Abstract
The ade6-M26 meiotic recombination hot spot of fission yeast is defined by a cyclic AMP-responsive element (CRE)-like heptanucleotide sequence, 5′-ATGACGT-3′, which acts as a binding site for the Atf1/Pcr1 heterodimeric transcription factor required for hot spot activation. We previously demonstrated that the local chromatin around the M26 sequence motif alters to exhibit higher sensitivity to micrococcal nuclease before the initiation of meiotic recombination. In this study, we have examined whether or not such alterations in chromatin occur at natural meiotic DNA double-strand break (DSB) sites in Schizosaccharomyces pombe. At one of the most prominent DSB sites, mbs1 (meiotic break site 1), the chromatin structure has a constitutively accessible configuration at or near the DSB sites. The establishment of the open chromatin state and DSB formation are independent of the CRE-binding transcription factor, Atf1. Analysis of the chromatin configuration at CRE-dependent DSB sites revealed both differences from and similarities to mbs1. For example, the tdh1+ locus, which harbors a CRE consensus sequence near the DSB site, shows a meiotically induced open chromatin configuration, similar to ade6-M26. In contrast, the cds1+ locus is similar to mbs1 in that it exhibits a constitutive open configuration. Importantly, Atf1 is required for the open chromatin formation in both tdh1+ and cds1+. These results suggest that CRE-dependent meiotic chromatin changes are intrinsic processes related to DSB formation in fission yeast meiosis. In addition, the results suggest that the chromatin configuration in natural meiotic recombination hot spots can be classified into at least three distinct categories: (i) an Atf1-CRE-independent constitutively open chromatin configuration, (ii) an Atf1-CRE-dependent meiotically induced open chromatin configuration, and (iii) an Atf1-CRE-dependent constitutively open chromatin configuration.
Eukaryotic chromosomal DNA is compacted with histones and other proteins to form chromatin, which helps in efficient storage of genetic information. However, this prevents many DNA-associated processes, such as transcription, replication, repair, and recombination, from accessing the DNA template (38). Homologous recombination contributes to gaining genetic diversity in the next generation as well as proper segregation of homologous chromosomes in meiosis. Meiotic recombination is a highly regulated process, with some loci that are elevated (hot spots) or suppressed (cold spots) (15, 18, 22, 34).
In the distantly related budding and fission yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, meiotic recombination hot spots are closely associated with sites of DNA double-strand breaks (DSBs), which are introduced by Spo11 (or its ortholog in S. pombe, Rec12) and are required for initiation of recombination (7, 22, 28, 30). Elevated sensitivity of chromatin to micrococcal nuclease (MNase) is found at meiosis-specific hot spots in both budding and fission yeasts (17, 21, 39).
The ade6-M26 hot spot of fission yeast is the only reported eukaryotic hot spot whose essential nucleotide sequence, 5′-ATGACGT-3′, has been precisely defined. The M26 hot spot confers a meiosis-specific elevation of recombination of up to 20-fold compared with other ade6 alleles (e.g., ade6-M375) (8, 23, 25). The ade6-M26 allele is a single G/T transversion at the 5′ end of the ade6 coding region (23, 31). This mutation creates a nonsense codon and cyclic AMP-responsive element (CRE)-like heptanucleotide sequence. The heptamer acts as a binding site for the Atf1/Pcr1 (also called Mts1/Mts2 or Gad7/Pcr1) heterodimeric transcription factor, which is required for hot spot activation (14, 35). We have demonstrated that local chromatin with the M26 sequence motif becomes more sensitive to MNase in the early stage of meiosis, suggesting active chromatin remodeling around M26 (17). Furthermore, we have shown that Atf1 facilitates such chromatin remodeling (40).
Although the molecular basis of chromatin remodeling in ade6-M26 has been analyzed, it remained unclear whether a similar mechanism occurs at natural hot spots of recombination. Recently, it was demonstrated that natural meiotic DSB sites defined by CRE-like sequences are present in the S. pombe genome and that one of the prominent DSB sites, the cds1+ locus, is a meiotic recombination hot spot (29).
Moreover, Smith and colleagues analyzed natural DSBs in the S. pombe genome and identified the prominent meiotic DSB sites in chromosome I (1, 42). One such prominent DSB site, the mbs1 locus (meiotic break site 1), consists of clusters of DSBs and is a meiotic recombination hot spot (2). However, the correlation of DSB formation and the chromatin structure has not been fully elucidated. In this study, we analyzed the chromatin structure of natural meiotic DSB sites, demonstrating that meiotic DSBs are introduced around the regions where the chromatin configuration is either constitutively open or induced to become open during meiosis. These results demonstrate that the CRE-mediated chromatin remodeling coupled to DSB formation is one of the intrinsic and general properties of natural CRE-related hot spots.
MATERIALS AND METHODS
Fission yeast strains, culture methods and media.
The S. pombe strains used in this study are listed in Table 1. General genetic procedures were carried out as described previously (9). To induce meiosis using diploid S. pombe strains, cells were cultured in MM medium (12) to ∼1 × 107 cells/ml. Cells were harvested and washed with distilled H2O twice then transferred to MM medium lacking nitrogen (NH4Cl) to induce meiosis. For synchronous meiosis, a pat1-114 mutant strain was cultured in MM medium containing nitrogen at 25°C, transferred to MM medium lacking nitrogen at a density of 0.6 × 107 cells/ml, and cultured further for 20 h to arrest the cell cycle at the G1 phase. An equal volume of MM-NH4Cl (0.1%) medium was warmed at 37°C and added to the G1 phase-arrested cell culture. The culture temperature was then raised to 34°C to induce meiosis.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype |
|---|---|
| PKH50 | h−ade6-M26 rad50s pat1-114 ura4-D18 rec12-Flag:KanMX |
| PKH52 | h−ade6-M26 rad50s pat1-114 ura4-D18 rad32-Flag:KanMX |
| PKH118 | h+ ade6-M26 rec12::ura4 rad50s pat1-114 ura4-D18 leu1-32 |
| PKH138 | h+ ade6-M26 pat1-114 rad50s ura4-D18 |
| PKH160 | h+ ade6-M26 pcr1::his3 pat1-114 rad50s his3-D1 |
| PKH163 | h+ ade6-M26 aft1::ura4 pat1-114 rad50s ura4-D18 his3-D1 |
| PKH338 | h−ade6-M375 rad50s pat1-114 rec12-Flag:KanMX leu1-32 |
| PKH339 | h−ade6-M375 rad50s pat1-114 rad32-Flag:KanMX leu1-32 |
| D12 | h+/h−ade6-M26/ade6-M26 rec12::ura4+/rec12::ura4+ ura4-D18/ura4-D18 his5-303/+ leu1-32/+ |
| D20 | h+/h−ade6-M26/ade6-M26 his5-303/+ leu1-32/+ |
| D55 | h+/h−ade6-M26/ade6-M26 rec7::ura4+/rec7::ura4+ ura4-D18/ura4-D18 his5-303/+ leu1-32/+ |
| D66 | h+/h−ade6-M26/ade6-M26 mei4::ura4+/mei4::ura4+ ura4-D18/ura4-D18 his5-303/+ leu1-32/+ |
| D67 | h+/h−ade6-M26/ade6-M26 rec6::ura4+/rec6::ura4+ ura4-D18/ura4-D18 his5-303/+ leu1-32/+ |
| D68 | h+/h−ade6-M26/ade6-M26 rec8::ura4+/rec8::ura4+ ura4-D18/ura4-D18 his5-303/+ leu1-32/+ |
| D69 | h+/h−ade6-M26/ade6-M26 rec10::ura4+/rec10::ura4+ ura4-D18/ura4-D18 his5-303/+ leu1-32/+ |
| D70 | h+/h−ade6-M26/ade6-M26 rec15::ura4+/rec15::ura4+ ura4-D18/ura4-D18 his5-303/+ leu1-32/+ |
| WSP779 | h+/h ade6-M26/ade6-M26 atf1::ura4/atf1::ura4 his3-D1/his3-D1 leu1-32/leu1-32 ura4D18/ura4D18 |
For the construction of strains expressing proteins with epitope tags, we followed a standard integration method using the integration vector int4, which was derived from int2 (10) by replacing the green fluorescent protein open reading frame (ORF) with Flag. The diploid strains expressing fusion protein (Rec12-Flag or Rad32-FLAG) can normally form viable spores, indicating that the fusion proteins are functional.
Northern blot analysis.
Total RNA was prepared from S. pombe cells by a method described elsewhere (5). For the Northern blot analysis, 10 μg of total RNA was denatured with formamide, separated on 1.5% agarose gels containing formaldehyde (24), and blotted on a charged nylon membrane (BioDyne B membrane; Pall, NY). The probe to detect the cds1+ transcript was the same probe used for Fig. 7. The probe to detect the tdh1+ transcript was prepared from a PCR-amplified DNA fragment using a random-priming kit (GE Healthcare, Little Chalfont, United Kingdom). The DNA fragment was amplified from S. pombe genomic DNA by PCR using the primer set ACGGTTTCGGTCGTATTGGA and CATGAGACCCTCCTCGATAC.
FIG. 7.
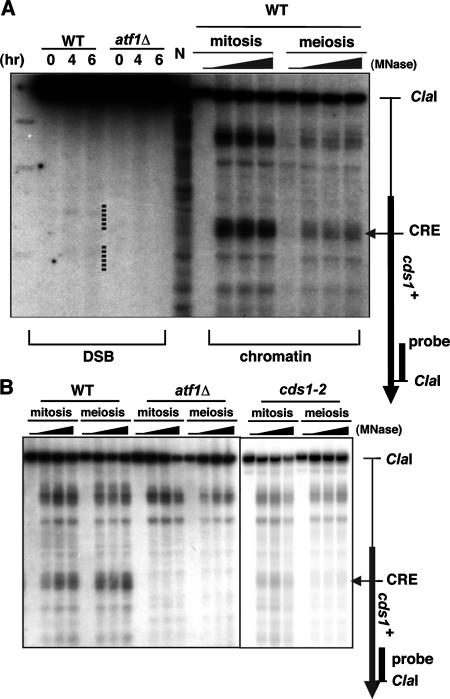
The chromatin structure at the CRE sequence in the cds1+ locus demonstrates a constitutively open state in an Atf1-dependent manner. (A) Meiotic DSB and chromatin structures were analyzed in the same gel. The ORF of cds1+ and its associated CRE sequence are indicated as in Fig. 2. Lane N, MNase-digested naked S. pombe genome DNA. Meiotic DSBs are indicated by dotted lines. (B) The chromatin structures in diploid strains D20 (wild type [WT]), WSP779 (atf1Δ), and D74 (cds1-2) were compared as for Fig. 1. The CRE sequence is indicated by an arrowhead.
Chromatin analysis.
Isolation of crude chromatin from cells and digestion with MNase were performed as described by Mizuno et al. (17) with slight modifications as described bellow. The samples of chromatin were prepared from a fixed amount of cells (0.5 g [wet weight]). These cells were suspended in preincubation buffer (0.7 M β-mercaptoethanol, 3 mM EDTA, 20 mM Tris-HCl [pH 8.0]), incubated at 30°C for 10 min, and washed once with 5 ml of ice-cold wash buffer (1.0 M sorbitol, 10 mM EDTA). Cells were centrifuged and resuspended in 2.5 ml of incubation buffer (0.75 M sorbitol, 37.5 mM Tris-HCl [pH 7.5], 1.25% glucose, 6.25 mM EDTA), and 2.5 ml of freshly prepared incubation buffer containing 2.5 mg/ml (or, for the atf1Δ strain, 1.25 mg/ml) of Zymolyase 100T (Seikagaku Cooperation, Japan) was added and mixed well. The cells were incubated with gentle agitation at 30°C for 5 min and washed once with 5 ml of ice-cold wash buffer. The resultant spheroplasts were suspended well by pipetting in 7 ml of lysis buffer (18% Ficoll 400, 10 mM KH2PO4, 10 mM K2HPO4, 1 mM MgCl2, 0.25 mM EGTA, 0.25 mM EDTA, 1 mM Pefabloc SC [Roche, Mannheim, Germany]). After centrifugation at 14,000 rpm for 30 min at 4°C, the crude nuclear pellet was resuspended in 4 ml of buffer A (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM KCl, 1 mM EDTA, 1 mM Pefabloc SC [Roche, Mannheim, Germany]). After addition of CaCl2 (5 mM final concentration), 1-milliliter aliquots of crude nuclear suspension were digested with different amounts of MNase (0, 10, 20, and 50 U/ml) at 37°C for 5 min. The reaction was terminated by adding 25 mM EDTA, and DNA was purified by incubation with 1% sodium dodecyl sulfate (SDS) and 20 μg of proteinase K (Merck, Darmstadt, Germany) at 55°C followed by phenol-chloroform extraction. The DNA samples were analyzed by Southern blotting as described below. To analyze chromatin around the tdh+, cds1+, and mbs1 loci, the MNase-treated DNA was digested with ApaLI/AflII, ClaI, and SpeI, respectively, and separated using agarose gel electrophoresis (40-cm-long gel) containing Tris-acetate-EDTA buffer. The separated DNA fragments were alkali transferred to charged nylon membranes (Biodyne B membrane; Pall, NY). The probe used for the indirect end labeling was prepared from PCR-amplified DNA fragments, and the DNA fragments were further labeled with 32P using a random-priming kit (GE Healthcare, Little Chalfont, United Kingdom). The DNA fragments were amplified from the S. pombe genome by PCR using the following primer sets: for tdh1+, CTAGCTAATCATCCCGATG and GAGATTACACAAGACTAC; for cds1+, GATGATAAAGTTGATATATGGAG and GATTCCCTCTTCTGAAATTTCG; and for mbs1, GAATACGCGACTTAACCGC and GACGATGTGGGAGGTGTG.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed according to the method of Yamada et al. (40) with slight modifications as described below. Fifty milliliters of culture was incubated with 1.4 ml of 37% formaldehyde solution for 20 min at room temperature, and then 2.5 ml of 2.5 M glycine was added and incubated for 5 min. After centrifugation, collected cells were washed twice with cold Tris-buffered saline (150 mM NaCl, 20 mM Tris HCl [pH 7.5]). The cells were mixed with 400 μl of lysis 140 buffer (0.1% Na-deoxycholate, 1 mM EDTA, 50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1% Triton X-100) supplemented with protease inhibitor cocktail (Complete Mini; Roche, Mannheim, Germany), and 0.6 ml of zirconia beads was added. After disruption of the cells using a multibead shocker (Yasuikikai, Osaka, Japan), the suspension was sonicated five times for 30 s each and centrifuged at 4°C, and the supernatant was collected as a whole-cell extract. The proper amount of antibody (anti-Flag M2; Sigma, St. Louis, MO), in accordance with the specifications provided by the manufacturer, and 40 μl of DYNA-protein G beads (DYNAL, Oslo, Norway) were mixed at 4°C overnight to conjugate antibody and beads, which were then washed twice with phosphate-buffered saline (138 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) containing 0.1% bovine serum albumin. Finally, 300 μl of whole-cell extract was mixed with the pretreated beads and allowed to immunoprecipitate at 4°C overnight. The precipitates were washed twice with lysis 140 buffer, once with lysis 500 buffer (0.1% Na-deoxycholate, 1 mM EDTA, 50 mM HEPES-KOH [pH 7.5], 500 mM NaCl, 1% Triton X-100) and further washed once with wash buffer (0.5% Na-deoxycholate, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 10 mM Tris-HCl [pH 8.0]), followed by one wash with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The well-washed precipitates were mixed with 150 μl of elution buffer (10 mM EDTA, 1% SDS, 50 mM Tris-HCl [pH 8.0]), and allowed to elute the immunoprecipitated protein-DNA complexes at 65°C for 15 min (IP sample). The IP sample or 3 μl of whole-cell extract was mixed with 250 μl or 397 μl of 1% SDS-containing TE buffer, 60 μg of proteinase K (Merck, Darmstadt, Germany) was added, and the mixture was incubated at 37°C for 8 h. The sample was then further incubated at 65°C overnight. The DNA present in the whole-cell extract and immunoprecipitates was purified and the DNA sample amplified by PCR using the following primer sets: for ade6-M26, CTCCGCACTAACTCACTAC and CGCTCATATTCGATGAAGTATG; for tdh1+, CGAAGCAACGACCATCTCGG and ATGCTAGGCACCGCCTGCCT; and for mbs1, TACGCCTCAGCGCGGAGACT and GTCAATCGCATCTACGCCTG.
The PCR products were then separated by electrophoresis, and the images were stored digitally. The amount of DNA was quantified using a Fast real-time PCR system 7300 (Applied Biosystems, Foster City, CA) and SYBER premix EX Taq (Takara, Japan). The IP efficiency (percent) was calculated as IP value/1% input value.
Detection of DSBs.
DNA samples were prepared in agarose plugs from cells of a synchronous culture, as described by Ogino et al. (20). The plugs were thoroughly equilibrated with appropriate restriction enzyme buffer and then heated to 65°C to melt the agarose. To detect DSBs in the tdh1+ locus, samples were held at 37°C, digested with BamHI and AflII, and separated by electrophoresis in a 1% agarose gel (40 cm long). The probe used for indirect end labeling was amplified from the S. pombe genome by PCR using the primer set AGCGGAGCCACGTTAC and CAATCGAGTTGGTTCATGG. When DSBs and chromatin structure were analyzed in the same gel, the restriction enzymes and probes were the same as those used in the chromatin assay described above.
RESULTS
CRE-independent natural DSBs form around a constitutively open chromatin region in mbs1.
To examine the relationship between meiotic DSB formation and chromatin structure at natural meiotic recombination hot spots, we first analyzed the chromatin structure at the natural meiotic recombination hot spot, mbs1. In this analysis, we employed an indirect end-labeling analysis using partial digestion of chromatin with MNase. Because MNase preferentially digests DNA that is not wrapped on nucleosomes such as linker DNA, chromatin remodeling is detected as changes in the MNase cleavage pattern.
Because DSBs in wild-type cells are repaired promptly, they are hard to detect (1). Therefore, we used a pat1-114 rad50s mutant strain, which allowed us to detect DSBs efficiently. In the rad50s mutant strains, DSBs are not repaired, and hence they accumulate (42). The pat1-114 allele encodes a temperature-sensitive form of Pat1 kinase, which normally serves to inhibit meiosis. Thus, when pat1-114 strains are raised to the nonpermissive temperature, cells in culture undergo a synchronous meiosis (11, 16). A pat1-114-induced meiosis is generally similar to diploid wild-type meiosis in terms of meiotic recombination frequency when it is induced from G1 phase (36). The pat1-114 haploid cells can also undergo meiosis without affecting the timing and efficiency of meiotic DSB formation (1, 2, 6, 20, 26, 28, 29, 41, 42), although the resulting spores have low viability due to chromosome missegregation (19). Thus, synchronous meiosis using pat1-114 is a widely used technique for analyzing DSBs in S. pombe (1, 2, 6, 20, 26, 28, 29, 41, 42).
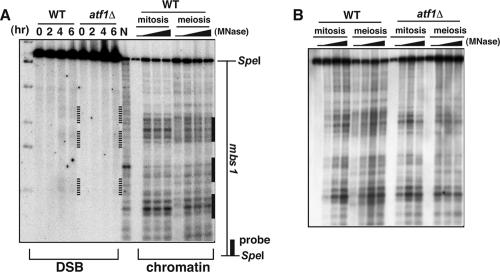
To investigate the chromatin structure around DSB sites precisely, we analyzed DNA from chromatin that had been partially digested with MNase and mapped the MNase-sensitive sites. We then compared the MNase-sensitive sites with the DSB sites observed in the pat1-114 rad50s strain in the same gel. At the mbs1 locus, at least three DSBs and some MNase-sensitive sites around the DSB sites were detected (Fig. 1A). The positions of the MNase-sensitive sites were unchanged in meiosis, although slight alterations in MNase sensitivity were detected at a few sites. These results suggest that DSBs are preferentially introduced around open chromatin regions at mbs1. The DSBs at the mbs1 locus were detected even in the atf1Δ strain, but the formation of DSBs was slightly delayed and the intensity was partially reduced (wild type [6 h], 1.45% DSBs/total lane; atf1Δ [6 h], 0.82% DSBs/total lane [∼44% reduction]). This probably reflects a partial defect in meiotic progression in atf1Δ strains (13, 27, 33, 37). These results suggest that the mbs1 DSB sites are not dependent upon Atf1 (Fig. 1A).
FIG. 1.
The chromatin structure around the DSB sites at the mbs1 locus shows a relatively open state in mitosis and meiosis. (A) Diploid strain D20 (wild type [WT]) was cultured in MM-NH4Cl medium (mitosis lanes). Cells were then transferred to MM medium lacking nitrogen and cultured further for 4 h (meiosis lanes). Chromatin isolated from the cells was digested with MNase and analyzed as described previously (17). To analyze the meiotic DSBs, haploid pat1-114 rad50s strains PKH138 (wild type) and PKH163 (atf1Δ) were cultured, and DNA was prepared as described in Materials and Methods. DNA samples from MNase-digested chromatins and synchronous meioses were digested with SpeI and analyzed in the same gel. Lane N, MNase-digested naked S. pombe genome DNA. Meiotic DSBs are indicated by dotted lines. Thick lines indicate MNase-sensitive regions. (B) MNase-digested chromatin DNA from diploid strains D20 (wild type) and WSP779 (atf1Δ) were analyzed in same gel to compare the chromatin structure.
To examine the atf1Δ effects on the chromatin structure around mbs1, we next examined the pattern of MNase-sensitive sites around mbs1 in the atf1Δ strain. As shown in Fig. 1B, the banding patterns of MNase-sensitive sites in the atf1Δ strain were similar to those in the wild type, except for slight alterations of band positions located apart from the mbs1 DSB site. This indicates that Atf1 is dispensable for the formation of an accessible chromatin configuration around DSB sites at the mbs1 locus (Fig. 1B). This notion is consistent with Atf1-independent DSB formation at mbs1.
Meiotic chromatin remodeling and Rec12-dependent DSBs are induced in the tdh1+ locus.
To test the generality of the meiotic chromatin remodeling observed at ade6-M26, we examined whether chromatin remodeling occurs around a natural CRE-related sequence in meiosis. We observed the chromatin structure at several loci containing CRE sequences (Table 2). Among them, drastic chromatin remodeling was observed at both the tdh1+ and cgs2+ loci in meiosis (3) (Fig. 2 and data not shown). As shown in Fig. 2, chromatin remodeling at tdh1+ was not observed in the atf1Δ mutant, indicating that the chromatin remodeling in tdh1+ requires Atf1, as in ade6-M26. Low levels of DNA breakages detected in meiotic samples without MNase treatment are probably due to degradation by endogenous nuclease activity in meiosis, since these breaks were independent of Rec12 around both ade6-M26 and tdh1+ (Fig. 3). It should be noted that meiotic chromatin remodeling was still observed in DSB-defective mutants such as the rec12Δ mutant, indicating that meiotic chromatin remodeling precedes, and hence may be prerequisite for, meiotic DSB formation.
TABLE 2.
Meiotic chromatin remodeling in the natural S. pombe genome
| Sitea | Enzyme used for DNA digestion | Chromatin-remodeling region:
|
Constitutive open configuration <0.5 kbp from CRE sequence | |
|---|---|---|---|---|
| <0.5 kbp from CRE sequence | >0.5 kbp from CRE sequence | |||
| tdh1(−334) | ApaLI/AflII | + | ||
| cgs2(−72) | ClaI/PstI | + | ||
| pyp2(−1147) | EcoRV/PstI | + | ||
| cds1(+530) | ClaI | + | ||
| cgs1(−734) | SphI | + | ||
| ptc1(−190) | SphI | + | ||
| fbp1(−2737) | EcoT22I | + | ||
The position of the CRE sequence from the first ATG is indicated in parentheses.
FIG. 2.
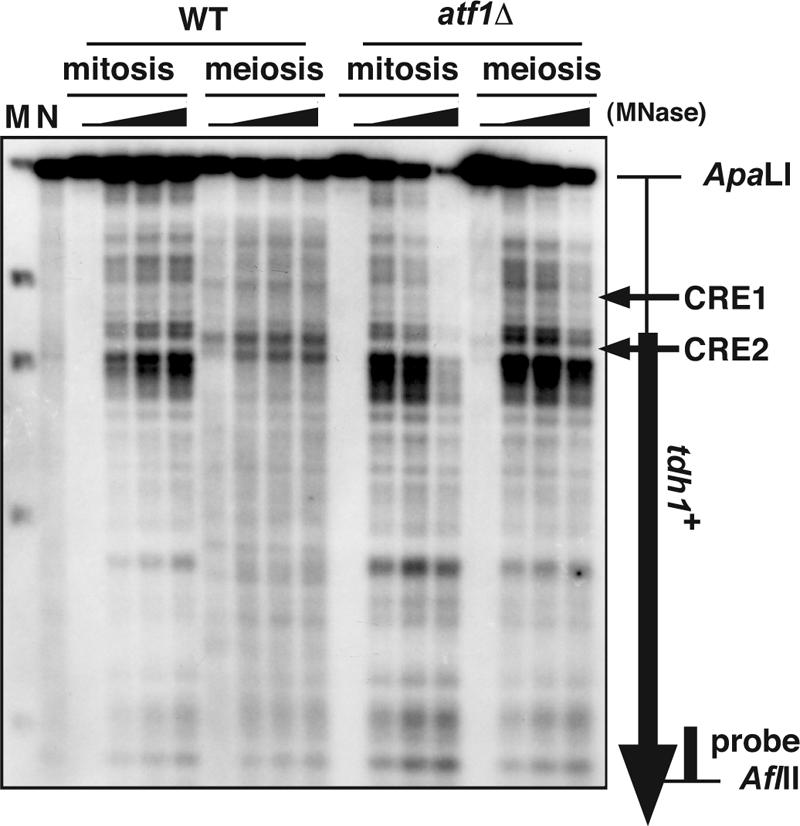
Meiotic chromatin remodeling in the tdh1+ locus depends on Atf1. MNase-digested chromatin DNAs from diploid strains D20 (wild type [WT]) and WSP779 (atf1Δ) were analyzed as in Fig. 1. The atf1Δ samples were slightly overdigested, since the atf1Δ mutant is more sensitive to Zymolyase treatment that allows increased permeation of MNase. The vertical and the horizontal arrows indicate the tdh1+ ORF and the position of the CRE sequences, respectively. Lane M, marker λ, EcoT14I digested (Takara); lane N, MNase-digested naked S. pombe genome DNA.
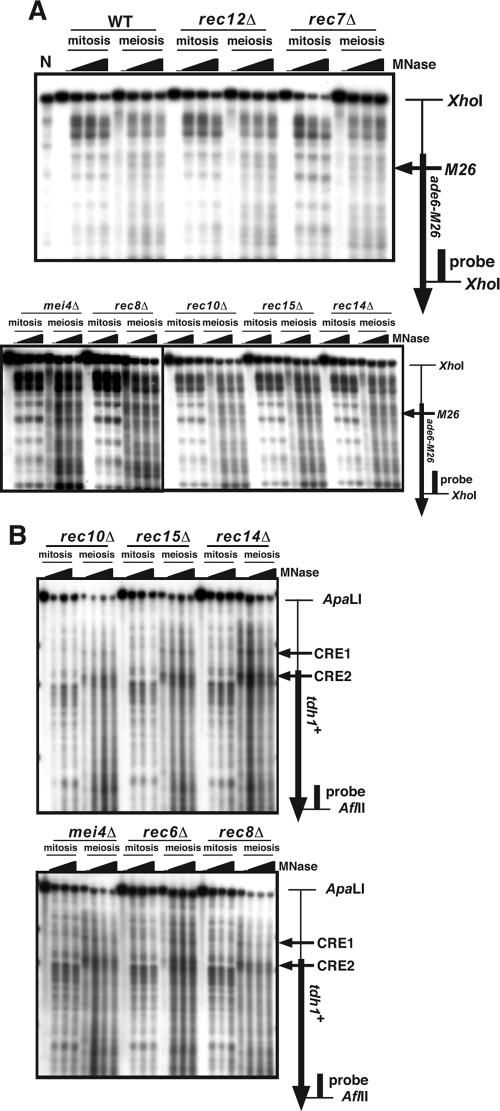
FIG. 3.
The chromatin structure around ade6-M26 and tdh1+ in mitosis and meiosis in DSB-defective mutants. Meiotic DSB formation is critically impaired in rec6Δ, rec7Δ, rec8Δ, rec10Δ, rec12Δ, rec14Δ, rec15Δ, and mei4Δ mutants (1, 4, 6, 41). We examined the chromatin structure around ade6-M26 (A) and tdh1+ (B) in these mutants. The M26 diploid strains D20 (wild type [WT]), D55 (rec7Δ), D68 (rec8Δ), D69 (rec10Δ), D12 (rec12Δ), D70 (rec15Δ), and D66 (mei4Δ) were cultured as in Fig. 1. Isolation of the chromatin fraction and treatment with MNase were done as described in Materials and Methods. Southern blot analysis was performed according to the method of Mizuno et al. (17). Lane N, partial digestion of naked DNA with MNase.
We previously reported that the Atf1-Pcr1-M26 complex links stress-activated mitogen-activated protein kinase and protein kinase A signaling pathways via chromatin remodeling in the cgs2+ promoter (3). In that case, we suggested that the chromatin remodeling facilitates transcriptional activation of cgs2+ (3). On the other hand, the transcription of tdh1+ was not induced at all in meiosis (Fig. 4). This result led us to speculate that the chromatin remodeling in tdh1+ is dedicated to the regulation of meiotic DSB formation. To test this notion, we next investigated DSB formation at tdh1+. Rec12-dependent DSB formation was detected at one of the CRE sequences (CRE2) in the tdh1+ locus at 4 h after the meiotic induction (Fig. 5A, upper panel). This meiotic DSB was not detected in the atf1Δ or pcr1Δ mutants (Fig. 5A, middle panel), while the genome-wide DSB frequency was not severely affected in those mutants (Fig. 5A, lower panel). These results indicate that meiotic DSB formation at the tdh1+ locus is regulated by the Atf1-Pcr1 complex, as observed at ade6-M26.
FIG. 4.
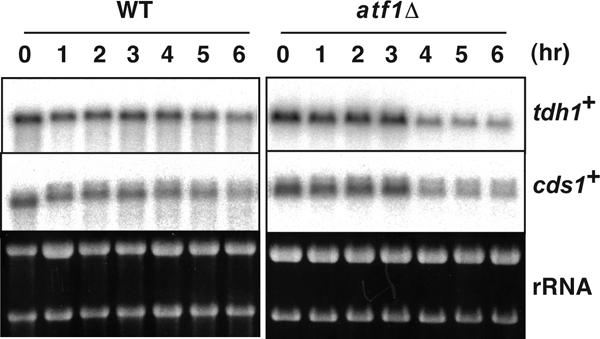
The transcription of tdh1+ and cds1+ is not dependent on Atf1 and is not induced during meiosis. The diploid strains D20 (wild type [WT]) and WSP779 (atf1Δ) were cultured as in Fig. 1. The cells were harvested at the indicated time points after the medium shift. Preparation of total RNA and Northern blot analysis were performed as described in Materials and Methods. rRNA was detected by ethidium bromide staining as a loading control.
FIG. 5.
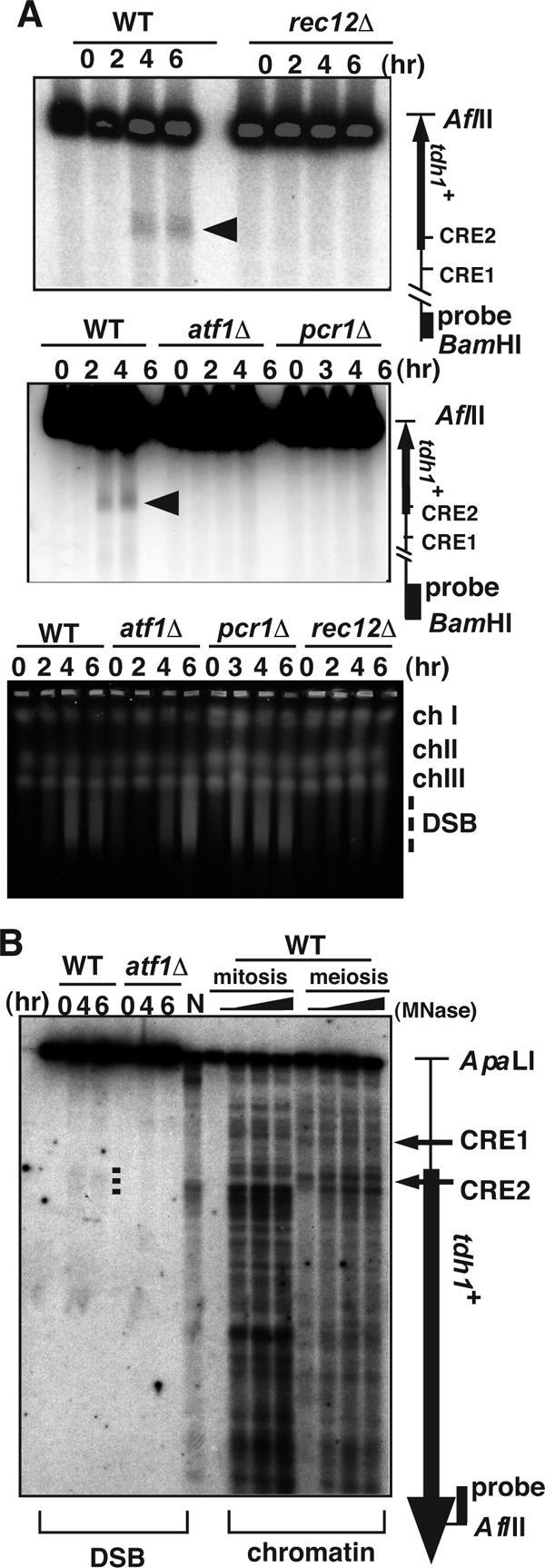
Formation of meiotic DSBs in the tdh1+ locus depends on Rec12 and the Atf1-Pcr1 complex. (A) Haploid pat1-114 rad50s strains PKH138 (wild type [WT]), PKH118 (rec12Δ), PKH163 (atf1Δ), and PKH160 (pcr1Δ) were cultured, and DNA was prepared as described in Materials and Methods. tdh1+ ORF and CRE sequences are indicated as in Fig. 2. An arrowhead indicates the DSB site. The genome-wide DSBs in the same samples were analyzed by pulsed-field gel electrophoresis. (B) DNA samples from the MNase-digested chromatins and synchronous meioses in Fig. 1 were digested with ApaLI and AflII and analyzed in the same gel. DSBs are indicated by a dotted line; tdh1+ ORF and CRE sequences are indicated as in Fig. 2.
To compare precisely the positions of the MNase-sensitive sites and DSBs, we examined them side by side on the same gel. As shown in Fig. 5B, the observed MNase-sensitive sites were located slightly downstream of the tdh1+ DSB site in mitosis. In meiosis new MNase-sensitive sites appeared around the DSB site (Fig. 5B), and these new sites are dependent on Atf1 (Fig. 2). This result is similar to the Atf1-dependent nucleosome repositioning observed at ade6-M26.
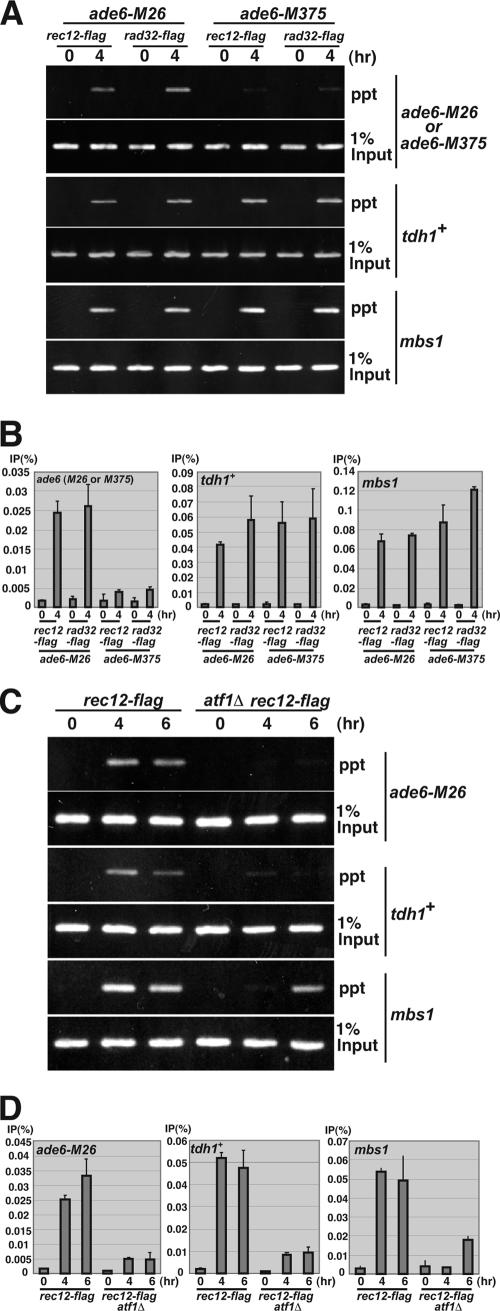
The similarity between tdh1+ and ade6-M26 suggests that the CRE-dependent meiotic remodeling of chromatin may be a prerequisite to the loading of Rec12 and other DSB-initiating proteins onto DNA at those sites. To test this idea, we examined the binding of Rec12 protein and Rad32 (the ortholog of Mre11 in S. pombe) in vivo by ChIP. Both Rec12-Flag and Rad32-Flag proteins bind meiotically to ade6-M26 but not to ade6-M375, which has an identical nonsense mutation at the codon adjacent to that carrying M26 (Fig. 6A and B). We also analyzed Rec12-Flag and Rad32-Flag binding to the tdh1+ and mbs1 DSB sites, as natural CRE-dependent and -independent DSB hot spots, respectively. Both Rec12-Flag and Rad32-Flag proteins bind to tdh1+ and mbs1 during meiosis (Fig. 6A and B).
FIG. 6.
Rec12 and Rad32 proteins bind to meiotic DSB sites ade6-M26, tdh1+ (CRE-mediated hot spots), and mbs1 (CRE-independent hot spot) in meiosis. (A) The binding of Rec12-Flag and Rad32-Flag proteins to the ade6 locus carrying the M26 or M375 allele was examined. The haploid pat1-114 rad50s strains PKH50 (rec12-flag ade6-M26), PKH52 (rad32-flag ade6-M26), PKH338 (rec12-flag ade6-M375), and PKH339 (rad32-flag ade6-M375) were cultured to induce meiosis (0 and 4 h) and fixed with formaldehyde. ChIP analysis was performed as described in Materials and Methods. The binding of Rec12-Flag and Rad32-Flag to the ade6, tdh1+, and mbs1 loci was detected using PCR. Whole genomic DNA from the 1% input sample was amplified at the same time. (B) ChIP efficiency in the M26 and M375 alleles was quantified by real-time PCR analysis as described in Materials and Methods. ChIP efficiency was calculated as IP sample/input material and represented as IP (percent) Error bars represent standard deviations. (C) The binding of Rec12-Flag protein to the ade6, tdh1+, and mbs1 loci was examined in wild-type and atf1Δ strains. The haploid ade6-M26 pat1-114 rad50s rec12-flag strains PKH50 (wild type) and PKH114 (atf1Δ) were cultured to induce meiosis (0, 4, and 6 h) and fixed with formaldehyde. The binding of Rec12-Flag to ade6, tdh1+, and mbs1 loci was detected as for panel A. (D) Quantification of ChIP efficiency from panel C. Error bars represent standard deviations.
We further examined the binding of Rec12-Flag to those DSB sites in the atf1Δ mutant, in which cellular CRE-mediated chromatin remodeling may be impaired. The binding of Rec12-Flag to ade6-M26 and tdh1+ was reduced five- to sevenfold relative to that in the atf1+ strain (Fig. 6C and D). On the other hand, the binding of Rec12-Flag to mbs1 was reduced only ∼2.7-fold. Notably, the Rec12-Flag binding to mbs1 increased 5.6-fold between 4 and 6 h of meiosis. Thus, the Rec12-Flag binding at mbs1 is partly reduced and delayed in the atf1Δ mutant, but the levels are still higher than those in the Atf1-dependent hot spots, ade6-M26 and tdh1+ (Fig. 6C and D). It is likely that the reduced and delayed DSB formation at mbs1 in the atf1Δ mutant is due to its impaired ability to proceed through meiosis. From these data, we conclude that Atf1-CRE-mediated chromatin alteration is a prerequisite for the binding of DSB-initiating proteins to DNA at ade6-M26 and tdh1+ but not at mbs1.
Chromatin structure around the cds1+ locus exhibits an Atf1-dependent constitutively open configuration.
In an independent study, it was demonstrated that natural meiotic DSB sites occur at many CRE-like sequences present naturally in the S. pombe genome (29). One of the prominent DSB sites, the cds1+ locus, was demonstrated to be an Atf1-Pcr1-dependent meiotic recombination hot spot (29). We analyzed the chromatin structure around the cds1+ locus and compared it to the observed DSB site (Fig. 7A). Surprisingly, MNase sensitivity was high at the CRE sequence, and the MNase cleavage patterns appeared to be virtually identical in mitosis and meiosis. In addition, the meiosis-specific DSBs observed at cds1+ occur adjacent to a site that is constitutively accessible in mitosis and meiosis.
Because DSB formation is dependent on the Atf1-Pcr1 complex in cds1+ (29) (Fig. 7A), we also analyzed chromatin structure in the atf1Δ mutant. Interestingly, the MNase cleavage sites around the DSB sites were abolished in both mitotic and meiotic chromatin by the atf1Δ mutation, indicating that formation of the open chromatin configuration in cds1+ depends on the Atf1 protein (Fig. 7B). In addition, we monitored the transcription of cds1+ during meiosis in the wild-type and atf1Δ strains. The expression of cds1+ was not induced during meiosis irrespective of presence or absence of Atf1, indicating that Atf1 bound on that site does not participate in the regulation of cds1+ transcription (Fig. 4). We also analyzed the chromatin structure at cds1-2, a point mutation that abolishes the CRE sequence in the cds1 gene (29). The chromatin structure around the cds1-2 mutation showed reduced accessibility compared to cds1+ in mitosis, and the accessibility was further reduced in meiosis (Fig. 7). Thus, it is likely that the role of Atf1 at this locus is to maintain a constitutively open chromatin configuration during mitosis and meiosis, rather than to induce an alteration in chromatin configuration as observed in ade6-M26 (29).
DISCUSSION
This study provides the first evidence that chromatin remodeling occurs in meiosis at natural meiotic DSB sites in S. pombe. In addition, we demonstrated that CRE-dependent DSB sites also show a CRE-dependent (or Atf1-Pcr1-dependent) open chromatin configuration. The open chromatin configuration at these sites may be constitutive (cds1+) or meiotically induced (tdh1+).
The regulation pattern of the chromatin configuration in natural meiotic recombination hot spots can be classified into at least three types.
We analyzed meiotic DSB formation at three distinct chromosomal loci and found that DSBs at those loci occur around regions with an open chromatin configuration. The chromatin structure around the DSB sites of mbs1 is accessible and changes little from mitosis to meiosis (Fig. 1). Chromatin around the CRE2 site of tdh1+ also shows an open chromatin configuration in mitosis, but significant Atf1-dependent chromatin remodeling is observed there during meiosis (Fig. 2). Chromatin at the CRE site of cds1+ has a constitutive open configuration, but unlike mbs1, this open chromatin configuration is dependent on Atf1 (Fig. 7). These results indicate that there are at least three types of regulation of chromatin configuration around meiotic DSB sites: (i) an Atf1-CRE-independent constitutively open chromatin configuration (mbs1), (ii) an Atf1-CRE-mediated meiotic chromatin alteration to a more accessible configuration (tdh1+), and (iii) an Atf1-CRE-dependent constitutively accessible chromatin configuration (cds1+). At those sites tested in S. cerevisiae, meiotic DSBs are formed around sites with a constitutively open chromatin configuration, such as the well-characterized DSB sites ARG4 and CYC3 (21). On the other hand, the results in this study suggest that the situation in S. pombe is more complex; e.g., meiotic DSB sites show distinct modes of chromatin configuration before meiotic DSB formation.
General roles of ATF1-CRE dependent chromatin alteration in meiotic recombination in S. pombe.
The S. pombe genome has number of CRE-related Atf1-dependent meiotic DSB sites (29). In this study, we report that tdh1+ is also an Atf1-CRE-dependent DSB site (Fig. 5). We further demonstrate that Atf1-dependent chromatin remodeling (i.e., changes in MNase-sensitive sites) occurs during meiosis around the DSB site in the tdh1+ locus (Fig. 2 and 5). In addition, the atf1Δ mutation severely affects meiotic chromatin remodeling in tdh1+, DSB formation, and the binding of DSB initiating proteins in tdh1+ (Fig. 2, 5, and 6), Thus, the regulation of chromatin structure and recombination at tdh1+ is very similar to the artificial CRE-dependent hot spot ade6-M26 (17).
The DSB site found in the cds1+ locus is another CRE-Atf1-dependent recombination hot spot (29). However, unlike ade6-M26 and tdh1+, the cds1+ locus shows an Atf1-dependent constitutively open configuration at the CRE site in both mitosis and meiosis (Fig. 7). Interestingly, mutation of the CRE sequence (cds1-2) results in a significant reduction in MNase sensitivity at that site in mitosis and an even greater reduction in meiosis (Fig. 7B). In meiosis, the chromatin configuration around the CRE consensus sequence in cds1-2 is very similar to that of the cds1+ atf1Δ mutant in meiosis. It is unclear why the cds1-2 mutant shows a partially open configuration in mitotic cells, but a residual binding of Atf1 to the mutated CRE sequence may be sufficient to form the partially open chromatin configuration in mitotic cells but not in meiotic cells. Alternatively, another CRE-related site, ∼350 bp upstream of the cds1-2 CRE, may contribute to the open chromatin configuration in mitosis but not in meiosis.
Formation of accessible chromatin is pivotal for the binding of DSB-initiating proteins to DSB sites in meiosis.
We demonstrated that Rec12 and Rad32 (the orthologs of Spo11 and Mre11 in S. pombe, respectively) bind to the ade6-M26 DSB site in meiosis (Fig. 6). Importantly, such binding of Rec12 and Rad32 to the ade6 locus is not observed in ade6-M375, a negative control allele. Moreover, we showed that Atf1 is required for the binding of Rec12 to ade6-M26 and tdh1+ (Fig. 6C and D). Since the alteration of chromatin structure is dependent on the CRE-Atf1 complex (15, 37), these results suggest that the M26 meiotic chromatin alteration may be pivotal for the binding of DSB-initiating proteins to DSB sites. An alternative but nonexclusive possibility is that Atf1 directly recruits Rec12 together with other DSB-initiating proteins to CRE-related sites, and then the chromatin alteration occurs as a consequence of the binding of those proteins. However, this notion seems unlikely, since the binding of DSB-initiating proteins to DSB sites is severely reduced in another type of chromatin alteration-defective mutants, hsk1-89, a temperature-sensitive allele of hsk1+ encoding the fission yeast CDC7 homolog (K. Hirota et al., unpublished results) (20, 32). In addition, at the ade6-M26 and tdh1+ loci, the chromatin remodeling is observed even in mutants lacking rec12+ and other genes (rec7+, rec10+, rec14+, rec15+, mei4+, and rec8+) involved in DSB formation (Fig. 3). From these results, we propose that meiotic chromatin remodeling is prerequisite for, but not a consequence of, the binding of DSB-initiating proteins at those sites.
Biological significance of CRE-related meiotic recombination and chromatin alteration.
We speculate that CRE-dependent meiotic recombination may account for a small portion of the total recombination events occurring in S. pombe, since the maximum frequency of whole DSBs is generally unaffected in atf1Δ or pcr1Δ strains (Fig. 5). Although their contribution may be limited, it is possible that Atf1-CRE-mediated meiotic recombination is conserved in other eukaryotes in which the Atf1-CRE-type transcriptional regulation units are present. Thus, it is possible that the Atf1-CRE complex acts as an additional regulatory level for meiotic recombination, in addition to its function as a transcription factor. Further investigation of CRE-related meiotic recombination hot spots may enlarge the definition of “CRE-dependent transcription factors”.
Acknowledgments
K.H. thanks all members of the Genetic System Regulation Laboratory and Cellular and Molecular Biology Laboratory in Riken for helpful discussion and K. Ogino and H. Masai for the hsk1-89 strain of fission yeast. We thank T. Yamada for critical advice regarding ChIP analysis. We also thank Y. Ichikawa and R. Nakazawa for DNA sequencing and Y. Sakuma for technical assistance.
This work was supported by basic research grants from the Bio-oriented Technology Research Advancement Institution (to T. Shibata and K. Ohta) and by grants-in-aid for scientific research on priority areas from the Ministry of Education, Science, Culture, & Sports, Japan (to K. Ohta). Walter Steiner was supported by a special fellowship (3230-05) from the Leukemia and Lymphoma Society and a grant (GM078065) from the National Institutes of Health.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Cervantes, M. D., J. A. Farah, and G. R. Smith. 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5:883-888. [DOI] [PubMed] [Google Scholar]
- 2.Cromie, G. A., C. A. Rubio, R. W. Hyppa, and G. R. Smith. 2005. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics 169:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson, M. K., H. K. Shandilya, K. Hirota, K. Ohta, and W. P. Wahls. 2004. Atf1-Pcr1-M26 complex links stress-activated MAPK and cAMP-dependent protein kinase pathways via chromatin remodeling of cgs2+. J. Biol. Chem. 279:50857-50863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis, L., and G. R. Smith. 2001. Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 98:8395-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elder, R. T., E. Y. Loh, and R. W. Davis. 1983. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc. Natl. Acad. Sci. USA 80:2432-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellermeier, C., and G. R. Smith. 2005. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 102:10952-10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan, Q., F. Xu, and T. D. Petes. 1995. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15:1679-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutz, H. 1971. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics 69:317-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe, p. 395-446. In R. D. King (ed.), Handbook of genetics, vol. 1. Plenum, New York, NY. [Google Scholar]
- 10.Hirota, K., K. Tanaka, Y. Watanabe, and M. Yamamoto. 2001. Functional analysis of the C-terminal cytoplasmic region of the M-factor receptor in fission yeast. Genes Cells 6:201-214. [DOI] [PubMed] [Google Scholar]
- 11.Iino, Y., and M. Yamamoto. 1985. Negative control for the initiation of meiosis in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 82:2447-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isshiki, T., N. Mochizuki, T. Maeda, and M. Yamamoto. 1992. Characterization of a fission yeast gene, gpa2, that encodes a G alpha subunit involved in the monitoring of nutrition. Genes Dev. 6:2455-2462. [DOI] [PubMed] [Google Scholar]
- 13.Kanoh, J., Y. Watanabe, M. Ohsugi, Y. Iino, and M. Yamamoto. 1996. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells. 1:391-408. [DOI] [PubMed] [Google Scholar]
- 14.Kon, N., M. D. Krawchuk, B. G. Warren, G. R. Smith, and W. P. Wahls. 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 94:13765-13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lichten, M., and A. S. Goldman. 1995. Meiotic recombination hotspots. Annu. Rev. Genet. 29:423-444. [DOI] [PubMed] [Google Scholar]
- 16.McLeod, M., and D. Beach. 1986. Homology between the ran1+ gene of fission yeast and protein kinases. EMBO J. 5:3665-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuno, K., Y. Emura, M. Baur, J. Kohli, K. Ohta, and T. Shibata. 1997. The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 11:876-886. [DOI] [PubMed] [Google Scholar]
- 18.Nachman, M. W. 2002. Variation in recombination rate across the genome: evidence and implications. Curr. Opin. Genet. Dev. 12:657-663. [DOI] [PubMed] [Google Scholar]
- 19.Nurse, P. 1985. Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol. Gen. Genet. 198:497-502. [Google Scholar]
- 20.Ogino, K., K. Hirota, S. Matsumoto, T. Takeda, K. Ohta, K. Arai, and H. Masai. 2006. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc. Natl. Acad. Sci. USA 103:8131-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohta, K., T. Shibata, and A. Nicolas. 1994. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 13:5754-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petes, T. D. 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2:360-369. [DOI] [PubMed] [Google Scholar]
- 23.Ponticelli, A. S., E. P. Sena, and G. R. Smith. 1988. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics 119:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Schuchert, P., M. Langsford, E. Kaslin, and J. Kohli. 1991. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 10:2157-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada, M., K. Nabeshima, T. Tougan, and H. Nojima. 2002. The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. EMBO J. 21:2807-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10:2276-2288. [DOI] [PubMed] [Google Scholar]
- 28.Steiner, W. W., R. W. Schreckhise, and G. R. Smith. 2002. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell 9:847-855. [DOI] [PubMed] [Google Scholar]
- 29.Steiner, W. W., and G. R. Smith. 2005. Natural meiotic recombination hot spots in the Schizosaccharomyces pombe genome successfully predicted from the simple sequence motif M26. Mol. Cell. Biol. 25:9054-9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, H., D. Treco, N. P. Schultes, and J. W. Szostak. 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338:87-90. [DOI] [PubMed] [Google Scholar]
- 31.Szankasi, P., W. D. Heyer, P. Schuchert, and J. Kohli. 1988. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6-M26. J. Mol. Biol. 204:917-925. [DOI] [PubMed] [Google Scholar]
- 32.Takeda, T., K. Ogino, K. Tatebayashi, H. Ikeda, K. Arai, and H. Masai. 2001. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol. Biol. Cell 12:1257-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida, and N. Jones. 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14:6193-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahls, W. P. 1998. Meiotic recombination hotspots: shaping the genome and insights into hypervariable minisatellite DNA change. Curr. Top. Dev. Biol. 37:37-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahls, W. P., and G. R. Smith. 1994. A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev. 8:1693-1702. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe, Y., S. Yokobayashi, M. Yamamoto, and P. Nurse. 2001. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature 409:359-363. [DOI] [PubMed] [Google Scholar]
- 37.Wilkinson, M. G., M. Samuels, T. Takeda, W. M. Toone, J. C. Shieh, T. Toda, J. B. Millar, and N. Jones. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10:2289-2301. [DOI] [PubMed] [Google Scholar]
- 38.Wolffe, A. 1997. Chromatin: structure and function, 3rd ed. Academic Press, San Diego, CA.
- 39.Wu, T. C., and M. Lichten. 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263:515-518. [DOI] [PubMed] [Google Scholar]
- 40.Yamada, T., K. I. Mizuno, K. Hirota, N. Kon, W. P. Wahls, E. Hartsuiker, H. Murofushi, T. Shibata, and K. Ohta. 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 23:1792-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young, J. A., R. W. Hyppa, and G. R. Smith. 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young, J. A., R. W. Schreckhise, W. W. Steiner, and G. R. Smith. 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9:253-263. [DOI] [PubMed] [Google Scholar]