Abstract
The SrrAB system regulates metabolism and virulence factors in Staphylococcus aureus. We sequenced the srrAB loci of 21 isolates and performed a phylogenetic analysis. Vaginal and bovine isolates clustered together, while skin isolates were genetically diverse. Few nucleotide polymorphisms were observed, and most were synonymous. Two strains (N2 and N19) with N-terminal truncations in SrrA displayed defects in growth and abnormally upregulated virulence factor expression under low-oxygen conditions.
Staphylococcus aureus is a gram-positive coccus that colonizes the skin and mucous membranes of humans and animals. Coordinated expression of virulence factors may result in serious infections, such as toxic shock syndrome (TSS), sepsis (10, 16, 20), and bovine mastitis (12). Differences in virulence factor regulation contribute to the variable pathogenic potential of the organism in humans or other animals. S. aureus encodes many global regulators of virulence, including a quorum-sensing system, the Sar family of virulence regulators, two-component systems, and transcriptional regulators (5-7, 13-15, 18, 22). The staphylococcal respiratory response (SrrAB) two-component system regulates energy metabolism as well as the genes tst (toxic shock syndrome toxin 1 [TSST-1]), spa (staphylococcal protein A), and icaR (intercellular adhesion locus repressor) in response to oxygen (17, 19, 25, 25a, 26). Although the effects of SrrAB have been investigated for four different S. aureus strains to date and the srrAB loci have been found in all sequenced isolates of S. aureus, the conservation of DNA and protein sequences from isolates of diverse origin is unknown. In order to ascertain the level of conservation of srrAB among human skin, human vaginal, and bovine udder isolates, PCR amplification and sequencing methods were used. The DNA and amino acid sequences were analyzed for phylogenetic relatedness. The strains and primers used in this study are described in Tables 1 and 2, respectively. For sequencing, S. aureus was grown in Todd-Hewitt medium (Difco Laboratories, Sparks, MD) in laboratory aerobic atmosphere with shaking. Genomic DNA was isolated by digestion with lysostaphin (Sigma-Aldrich Corp., St. Louis, MO), followed by purification using the DNeasy tissue kit (QIAGEN Corp., Valencia, CA). srrAB was PCR amplified using a high-fidelity enzyme (ABgene, Rochester, NY). For each strain, 30 srrAB sequence reads were performed using the primers shown in Table 2, and the sequences were assembled with DNASTAR SeqMan (LaserGene, Madison, WI). Assemblies were analyzed for weak or disparate residues and manually corrected, trimmed, and aligned by nucleotide and amino acid homology using ClustalW. Alignments were analyzed by parsimony analysis using PAUP with hierarchical clustering and a bootstrap value of 1000 (Sinauer Associates, Sunderland, MA) (23).
TABLE 1.
S. aureus strains sequenced
| Strain name(s) | Source or reference | Yr isolated | Type of infectiona |
|---|---|---|---|
| 1926 (26665) | Dennis W. Watson Culture Collection | 1926 | Mild skin infection |
| 1956 | William Altemeier (1) | 1956 | Typical vaginal isolate (nonmenstrual) from a healthy person |
| 2000 | Patrick M. Schlievert | 2004 | Toxic shock syndrome (menstrual) |
| USA400 | Patrick M. Schlievert (10) | 2000 | Purpura fulminans caused by S. aureus |
| K15A | Patrick M. Schlievert | 1981 | Axillary culture from a healthy child |
| T35 | Patrick M. Schlievert (21) | 1981 | Typical menstrual isolate from a healthy person |
| N2, N6, N7, N9, N12, N19 | N Study | 2003 | Persistent skin infections in patients with atopic dermatitis |
| PSA 6, PSA 10, PSA 20 | Vivek Kapur | 1992 | Bovine mastitis strains from the United States |
| COL | 8 | Early 1960s | Early MRSA from a wound infection in the United Kingdom |
| MW2 | 2 | 1998 | Community-acquired MRSA, septicemia, and septic arthritis in North Dakota patient |
| N315 | 11 | 1982 | MRSA pharyngeal isolate from a Japanese patient |
| MRSA252 | 9 | 1997 | Hospital-acquired MRSA from a case of postoperative septicemia in the United Kingdom |
| MSSA276 | 9 | 1998 | Invasive community-acquired MSSA from a case of osteomyelitis and bacteremia in the United Kingdom |
| Mu50 | 11 | 1997 | MRSA, surgical wound infection in a Japanese patient |
MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus.
TABLE 2.
Primers used in srrAB sequencing
| Primer | Sequence |
|---|---|
| srrAB sense | 5′-ATGTATTTATCACAAAGTTTGA-3′ |
| srrAB antisense | 5′-ATTTAATAGTTGATATTCGCAA-3′ |
| Forward 1 | 5′-GTGAAGAAACAAACCGTGTTGAAGG-3′ |
| Forward 2 | 5′-GTTAAATCGTGTGTCTAGTGAAGCTG-3′ |
| Forward 3 | 5′-TGACGATGTGTTTGATAAAGGTAAATCTG-3′ |
| Forward 4 | 5′-TTGGTCAATTATCGCAGGCATTTA-3′ |
| Forward 5 | 5′-TGTTGTTGTGACAGTTCGTGATATG-3′ |
| Forward 6 | 5′-GAAGCGTGTTTGGAGTTATGATATGGA-3′ |
| Forward 7 | 5′-TTCAGAACGCATGTTATTGATTAGAGA-3′ |
| Forward 8 | 5′-GAAAAGCATGTGTGGGAGGTATGA-3′ |
| Forward 9 | 5′-AAGAAGAACGCAATCTACAACTG-3′ |
| Forward 10 | 5′-TGATGAGCCGGCTAAATAGTGTCG-3′ |
| Forward 11 | 5′-GGCTATCCAACAAAAGCACAGA-3′ |
| Forward 12 | 5′-GCATGTCGACGCATTATCCA-3′ |
| Forward 13 | 5′-CGTACACCGATATCATTACTTCAAGG-3′ |
| Forward 14 | 5′-ATACGAAACCTGGAGATGAAAT-3′ |
| Forward 15 | 5′-TTTTTGATTGATGTGGGGAAT-3′ |
| Reverse 1 | 5′-GGTGCAATGCCTGTACCTGTATCTT-3′ |
| Reverse 2 | 5′-CAGCTTCACTAGACACACGATTTAAC-3′ |
| Reverse 3 | 5′-CAGATTTACCTTTATCAAACACATCGTCA-3′ |
| Reverse 4 | 5′-TAAATGCCTGCGATAATTGACCAA-3′ |
| Reverse 5 | 5′-CATATCACGAACTGTCACAACAACA-3′ |
| Reverse 6 | 5′-TCCATATCATAACTCCAAACACGCTTC-3′ |
| Reverse 7 | 5′-TCTCTAATCAATAACATGCGTTCTGAA-3′ |
| Reverse 8 | 5′-TCATACCTCCCACACATGCTTTTC-3′ |
| Reverse 9 | 5′-CAGTTGTAGATTGCGTTCTTCTT-3′ |
| Reverse 10 | 5′-CGACACTATTTAGCCGGCTCATCA-3′ |
| Reverse 11 | 5′-TCTGTGCTTTTGTTGGATAGCC-3′ |
| Reverse 12 | 5′-TGGATAATGCGTCGACATGC-3′ |
| Reverse 13 | 5′-CCTTGAAGTAATGATATCGGTGTACG-3′ |
| Reverse 14 | 5′-ATTTCATCTCCAGGTTTCGTAT-3′ |
| Reverse 15 | 5′-ATTCCCCACATCAATCAAAAA-3′ |
Strain growth as well as hemolysin activity and TSST-1 expression in srrA mutants and two wild-type strains (MN8 and CDC587) was determined as follows: S. aureus was grown in beef heart medium at 37°C under aerobic conditions (with shaking at 200 rpm), low-oxygen conditions (<0.3% oxygen, without shaking), or anaerobically in BBL GasPak jars (Becton Dickinson and Company, Franklin Lakes, NJ) without shaking. Cell densities after 24 h were determined by plate counts, hemolysin activity was determined by bioassay (lysis of rabbit erythrocytes incorporated into 0.8% agarose) (21), and TSST-1 expression was determined by quantitative Western immunoblotting (3). Strain N19 was complemented with pJMY11, a multicopy plasmid with wild-type srrAB (26). The resultant strain was also assayed for growth and hemolysin production.
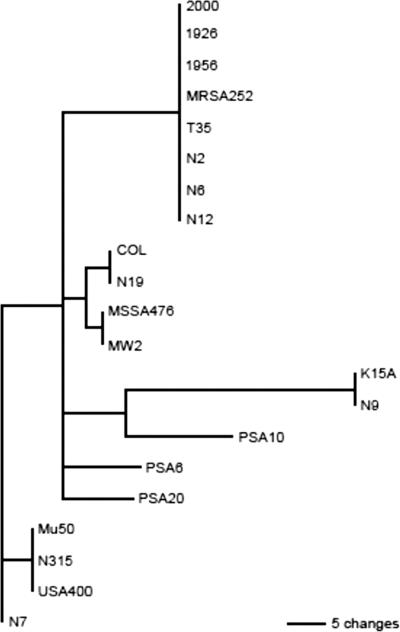
The results of the srrAB nucleotide sequence comparison appear in the phylogenetic tree shown in Fig. 1. Nucleotide sequences were compared due to the large proportion of synonymous mutations compared to nonsynonymous mutations among the sequences. Two clusters are immediately apparent. The bovine mastitis isolates (PSA6, PSA10, PSA20) cluster together, as do the vaginal isolates (1956, 2000, T35). The skin isolates appear scattered throughout the phylogram. MRSA252, an isolate from the United Kingdom, clusters near the U.S. vaginal isolates. This is not surprising, as the genomic backbone of MRSA252 is conserved relative to a recently sequenced TSS-associated isolate from the United States (Lisa Herron-Olson, personal communication). The oldest isolate, from a mild skin infection in 1926, clusters closely with several of the more recent vaginal and skin isolates.
FIG. 1.
srrAB phylogenetic tree. The nucleotide coding sequences of the 21 srrAB loci were used to create a phylogenetic tree. Distinct clusters include the vaginal isolates (1956, 2000, and T35) and the bovine mastitis isolates (PSA 6, 10, 20). Human skin isolates are scattered throughout the phylogram.
Alignments of SrrAB amino acids show that the protein sequences are well conserved among diverse isolates. Most of the DNA polymorphisms resulted in synonymous mutations; of the six nonconserved residues in SrrAB, only two (SrrB A/T322 and A/D502) resulted in a change to a dissimilar amino acid. Of note, strains N2 and N19 have N-terminal truncations in their srrA sequences. The first 22 amino acids of the N2 SrrA sequence are absent, with the majority of the phosphate receiver domain still present. In strain N19, the first 78 amino acids are absent, including an aspartate residue at position 56 that is predicted to be the site of phosphorylation in SrrA (4, 24). Due to this truncation, N19 SrrA may be incapable of acting as a phosphoacceptor or may exhibit unregulated DNA-binding activity. Although both N2 and N19 have ribosomal binding sites upstream of their SrrA translational start sites, it is not known if SrrA is translated in these strains. We do not predict that SrrB translation is altered in N2 and N19, as these strains exhibit no sequence changes near the SrrB translational start site.
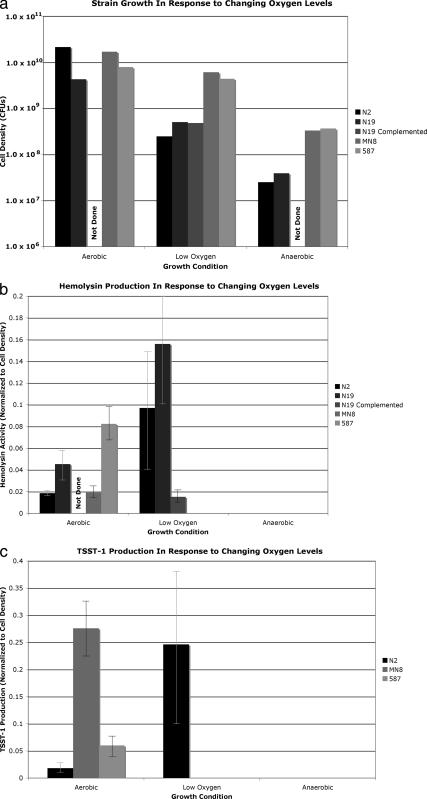
The effect of srrA mutation on growth and virulence factor production was assessed for strains N2 and N19. Strains N2 and N19 have growth defects under low-oxygen and anaerobic conditions, in comparison to MN8 and CDC587, two strains with intact SrrAB that are capable of expressing hemolysin and TSST-1 (Fig. 2a). N2 and N19 display normal hemolysin activity under aerobic conditions and increased activity under low-oxygen conditions; wild-type strains MN8 and 587 did not express hemolysin under low-oxygen conditions. Complementation of N19 with a multicopy plasmid containing wild-type srrAB resulted in a dramatic repression of hemolysin production. This suggests that while the wild-type SrrAB system represses hemolysin under low-oxygen conditions, the truncated SrrAB system in strains N2 and N19 is unable to repress hemolysin production under low-oxygen conditions. None of the strains produced hemolysin under anaerobic conditions (Fig. 2b). N2 demonstrates TSST-1 production under aerobic conditions with enhanced TSST-1 production under low oxygen. Strains MN8 and 587 demonstrated no TSST-1 production under low-oxygen conditions. These findings suggest that the N2 SrrAB system is incapable of repressing TSST-1 production under low-oxygen conditions. N19 lacks the gene for TSST-1 and is therefore unable to express it. No strain produced TSST-1 under anaerobic conditions (Fig. 2c). The SrrAB system has been shown to repress virulence factors such as hemolysin and TSST-1 under low-oxygen conditions. N2 and N19 display an increase in hemolysin and TSST-1 production under low-oxygen conditions that is consistent with a loss of repression due to a nonfunctional SrrA. Complementation of N19 with wild-type srrAB resulted in restoration of hemolysin repression under low-oxygen conditions. Both N2 and N19 were isolated from patients with chronic skin infections in the setting of atopic dermatitis. This superficial and chronic skin infection environment may favor strains that are deficient in sensing oxygen.
FIG. 2.
N-terminal truncations of srrA affect growth and virulence factor production. All data points represent an average of results of four separate experiments. (a) Strain growth in response to change in oxygen levels. Strains N2 and N19 demonstrate growth defects under low-oxygen and anaerobic conditions, in comparison to control strains MN8 and CDC587. Complementation of N19 with wild-type srrAB did not affect growth. (b) Hemolysin activity. Strains N2 and N19 demonstrate increased hemolysin levels under low-oxygen conditions, while control strains demonstrate no production. Complementation of N19 with wild-type srrAB repressed hemolysin production. No strains produced hemolysin during anaerobic growth. Activity is reported in micrograms of hemolysin/1.0 × 108 CFU. (c) TSST-1 activity. Strain N2 demonstrates increased production of TSST-1 under low-oxygen conditions, while neither control strain is able to make TSST-1 under low-oxygen conditions. Strain N19 lacks the gene for TSST-1 and is therefore unable to make TSST-1. TSST-1 was not made by any strain under anaerobic conditions. Activity is reported in micrograms of TSST-1/1.0 × 108 CFU.
In summary, our alignments of srrAB sequences from disparate isolates demonstrate relatively few changes in the sequences at the nucleotide level and no focal points of increased polymorphism. The phylogenetic tree demonstrates clustering of bovine mastitis isolates and clustering of human vaginal isolates, while human skin isolates do not cluster. The separate niches inhabited by the vaginal and bovine strains may account for the divergence. In this study, truncations in srrA affected growth and virulence factor regulation. N2 demonstrated decreased growth and increased hemolysin and TSST-1 activity with oxygen limitation, while N19 showed decreased growth and increased hemolysin activity with oxygen limitation. Complementation of N19 with wild-type srrAB resulted in restoration of hemolysin repression under low-oxygen conditions. These findings are consistent with a loss of SrrAB-mediated virulence factor repression under low-oxygen conditions in strains N2 and N19.
Acknowledgments
A.A.P. was supported by an NIAID predoctoral fellowship (T32 AI 07421). L.H.-O. was supported by the University of Minnesota Martha Kunze Graduate School Fellowship in Biological Sciences.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Altemeier, W. A., S. Lewis, P. M. Schlievert, and H. S. Bjornson. 1981. Studies of the staphylococcal causation of toxic shock syndrome. Surg. Gynecol. Obstet. 153:481-485. [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Blake, M. S., K. H. Johnston, G. J. Russell-Jones, and E. C. Gotschlich. 1984. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal. Biochem. 136:175-179. [DOI] [PubMed] [Google Scholar]
- 4.Brissette, R. E., K. L. Tsung, and M. Inouye. 1991. Suppression of a mutation in OmpR at the putative phosphorylation center by a mutant EnvZ protein in Escherichia coli. J. Bacteriol. 173:601-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kravitz, G. R., D. J. Dries, M. L. Peterson, and P. M. Schlievert. 2005. Purpura fulminans due to Staphylococcus aureus. Clin. Infect. Dis. 40:941-947. [DOI] [PubMed] [Google Scholar]
- 11.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 12.Lammers, A., E. Kruijt, C. van de Kuijt, P. J. Nuijten, and H. E. Smith. 2000. Identification of Staphylococcus aureus genes expressed during growth in milk: a useful model for selection of genes important in bovine mastitis? Microbiology 146:981-987. [DOI] [PubMed] [Google Scholar]
- 13.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manna, A. C., S. S. Ingavale, M. Maloney, W. van Wamel, and A. L. Cheung. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J. Bacteriol. 186:5267-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novick, R. P. 1990. Molecular biology of the staphylococci, p. 1-40. VCH Publishers, Inc., New York, NY.
- 17.Pragman, A. A., Y. Ji, and P. M. Schlievert. 2007. Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry 46:314-321. [DOI] [PubMed] [Google Scholar]
- 18.Pragman, A. A., and P. M. Schlievert. 2004. Virulence regulation in Staphylococcus aureus: the need for in vivo analysis of virulence factor regulation. FEMS Immunol. Med. Microbiol. 42:147-154. [DOI] [PubMed] [Google Scholar]
- 19.Pragman, A. A., J. M. Yarwood, T. J. Tripp, and P. M. Schlievert. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, NY.
- 21.Schlievert, P. M., M. T. Osterholm, J. A. Kelly, and R. D. Nishimura. 1982. Toxin and enzyme characterization of Staphylococcus aureus isolates from patients with and without toxic shock syndrome. Ann. Intern. Med. 96:937-940. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swofford, D. L., P. J. Waddell, J. P. Huelsenbeck, P. G. Foster, P. O. Lewis, and J. S. Rogers. 2001. Bias in phylogenetic estimation and its relevance to the choice between parsimony and likelihood methods. Syst. Biol. 50:525-539. [PubMed] [Google Scholar]
- 24.Tanaka, T., S. K. Saha, C. Tomomori, R. Ishima, D. Liu, K. I. Tong, H. Park, R. Dutta, L. Qin, M. B. Swindells, T. Yamazaki, A. M. Ono, M. Kainosho, M. Inouye, and M. Ikura. 1998. NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature 396:88-92. [DOI] [PubMed] [Google Scholar]
- 25.Throup, J. P., F. Zappacosta, R. D. Lunsford, R. S. Annan, S. A. Carr, J. T. Lonsdale, A. P. Bryant, D. McDevitt, M. Rosenberg, and M. K. Burnham. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392-10401. [DOI] [PubMed] [Google Scholar]
- 25a.Ulrich, M., M. Bastian, S. E. Cramton, K. Ziegler, A. A. Pragman, A. Bragonzi, G. Memmi, C. Wolz, P. M. Schlievert, A. Cheung, and G. Doring. 2007. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic conditions. Mol. Microbiol. 65:1276-1287. [DOI] [PubMed] [Google Scholar]
- 26.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]