproducible upregulation in response to TcUBP1 and TbUBP2 expression are listed in Table 2. This list is not exhaustive since we selected only the spots showing the strongest regulation for sequencing. Of the 19 different gene loci identified, 5 were represented by two or more array spots, and the regulation of two more was confirmed by Northern blotting (see Table 2 and see Fig. S11 in the supplemental material). Most fall into the class “conserved hypothetical,” which means that the open reading frame is present in each of the three available complete kinetoplastid genomes, and the sequence provides little if any functional clues. Notable among the targets were genes encoding proteins potentially interacting with DNA or RNA. Two products, one of which has an endo/excinuclease amino-terminal domain, might be involved in DNA repair, and there was also a predicted DNA topoisomerase. In addition, there was a putative eukaryotic translation initiation factor 2-alpha kinase, a protein with two zinc fingers, and a subunit of RNA polymerase II. Northern blot results (not shown) confirmed that three mRNAs containing destabilizing U-rich regions—EP, PGKB, and PPDK—were unaffected, but a slight decrease in the stable PGKC mRNA was apparent.
TABLE 2.
Identities of clones showing the greatest reproducible regulation after overexpression of TbUBP2
| Well | Fold regulationa
|
Gene annotation and datab | |
|---|---|---|---|
| TbUBP2 | TcUBP1 | ||
| 35G1 | 2.00 | 1.13 | Tb927.1.1500 hypothetical conserved, GPI signal; predicted mRNA is 11 kb with a 6-kb 3′-UTR which contains many poly(U) tracts exceeding 17/20 residues |
| 06F13 | 1.84 | 1.71 | Tb927.1.1500 (see 35G1) |
| 10F17 | 2.07 | 1.86 | Tb927.1.1500 (see 35G1) |
| 58D11 | 2.37 | 1.50 | Tb927.1.3450 hypothetical conserved, protein with PS50325; threonine-rich region profile (all three clone sequences are identical); the predicted size of the RNA is around 4.5 kb; a band of about 10 kb appears upon UBP1/2 overexpression; the RNA of the expected size is not regulated |
| 50I8 | 6.47 | 3.41 | Tb927.1.3450 (see 58D11) |
| 50I16 | 2.73 | 1.20 | Tb927.1.3450 (see 58D11) |
| 16L6 | 6.55 | 2.83 | Tb927.1.4560-Tb927.1.4650 cyclin F-box protein array; regulation confirmed, mediated by 3′-UTR (see the text) |
| 30L21 | 6.52 | 2.96 | Tb927.1.4560-Tb927.1.4650 (see 16L6) |
| 20P20 | 6.10 | 1.49 | Tb927.1.4560-Tb927.1.4650 (see 16L6) |
| 19F12 | 5.10 | 1.67 | Tb927.1.4560-Tb927.1.4650 (see 16L6) |
| 39F24 | 6.06 | 2.65 | Tb927.1.4560-Tb927.1.4650 (see 16L6) |
| 08A12 | 3.97 | 3.17 | Tb927.1.4560-Tb927.1.4650 (see 16L6) |
| 12D15 | 2.63 | 1.31 | Tb927.3.1220 conserved hypothetical endo/excinuclease amino-terminal domain … Tb927.3.1210 3′-UTR of protein transport protein Sec24C |
| 10J21 | 1.86 | 2.00 | Tb927.3.1220 (see 12D15) |
| 50I12 | 4.48 | 2.21 | Tb927.4.3920 hypothetical, 122 kDa, 12 transmembrane helices; predicted RNA of 4.5 kb, best poly(U) is 18/22; Northern blots indicated twofold regulation |
| 36G14 | 2.16 | −1.33 | Tb927.4.2500 putative eukaryotic translation initiation factor 2-alpha kinase … Tb927.4.24903; UTR conserved hypothetical |
| 28N6 | 2.30 | 1.11 | Tb927.4.1910 hypothetical conserved, noncoding repeats |
| 17K17 | 2.00 | 2.28 | Tb927.4.500 conserved hypothetical, forkhead domain |
| 22F9 | 2.38 | 1.09 | Tb927.4.1000 hypothetical conserved, Tb927.4.1010 hypothetical, Tb927.4.1020 serine-palmitoyl-coenzyme A transferase (putative) (very short overlap) |
| 50F22 | 3.26 | 2.22 | Tb927.7.3590 hypothetical conserved … Tb927.7.3600 hypothetical conserved |
| 06K14 | 2.24 | 1.97 | Tb09.211.3040 DNA-repair (putative) … Tb09.211.3050 hypothetical conserved |
| 06D20 | 2.11 | 1.41 | Tb10.6k15.2720 l-isoaspartate O-methyltransferase (putative) and 3′-UTR of hypothetical |
| 05M11 | 2.09 | 1.70 | Tb10.70.2020, conserved hypothetical, two Zn fingers; predicted RNA about 10 kb |
| 17G18 | 2.39 | 1.82 | Tb10.70.2020 (see 05M11) |
| 13O16 | 2.00 | 1.37 | Tb10.70.4080 hypothetical conserved |
| 19F16 | 5.06 | 2.08 | Tb11.01.0820 hypothetical conserved |
| 12D23 | 2.06 | 1.62 | Tb11.01.2580 hypothetical conserved |
| 21K4 | 2.33 | 1.53 | Tb11.01.3390 DNA topoisomerase II (putative) (predicted 4.9-kb RNA); Tb11.01.3380 histone acetyltransferase (putative) (very short overlap, predicted 2.5-kb mRNA); clone hybridizes with both RNAs, the larger one is upregulated by UBP1/2 overexpression; predicted 3′-UTR about 700 nt, several poly(U) tracts |
| 12F14 | 2.03 | 1.11 | Tb11.02.5190 pantothenate kinase subunit (putative) … Tb11.02.5180 DNA-directed RNA polymerase II subunit 9 (putative) (both clone sequences identical) |
| 14F1 | 2.05 | 1.73 | Tb11.02.5190 (see 12F14) |
| 31M3 | 5.63 | 2.61 | Tb11.03.069 hypothetical conserved |
See Materials and Methods. The results here were reproduced for three different biological replicates, each with dye-swap. The effect of overexpression of TcUBP1 is also shown. Annotations were performed as previously described (7); the identities of the open reading frames given correspond to the mRNAs that are predicted to hybridize with the clone.
GPI, glycosyl phosphatidylinositol anchor.
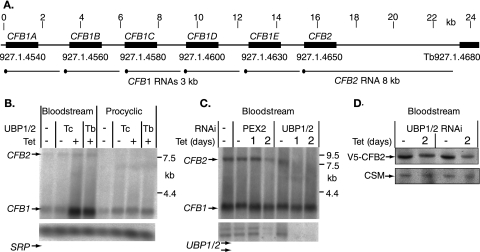
The strongest differential signals originated from a single locus on chromosome I, containing six open reading frames organized as a direct tandem repeat: Tb927.1.4540 (CFB1A), Tb927.1.4560 (CFB1B), Tb927.1.4580 (CFB1C), Tb927.1.4600 (CFB1D), Tb927.1.4630 (CFB1E), and Tb927.1.4650 (CFB2) (Fig. 5A). The CFB1 open reading frames are predicted to encode virtually identical 58-kDa proteins containing a cyclin F-box motif. CFB2 is predicted to encode a 52-kDa protein that is almost identical to the others in residues 111 to 307 of CFB1 (131 to 327 of CFB2) containing the F-box, but which has divergent N and C termini. There is evidence that CFB1 and CFB2 might be involved in cell cycle regulation (3), so we chose this locus for further study.
FIG. 5.
(A) Map of the region of chromosome 1 containing the CFB genes. Black blocks represent the coding regions, indicated by gene names above and locus numbers below. The scale above is in kilobases. The predicted mRNAs are indicated below the open reading frames, the spliced leader being indicated by a black dot. (B) Regulation of CFB mRNAs after overexpression of TbUBP2 or expression of TcUBP1. Northern blots of bloodstream or procyclic trypanosomes with tetracycline-inducible TcUBP1 or TbUBP2 were hybridized with DNA derived from positive array spots (see Table 1). Signal recognition particle (SRP) RNA served as a loading control. Cells with overexpression (+) were incubated with tetracycline for 24 h. (C) Effects of RNA interference downregulation of either PEX2 or TbUBP1/2. The cultured cells containing tetracycline-inducible constructs were incubated for either 24 h (1 day) or 48 h (2 days) with tetracycline. Note that after 2 days of PEX2 RNAi, the cells were dying and mRNA was disappearing. (D) Effect of TbUBP1/2 RNAi on the level of V5-tagged CFB2 in bloodstream cells. The loading control is a cytosolic marker protein. The results from two independent V5-tag clones are shown.
The polyadenylated CFB1 mRNAs are 3.0 to 3.2 kb long and are expressed at similar levels in both life cycle stages; the CFB2 mRNA is 8 kb and is very difficult to detect in procyclic forms (Fig. 5A) (3). Northern blot experiments confirmed that in bloodstream forms, CFB1 mRNA increased in abundance upon expression of TcUBP1 or overexpression of TbUBP2, while the CFB2 mRNA was unaffected (Fig. 5B). After the downregulation of TbUBP1/2 expression in bloodstream forms, the CFB1 mRNAs were unaffected, but the CFB2 mRNA disappeared (Fig. 5C). CFB1 mRNA was affected by TcUBP1 expression or TbUBP2 overexpression in procyclics; the effects, if any, on CFB2 mRNA in procyclics are unknown since we could not detect it.
To check that the effects of UBP1/2 expression on CFB mRNAs in bloodstream forms were not simply due to growth arrest, we examined the mRNAs after downregulation of the essential glycosomal membrane protein PEX2 (Fig. 5C) (28). In these growth-inhibited cells, no effects on CFB1 mRNAs were observed (the effects at 48 h are probably due to cell death since TbUBP RNAs decreased as well). To determine whether RNAi also affected protein levels, we used cells in which the CFB2 gene bore a V5 tag (3). Two days after TbUBP1/2 RNAi induction, there was a decrease in the abundance of the V5-tagged CFB2, as expected (Fig. 5D). We have thus far been unable to detect CFB1 protein by any method (3).
The CFB1 mRNAs contain a UBP1/2-responsive element in the 3′-untranslated region.
The intergenic regions downstream of the CFB1 genes show only minor (mostly single-nucleotide) differences, and the 280 bp upstream of the genes, which presumably supply trans splicing signals and the 5′-UTR, are similar for all copies. The CFB1 3′-UTR has many U-rich tracts, and several of them are predicted to be unstructured using the MFOLD program (not shown).
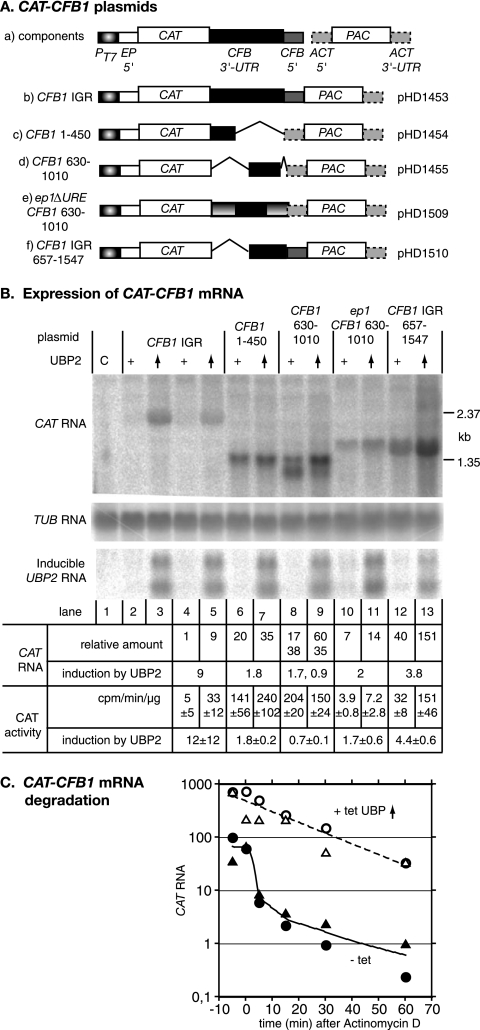
To find out whether the CFB1 3′-UTR was sufficient for UBP1/2-mediated regulation, we used a plasmid designed for expression of a CAT gene and the PAC selectable marker (30). The CFB1 intergenic region (CFB1-IGR) was cloned between the CAT and PAC open reading frames (Fig. 6). The plasmid is designed for integration into the tubulin gene array and should, after integration, be transcribed by RNA polymerase II. PAC mRNA produced from this plasmid (plasmid b in Fig. 6A) should be trans spliced using the CFB1 signal and polyadenylation of the CAT mRNA should occur in the normal position(s) for CFB1.
FIG. 6.
The CFB1B 3′-UTR affects mRNA levels. Various plasmids containing a CAT gene linked to a PAC selectable marker were transfected into bloodstream trypanosomes, and permanent cell lines were selected. The constructs are designed for integration into the tubulin locus and RNA polymerase II transcription. Panel A shows the relevant parts of the constructs, not drawn to scale (plasmid backbone not shown). (a) Key showing the two transgenes with surrounding intergenic regions: the CAT gene with a complete CFB intergenic region and the PAC gene with an actin 5′-UTR; (b) construct with the complete CFB intergenic region (IGR) separating the two open reading frames; (c) CFB 1-450, followed by the ACT 5′ splice site; (d) CFB 630-1010 followed by ACT 5′ splice site; (e) epΔURE 3′ UTR with inserted CFB 630-1010; (f) CFB intergenic region with deletion of the first 657 nt. Panel B shows a representative Northern blot hybridized with various probes. The sources of RNA are indicated above the lanes. Lane C is control bloodstream-form trypanosome RNA. For the remaining lanes, the nature of the intergenic region downstream of the CAT gene is indicated, corresponding to the plasmid maps below. The “+” indicates normal levels of TbUBP1/2 and the upward-oriented arrowhead indicates overexpression of TbUBP2. Expression of the CAT transgene is shown in the uppermost panel. Below this is a tubulin (TUB) loading control. The third blot panel shows expression of the TbUBP2 transgene mRNAs: the two transcripts are presumably produced by alternative processing. Beneath the blots are quantitations. The RNA quantitations are for this blot (others gave similar results), with an average for lanes 2 to 5. The CAT activities are the average for three to four independent experiments with standard deviations. (C) Effect of TbUBP2 overexpression on decay of CAT-CFB1 mRNA. Bloodstream trypanosomes expressing the CAT-CFB1 IGR mRNA and harboring the tetracycline-inducible TbUBP2 transgene were incubated for 24 h with or without tetracycline and then treated with Sinefungin (2 μg/ml) for 5 min before the addition of actinomycin D (10 μg/ml) at time zero. RNA was collected at the times indicated and the CAT mRNA was quantitated by real-time PCR, with SRP as a control. The results are for two experiments (with different symbols), expressed in arbitrary units relative to the SRP control. A third experiment gave similar degradation kinetics for cells with normal levels of TbUBP2, while for cells overexpressing TbUBP2 degradation only started after 30 min.
The CAT-CFB1-IGR-PAC plasmid was transfected into bloodstream trypanosomes with inducible TbUBP2 overexpression, and clones were selected. The cell lines retained the growth inhibition phenotype after tetracycline addition (not shown) and showed induction of the TbUBP2 transgene that was similar to that seen in the parent line (Fig. 6B). The results for two clones are shown in Fig. 6B (lanes 2 to 5). A single CAT mRNA of about 2.4 kb was expressed (lanes 2 and 4); the expected size was about 2.3 kb [2.1 + the poly(A) tail]. The abundance of this RNA increased 10-fold upon induction of TbUBP2 overexpression (lanes 3 and 5). The CAT activity increased 12-fold in parallel with mRNA, showing that the extra mRNA was translated. This result showed that the CFB1 intergenic region contained sufficient information to cause regulation of expression of a reporter gene by TbUBP2. In contrast and as expected, RNAi depletion of TbUBP1/2 had no effect on the CAT-CFB1-IGR mRNA or the resulting CAT activity (not shown).
To localize the sequences in the 3′-UTR required for TbUBP2-responsiveness, several deletions were designed in such a way that the predicted secondary structure might be retained. The simplest construct retained the intergenic region from nt 657 onward (CFB1 IGR 657-1647; Fig. 6A, plasmid f). The amount of CAT mRNA from this construct was 40 times higher than for the complete CFB1 3′-UTR, and the CAT activity was 11-fold higher (Fig. 6B, lane 12), suggesting either that the CFB1 1-657 region contains a negative element or that the deletion disrupts a negatively regulating secondary structure elsewhere in the 3′-UTR. Upon TbUBP2 overexpression, both mRNA and CAT activity from the CFB1 IGR 657-1647 construct increased fourfold (lane 13); thus, a UBP-responsive element might be present downstream of nt 657. Plasmid c was predicted to yield an mRNA containing 1 to 450 nt of the CFB1 3′-UTR. Once again, the amount of RNA and CAT activity was 20- to 40-fold that obtained with the whole IGR (Fig. 6B, lane 6). Ignoring potential secondary-structure effects, this would place the negative element between nt 450 and 657. Plasmid d was designed to give a CAT RNA containing nt 630 to 1010 of the CFB1 3′-UTR, but instead of a single transcript we detected two RNAs, only the longer of which had a size consistent with the predicted polyadenylation site (Fig. 6B, lane 8). The shorter RNA is probably polyadenylated about 200 nt upstream, but attempts to map the sites by RT-PCR have failed. Upon TbUBP2 induction the amount of the longer transcript increased at the expense of the shorter one (Fig. 6B, lane 9): this could have been due to effects on either RNA processing or stability. The simplest interpretation of the results thus far was that nt 451 to 656 contain a negative element and the TbUBP2-responsive region is downstream of nt 657.
We next investigated whether the CFB1 630-1010 region could function in another context. The EP1 3′-UTR contains a U-rich 26mer which destabilizes the RNA in bloodstream forms. We tested a construct in which the 26mer had been replaced by CFB1 630-1010 (Fig. 6A, plasmid e). A single RNA was obtained, but induction by TbUBP2 overexpression was only twofold (Fig. 6B, lanes 10 and 11). This indicated that the function of the 630-1010 region is context dependent.
Effects of TbUBP2 overexpression on CAT-CFB1 mRNA degradation.
Since the CAT mRNAs were predicted to be identical apart from the 3′ untranslated regions, the effects of TbUBP2 overexpression on CFB1 and CAT-CFB1 mRNA were likely to be caused either by improved mRNA processing or by inhibition of degradation. To find out the basis for the increase in CAT-CFB1 RNA after TbUBP2 overexpression, we therefore measured the half-life of the mRNA after inhibition of transcription using Sinefungin and actinomycin D (12). CAT-CFB1 RNA was quantitated by real-time PCR, using CAT primers. SRP served as the control. In three experiments the increase in CAT mRNA after TbUBP2 overexpression was estimated to be between 7.5- and 30-fold. The CAT-CFB1 mRNA was unstable in cells expressing normal amounts of TbUBP2, with extremely rapid degradation over the first 5 to 10 min and reduction below quantifiable limits after 15 min. In cells overexpressing TbUBP2, in contrast, the half-life was around 13 min (Fig. 6C). This confirmed that overexpression of TbUBP2 increases the half-life of CFB1 mRNA.
Association of TbUBP1 and TbUBP2 with proteins and RNA.
In T. cruzi, TcUBP1 is associated in vivo with TcUBP2 and with poly(A) binding protein (19). Also, both TcUBPs show some preference for binding to poly(U) sequences. In vitro binding studies may give very little insight into the situation within cells, where many different proteins with various affinities can compete for binding to the same RNA. We therefore investigated which proteins and RNAs were associated with the T. brucei proteins in vivo. We expressed C-terminally TAP-tagged TbUBP1 and TbUBP2 in bloodstream forms; both proteins were readily detected, and their expression did not affect the level of untagged TbUBP2 (Fig. 7A). The C-terminally tagged TbUBP2 was present as a major band and a smaller, minor band which reacted with both the anti-TcUBP1 antibody and antibody to the C-terminal part of the tag. The smaller band could, therefore, be a product of N-terminal proteolysis.
First, we subjected extracts from both cell lines to tandem affinity purification to find associated proteins. Only highly abundant proteins which frequently contaminate protein preparations were found (not shown). Western blots showed that the yields of TAP-tagged TbUBP1 or TbUBP2 were very poor (<10%), even from the first column after TEV cleavage; for both proteins, a single band was obtained (not two for TbUBP2). No copurification whatsoever of TbUBP1 with TbUBP2 (or vice versa) or with poly(A) binding protein was seen (Fig. 7A, lanes 11 to 13, and data not shown). Since the rather large C-terminal TAP tag might disrupt interactions, we also tried immunoprecipitation of the N-terminally V5-tagged proteins from either procyclic or bloodstream forms. This also failed to reveal specific binding partners, and again the untagged TbUBPs did not copurify with the N-terminally V5-tagged versions (data not shown and see Fig. S12 in the supplemental material). We therefore have no evidence that either TbUBP1 or TbUBP2 form homo- or heterodimers or make stoichiometric, stable complexes with other proteins. More transient or substoichiometric associations would not have been detected by this approach.
We next sought to determine whether CFB1 mRNA was specifically bound by TbUBP1 or TbUBP2 in vivo. The results for V5-UBP1 in procyclic forms suggested that TbUBP1 is an RNA-binding protein, but no reproducible specificity was apparent in procyclic trypanosomes for the RNAs tested (see Fig. S12 in the supplemental material). For bloodstream trypanosomes, we used IgG Sepharose to bind TAP-tagged TbUBP1 and TbUBP2 and released the complexes with TEV protease. We then compared the relative abundances of the CFB1 RNA in the bound fraction and the flowthrough fraction by RT-PCR. TUB and Tb11.01.6470 mRNAs, which are unaffected by TbUBP2 overexpression (Fig. 6 and array results) served as controls. Using extracts from cells expressing the TAP tag alone (Fig. 7D), ca. 1% of each RNA was eluted from the column. These values were reproducibly exceeded for extracts from cells expressing TbUBP1-TAP or TbUBP2-TAP, 3 to 5% of both control RNAs (TUB or Tb11.01.6470) usually being found in the eluates (Fig. 7B and C). Binding of CFB1 mRNA was consistently about five times higher than binding of the control mRNAs, at 12 to 45%. From these experiments, we concluded that TbUBP1 and TbUBP2 are able to bind RNAs with relatively little sequence specificity but that there might nevertheless be some preferential binding to the CFB1 mRNA. We cannot, however, rule out the possibility that some binding of TbUBP1/2-TAP to random RNAs could have occurred during cell lysis (43).
DISCUSSION
The results presented in this study indicate that the small RRM-motif proteins TbUBP1 and TbUBP2 are essential for normal trypanosome growth and that the precise levels of these proteins in the parasites are critical for their correct function. We found no evidence for developmental regulation of TbUBP1 or TbUBP2 mRNA expression, but TbUBP2 was present on Western blots as two bands, whose abundances varied between bloodstream and procyclic forms. Despite extensive investigations, the relationship between these two bands was not resolved. There were also indications that TbUBP1 might be posttranslationally modified in bloodstream forms, but since the effect was strain specific, its biological significance was questionable.
Overexpression of TbUBP2 changed the expression of several mRNAs encoding proteins of unknown function. The domains present in the proteins encoded by these potential target mRNAs suggest that several of them might be involved in growth control. We have to consider two possibilities: either the identified mRNAs are directly regulated by TbUBP2, or they are increased as a consequence of growth inhibition alone. We concentrated here on regulation of the CFB1 and CFB2 mRNAs, which encode proteins with a cyclin F-box. F-box proteins are components of SCF E3 ubiquitin ligase complexes; they act as bridging factors between unstable proteins and the ubiquitination machinery and are important in cell cycle control (53). The CFB1 mRNA was strongly upregulated after sixfold overexpression of TbUBP2, which inhibited cell growth, but was unaffected by threefold overexpression of C-terminally TAP-tagged or N-terminally myc-tagged TbUBP2, which did not inhibit cell growth. The fact that CFB1 mRNA also accumulated after depletion of the signal recognition particle (S. Michaeli and S. Brems, unpublished results) or of 14-3-3 protein (C. Benz and C. Clayton, unpublished data) suggests that this mRNA accumulates simply as a consequence of growth inhibition. CFB1 mRNA did not, however, accumulate under a variety of other growth-inhibitory conditions: RNAi against PEX2 (the present study) or against two other RNA-binding proteins (A. Estevez, Instituto de Parasitologia y Biomedicina “Lopez-Neyra,” Granada, Spain, unpublished data). This, combined with the fact that CFB1 mRNA was not affected by growth-inhibitory depletion of TbUBP1 and TbUBP2 (the present study) and that both TbUBPs showed some preferential binding to CFB1 mRNA suggests that overexpression of TbUBP2 affects CFB1 mRNA directly. The results of experiments using reporter constructs indicated that the increase in CFB1 mRNA after TbUBP2 overexpression was mediated via the 3′-UTR and that it was caused by stabilization.
Overexpression of TbUBP2—and depletion of both TbUBPs—may inhibit trypanosome growth because target RNAs are dysregulated. The CFB2 mRNA was not affected by TbUBP2 overexpression but depleted after TbUBP1/2 RNAi. We have recently found that overproduction of CFB1 inhibits growth and depletion of CFB2 prevents cytokinesis (3). Thus, these effects alone may suffice to explain the growth. Since, however, we observed dysregulation of several other transcripts, cumulative effects most likely contribute to the phenotype.
It was previously reported that overexpression of TcUBP1 in epimastigotes decreases the stability of a mucin mRNA, that TcUBP1 and TcUBP2 bind to both U-rich and G-rich oligonucleotides, and that they associate specifically with different mRNAs in vivo (13). When we purified (overexpressed) tagged TbUBP1 and TbUBP2 from bloodstream-form trypanosomes, we indeed found that 20 to 50% of CFB1 mRNA was pulled down together with the proteins. We also, however, found more limited association of all other mRNAs tested. This is not totally unexpected since TcUBP1 bound 3 of 10 fairly randomly chosen RNAs (the proportion bound was not measured), and TcUBP1 and TcUBP2 were even less specific in in vitro RNA binding. Investigations of HuR (= HuD) specificity in binding AU-rich elements indeed revealed that at least two RRMs were required for high-affinity binding (45); the first RRM interacts with just six nucleotides, and the second one interacts with two more (56).
Although we cannot be sure that TbUBP1 and TbUBP2 have low specificity in intact cells since some association with mRNAs could have occurred after cell lysis (44), general mRNA binding would be consistent with the protein abundance. Measurements of total RNA indicate that there are approximately 20,000 mRNAs, a million tRNAs and 125,000 rRNAs in a bloodstream trypanosome (J. Haanstra et al, unpublished data), while we found about 600,000 molecules of TbUBP2. This means that the molar TbUBP:mRNA ratio is 30:1. A specific role for TbUBP1 or TbUBP2 in regulating the abundance of a small number of mRNAs does not, therefore, seem very likely.
Recently, both UBP1 and UBP2 were found to be associated with poly(A)+ RNAs in stress granules after starvation of procyclic T. brucei or epimastigote T. cruzi (9). The two proteins did not, however, colocalize with either DHH1 or RRP6 in unstressed cells (the present study). This, together with increases in a small subset of mRNAs after overexpression, argues against roles in promoting mRNA degradation. Most TbUBP1 and TbUBP2 was not associated with polysomes; also, after TbUBP2 overexpression, the amount of CAT activity obtained in cells containing CAT-CFB1 mRNAs mirrored the relative amount of mRNA, implying that TbUBP2 neither suppresses nor enhances translation.
Both proteins sedimented in a very broad area of sucrose gradients and gave granular immunofluorescence staining. This suggests that in vivo they do associate with other macromolecules. Despite this, neither TbUBP1 nor TbUBP2 makes a stoichiometric, stable complex with other proteins, as judged by coimmunoprecipitation analyses or affinity purification. It is possible that TbUBP1 and TbUBP2 interact with many different proteins and that consequently no obvious bands appeared above background on one-dimensional gels; alternatively, there may be interactions that did not survive the wash steps.
Work in mammalian cells suggests that the final levels of mRNAs are determined by the relative amounts of several RNA-binding proteins which compete for target sequences (reviewed in reference 2; see also references 39 and 48). Thus, overexpression may enable UBP1 or UBP2 to compete with more specific mRNA-binding factors which either promote or inhibit degradation. Altogether, our results are consistent with a general role of UBP1 and UBP2 as relatively nonspecific, constitutive components of many RNPs, perhaps acting as “RNA chaperones.”
Supplementary Material
Acknowledgments
This study was funded by the Deutsche Forschungsgemeinshaft (C.E.C., C.B., S.B., and J.H.; SFB 544 project B13 and CL112/8), Wellcome Trust (M.C. and M.S.), the National Institutes of Health AI060645-01 (A.C.C.F.), and a Howard Hughes Medical Institute International Research Scholars Grant (A.C.C.F.).
We thank Stuart Archer (Zentrum für Molekulare Biologie der Universität Heidelberg) for the TAP RNP purification protocol and Antonio Estévez (Instituto de Parasitologia y Biomedicina “Lopez-Neyra,” CSIC, Granada, Spain) for communicating unpublished results. We also thank Shula Michaeli for help with the trans splicing assay.
Footnotes
Published ahead of print on 1 September 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alibu, V. P., L. Storm, S. Haile, C. Clayton, and D. Horn. 2004. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 139:75-82. [DOI] [PubMed] [Google Scholar]
- 2.Barreau, C., L. Paillard, and H. Osborne. 2006. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33:7138-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benz, C., and C. Clayton. The cyclin F box protein CFB2 is required for cytokinesis of bloodstream-form Trypanosoma brucei. Mol. Biochem. Parasitol., in press. [DOI] [PubMed]
- 4.Benz, C., D. Nilsson, B. Andersson, C. Clayton, and D. L. Guilbride. 2005. Messenger RNA processing sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 143:125-134. [DOI] [PubMed] [Google Scholar]
- 5.Berriman, M. E. A. 2005. The genome of the African trypanosome, Trypanosoma brucei. Science 309:416-422. [DOI] [PubMed] [Google Scholar]
- 6.Biebinger, S., L. E. Wirtz, and C. E. Clayton. 1997. Vectors for inducible overexpression of potentially toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 85:99-112. [DOI] [PubMed] [Google Scholar]
- 7.Brems, S., D. L. Guilbride, D. Gundlesdodjir-Planck, C. Busold, V. D. Luu, M. Schanne, J. Hoheisel, and C. Clayton. 2005. The transcriptomes of Trypanosoma brucei Lister 427 and TREU927 bloodstream and procyclic trypomastigotes. Mol. Biochem. Parasitol. 139:163-172. [DOI] [PubMed] [Google Scholar]
- 8.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell Mol. Life Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassola, A., J. De Gaudenzi, and A. Frasch. 2007. Recruitment of mRNAs to cytoplasmic ribonucleoprotein granules in trypanosomes. Mol. Microbiol. 65:655-670. [DOI] [PubMed] [Google Scholar]
- 10.Clayton, C. E. 2002. Developmental regulation without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colasante, C., V. P. Alibu, S. Kirchberger, J. Tjaden, C. Clayton, and F. Voncken. 2006. Characterisation and developmentally regulated localization of the mitochondrial carrier protein homologue MCP6 from Trypanosoma brucei. Eukaryot. Cell 5:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colasante, C., A. Robles, C.-H. Li, A. Schwede, C. Benz, F. Voncken, D. L. Guilbride, and C. Clayton. 2007. Regulated expression of glycosomal phosphoglycerate kinase in Trypanosoma brucei. Mol. Biochem. Parasitol. 151:193-204. [DOI] [PubMed] [Google Scholar]
- 13.De Gaudenzi, J. G., I. D'Orso, and A. C. C. Frasch. 2003. RNA recognition motif-type RNA-binding proteins in Trypanosoma cruzi form a family involved in the interaction with specific transcripts in vivo. J. Biol. Chem. 21:18884-18894. [DOI] [PubMed] [Google Scholar]
- 14.De Gaudenzi, J. G., A. C. C. Frasch, and C. Clayton. 2006. RNA-binding domain proteins in Kinetoplastids: a comparative analysis. Eukaryot. Cell 4:2106-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diehl, S., F. Diehl, N. M. El-Sayed, C. E. Clayton, and J. D. Hoheisel. 2002. Analysis of stage-specific gene expression in the bloodstream and the procyclic form of Trypanosoma brucei using a genomic DNA microarray. Mol. Biochem. Parasitol. 123:115-123. [DOI] [PubMed] [Google Scholar]
- 16.Di Noia, J. M., I. D'Orso, D. O. Sánchez, and A. C. C. Frasch. 2000. AU-rich elements in the 3′-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J. Biol. Chem. 275:10218-10227. [DOI] [PubMed] [Google Scholar]
- 17.Djikeng, A., C. Agufa, J. E. Donelson, and P. A. Majiwa. 1998. Generation of expressed sequence tags as physical landmarks in the genome of Trypanosoma brucei. Gene 221:93-106. [DOI] [PubMed] [Google Scholar]
- 18.D'Orso, I., and A. C. C. Frasch. 2001. TcUBP-1, a developmentally regulated U-rich RNA-binding protein involved in selective mRNA destabilization in trypanosomes. J. Biol. Chem. 276:34801-34809. [DOI] [PubMed] [Google Scholar]
- 19.D'Orso, I., and A. C. C. Frasch. 2002. TcUBP-1, an mRNA destabilizing factor from trypanosomes, homodimerizes and interacts with novel AU-rich element- and poly(A)-binding proteins forming a ribonucleoprotein complex. J. Biol. Chem. 277:50520-50528. [DOI] [PubMed] [Google Scholar]
- 20.Drozdz, M., L. Quijada, and C. E. Clayton. 2002. RNA interference in trypanosomes transfected with sense and antisense plasmids. Mol. Biochem. Parasitol. 121:149-152. [DOI] [PubMed] [Google Scholar]
- 21.Durand-Dubief, M., L. Kohl, and P. Bastin. 2003. Efficiency and specificity of RNA interference generated by intra- and intermolecular double-stranded RNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 129:11-21. [DOI] [PubMed] [Google Scholar]
- 22.Duttagupta, R., B. Tian, C. Wilusz, D. Khounh, P. Soteropoulos, M. Ouyang, J. Dougherty, and S. Peltz. 2005. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol. Cell. Biol. 25:5499-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Sayed, N. M., E. Ghedin, J. Song, A. MacLeod, F. Bringaud, C. Larkin, D. Wanless, J. Peterson, L. Hou, S. Taylor, A. Tweedie, N. Biteau, H. G. Khalak, X. Lin, T. Mason, L. Hannick, E. Caler, G. Blandin, D. Bartholomeu, A. J. Simpson, S. Kaul, H. Zhao, G. Pai, A. Van, S. T. Utterback, B. Haas, H. L. Koo, L. Umayam, B. Suh, C. Gerrard, V. Leech, R. Qi, S. Zhou, D. Schwartz, T. Feldblyum, S. Salzberg, A. Tait, C. M. Turner, E. Ullu, O. White, S. Melville, M. D. Adams, C. M. Fraser, and J. E. Donelson. 2003. The sequence and analysis of Trypanosoma brucei chromosome II. Nucleic Acids Res. 31:4856-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Estévez, A., T. Kempf, and C. E. Clayton. 2001. The exosome of Trypanosoma brucei. EMBO J. 20:3831-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Estévez, A. M., B. Lehner, C. M. Sanderson, T. Ruppert, and C. Clayton. 2003. The roles of inter-subunit interactions in exosome stability. J. Biol. Chem. 278:34943-34951. [DOI] [PubMed] [Google Scholar]
- 26.Fellenberg, K., N. C. Hauser, B. Brors, J. D. Hoheisel, and M. Vingron. 2002. Microarray data warehouse allowing for the statistical analysis of experiment annotations. Bioinformatics 18:423-433. [DOI] [PubMed] [Google Scholar]
- 27.Fellenberg, K., N. C. Hauser, B. Brors, A. Neutzner, J. D. Hoheisel, and M. Vingron. 2001. Correspondence analysis applied to microarray data. Proc. Natl. Acad. Sci. USA 98:10781-10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerra-Giraldez, C., L. Quijada, and C. E. Clayton. 2002. Compartmentation of enzymes in a microbody, the glycosome, is essential in Trypanosoma brucei. J. Cell Sci. 115:2651. [DOI] [PubMed] [Google Scholar]
- 29.Haile, S., M. Cristodero, C. Clayton, and A. Estévez. 2007. The subcellular localization of trypanosome RRP6 and its association with the exosome. Mol. Biochem. Parasitol. 151:52-58. [DOI] [PubMed] [Google Scholar]
- 30.Haile, S., A. M. Estévez, and C. Clayton. 2003. A role for the exosome in the initiation of degradation of unstable mRNAs. RNA 9:1491-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall, N., M. Berriman, N. J. Lennard, B. R. Harris, C. Hertz-Fowler, E. N. Bart-Delabesse, C. S. Gerrard, R. J. Atkin, A. J. Barron, S. Bowman, S. P. Bray-Allen, F. Bringaud, L. N. Clark, C. H. Corton, A. Cronin, R. Davies, J. Doggett, A. Fraser, E. Gruter, S. Hall, A. D. Harper, M. P. Kay, V. Leech, R. Mayes, C. Price, M. A. Quail, E. Rabbinowitsch, C. Reitter, K. Rutherford, J. Sasse, S. Sharp, R. Shownkeen, A. MacLeod, S. Taylor, A. Tweedie, C. M. Turner, A. Tait, K. Gull, B. Barrell, and S. E. Melville. 2003. The DNA sequence of chromosome I of an African trypanosome: gene content, chromosome organisation, recombination, and polymorphism. Nucleic Acids Res. 31:4864-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Hartmann, C., and C. Clayton. Regulation of a transmembrane protein gene family by the small RNA binding proteins TbUBP1 and TbUBP2. Mol. Biochem. Parasitol., in press. [DOI] [PubMed]
- 32.Hendriks, E. F., A. Abdul-Razak, and K. R. Matthews. 2003. TbCPSF30 depletion by RNA interference disrupts polycistronic RNA processing in Trypanosoma brucei. J. Biol. Chem. 278:26870-26878. [DOI] [PubMed] [Google Scholar]
- 33.Holetz, F., A. Correa, A. Avila, C. Nakamura, M. Krieger, and S. Goldenberg. 2007. Evidence of P-body-like structures in Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 356:1062-1067. [DOI] [PubMed] [Google Scholar]
- 34.Hotz, H.-R., C. Hartmann, K. Huober, M. Hug, and C. E. Clayton. 1997. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a trypanosome surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 25:3017-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jäger, A., J. De Gaudenzi, A. Cassola, I. D'Orso, and A. Frasch. 2007. mRNA maturation by two-step trans-splicing/polyadenylation processing in trypanosomes. Proc. Natl. Acad. Sci. USA 104:2035-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, A., A. Faldas, A. Foucher, E. Hunt, A. Tait, J. Wastling, and C. Turner. 2006. Visualisation and analysis of proteomic data from the procyclic form of Trypanosoma brucei. Proteomics 6:259-267. [DOI] [PubMed] [Google Scholar]
- 37.Liang, X., A. Haritan, S. Uliel, and S. Michaeli. 2003. Trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang, X.-H., Q. Liu, L. Liu, C. Tschudi, and S. Michaeli. 2006. Analysis of spliceosomal complexes in Trypanosoma brucei and silencing of two splicing factors Prp31 and Prp43. Mol. Biochem. Parasitol. 145:29-39. [DOI] [PubMed] [Google Scholar]
- 39.Liao, B., Y. Hu, and G. Brewer. 2007. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 14:511-519. [DOI] [PubMed] [Google Scholar]
- 40.Luu, V. D., S. Brems, J. Hoheisel, R. Burchmore, D. Guilbride, and C. Clayton. 2006. Functional analysis of Trypanosoma brucei PUF1. Mol. Biochem. Parasitol. 150:340-349. [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Calvillo, S., D. Nguyen, K. Stuart, and P. J. Myler. 2004. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot. Cell 3:506-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Calvillo, S., S. Yan, D. Nguyen, M. Fox, K. Stuart, and P. J. Myler. 2003. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell 11:1291-1299. [DOI] [PubMed] [Google Scholar]
- 43.Mili, S., and J. A. Steitz. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10:1692-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myler, P. J., L. Audleman, T. deVos, G. Hixson, P. Kiser, C. Lemley, C. Magness, E. Rickel, E. Sisk, S. Sunkin, S. Swartzell, T. Westlake, P. Bastien, G. Fu, A. Ivens, and K. Stuart. 1999. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl. Acad. Sci. USA 96:2902-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, S., D. G. Myszka, M. Yu, S. J. Littler, and I. A. Laird-Offringa. 2000. HuD RNA recognition motifs play distinct roles in the formation of a stable compex with AU-rich RNA. Mol. Cell. Biol. 20:4765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635-646. [DOI] [PubMed] [Google Scholar]
- 47.Quijada, L., C. Hartmann, C. Guerra-Giraldez, M. Drozdz, H. Irmer, and C. E. Clayton. 2002. Expression of the human RNA-binding protein HuR in Trypanosoma brucei induces differentiation-related changes in the abundance of developmentally regulated mRNAs. Nucleic Acids Res. 30:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkar, B., Q. Xi, C. He, and R. J. Schneider. 2003. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 23:6685-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segal, S., T. Dunckley, and R. Parker. 2006. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:5120-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen, S., G. K. Arhin, E. Ullu, and C. Tschudi. 2001. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113:171-173. [DOI] [PubMed] [Google Scholar]
- 52.Shi, H., A. Djikeng, T. Mark, E. Wirtz, C. Tschudi, and E. Ullu. 2000. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 6:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tyers, M., and P. Jorgensen. 2000. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev. 10:54-64. [DOI] [PubMed] [Google Scholar]
- 54.van Deursen, F. J., S. H. Shahi, T. C. M. R., C. Hartmann, C. Guerra-Giraldez, K. R. Matthews, and C. E. Clayton. 2001. Characterisation of the growth and differentiation in vivo and in vitro of bloodstream-form Trypanosoma brucei strain TREU 927. Mol. Biochem. Parasitol. 112:163-172. [DOI] [PubMed] [Google Scholar]
- 55.Volpon, L., I. D'Orso, C. R. Young, A. C. Frasch, and K. Gehring. 2005. NMR structural study of TcUBP1, a single RRM domain protein from Trypanosoma cruzi: contribution of a beta hairpin to RNA binding. Biochemistry 44:3708-3717. [DOI] [PubMed] [Google Scholar]
- 56.Wang, X., and T. M. Tanaka Hall. 2001. Structural basis for the recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 8:141-145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.