Abstract
Most strains of Staphylococcus aureus produce one type of capsular polysaccharide that belongs to either type 5 or type 8. The production of these capsules has been shown to be regulated by various regulators. Here we report that the sbcD and sbcC genes are involved in the repression of type 5 capsule production. Chromosomal deletions in the sbcDC genes resulted in increased capsule promoter activity, capsule gene transcripts, and capsule production. The survival rates of the sbcDC deletion mutant were reduced upon UV irradiation compared to those for the wild-type strain Newman, suggesting that the genes are involved in DNA repair in S. aureus. The two genes were organized as an operon and were expressed very early in the exponential growth phase. A subinhibitory concentration of ciprofloxacin or mitomycin C induced sbcDC transcription but repressed the capsule promoter activity, suggesting that the sbcDC genes and the capsule genes are part of the SOS regulon. By reporter gene fusion and Northern blotting, we found that sbcDC regulated capsule by downregulating arl and mgr. Further genetic studies indicate that sbcDC functions upstream of arl and mgr in capsule regulation. Collectively, our results indicate that sbcDC, upon the SOS response, represses type 5 capsule production through an arl-mgr pathway. To our knowledge, this is the first demonstration that an SbcDC homolog was involved in transcriptional regulation.
Staphylococcus aureus is a major human opportunistic pathogen responsible for a broad spectrum of infections ranging from food poisoning and superficial skin abscesses to more serious diseases such as pneumonia, meningitis, endocarditis, septicemia, and toxic shock syndrome. The organism is capable of producing an array of virulence factors, including surface-associated adhesions, secreted exoproteins, toxins, and capsular polysaccharides (CP), which contribute to the pathogenesis of the organism (18). Most S. aureus strains produce one type of CP belonging to either type 5 or type 8 (CP5 or CP8, respectively) (reviewed in references 16 and 24). The cap5 and cap8 genes required for the synthesis of CP5 and CP8, respectively, are organized as an operon in which the polycistronic message is controlled primarily by the promoter located at the beginning of the operon. The cap5 and cap8 operons are allelic, sharing common genes that flank the central specific genes (26). The sequences in the promoter regions between the cap5 and cap8 operons are nearly identical, indicating that the regulation mechanisms between CP5 and CP8 are similar (12).
The production of CP5 and CP8 is highly regulated by various environmental cues, such as carbon dioxide, iron concentration, in vivo growth, and specific nutrients (16, 24). We have shown previously that global regulators agr and mgr positively regulate capsule production transcriptionally (20, 21). In S. aureus, virulence factors are controlled by a complex regulatory network (reviewed in references 4, 5, and 23). To further understand how capsule is regulated, we screened a transposon library and identified seven additional regulatory genes that affected capsule promoter activity by using a reporter gene fusion system (19). One of these genes identified by this strategy, arlR, has been characterized further and shown to upregulate type 5 capsule mostly through an mgr-dependent pathway but also by an independent pathway (19). In this communication, we report the regulation of capsule by sbcDC. We found that sbcDC represses cap5 genes through arl and mgr and that the regulatory pathway is part of the SOS regulon.
MATERIALS AND METHODS
Strains, plasmids, phage, and growth conditions.
The S. aureus strains and plasmids used in this study are listed in Table 1. Escherichia coli strain XL1-Blue was used as the host strain for plasmid construction. S. aureus and E. coli strains were routinely cultivated in Trypticase soy medium (Difco Laboratories, Detroit, MI) and Luria-Bertani medium (Difco), respectively, with appropriate antibiotic selection when necessary. Antibiotics used for selection were ampicillin, 100 μg/ml, or spectinomycin, 50 μg/ml, for E. coli and tetracycline, 3 μg/ml, chloramphenicol, 10 μg/ml, and erythromycin, 10 μg/ml, for S. aureus. Plasmids were first electroporated into S. aureus RN4220 by the procedure of Kraemer and Iandolo (13). Phage 52A, 80α, or ϕ11 was used for plasmid and chromosomal DNA transduction between S. aureus strains.
TABLE 1.
Plasmids and strains
| Plasmid or strain | Description of construction | Reference or source |
|---|---|---|
| Plasmids | ||
| pLL28 | Temperature-sensitive vector | 20 |
| pLL33 | Translational blaZ fusion vector | 19 |
| pLL35 | Transcriptional blaZ fusion vector | 19 |
| pLL38 | Transcriptional xylE fusion vector | This study |
| pZC3618 | Pcap5::xylE transcriptional fusion in pLL38 | This study |
| pZC3419 | Psbc::xylE transcriptional fusion in pLL38 | This study |
| pZC3624 | pZC3618 carrying sbcDC operon | This study |
| pCL8074 | Pcap5/8::xylE transcription fusion in pLC4 | 25 |
| pAM3175 | Pcap5/8::blaZ translational fusion in pLL33 | 19 |
| pAM3176 | Pcap5/8::blaZ transcriptional fusion in pLL35 | 19 |
| pTL3562 | Parl-blaZ transcriptional fusion in pLL35 | This study |
| pTL3564 | Pagr-3::blaZ transcriptional fusion in pLL35 | This study |
| pTL3580 | PmgrA-blaZ transcriptional fusion in pLL35 | This study |
| pTL3573 | pCL15 containing arlR | 19 |
| Strains | ||
| RN4220 | 8325r− | 14 |
| Newman | CP5 strain | T. Foster |
| CYL6619 | Newman ΔsbcD::cat | This study |
| CYL6620 | Newman ΔsbcC::cat | This study |
| CYL6621 | Newman ΔsbcDC::cat | This study |
| CYL1164 | Newman ΔarlR | 19 |
| CYL6851 | Newman ΔsbcDC::catΔarlR | This study |
Plasmid construction.
Plasmid pLL38 was constructed by replacing the blaZ gene with the xylE reporter gene in pLL33. Plasmid pZC3618 was constructed by ligating a 627-bp fragment containing the cap5 promoter (19) to the xylE gene in pLL38 at the EcoRI-HindIII sites. To construct sbcDC promoter fusion plasmids, we first mapped the promoter of the sbcDC operon by reverse transcriptase PCR (RT-PCR). It was found that the promoter was within 362 bp upstream of the SbcD start codon (not shown). To construct the sbcD promoter fusion with xylE, an 872-bp fragment including 603 bp upstream of the sbcD start codon was amplified by PCR using primers sbcFP1 and sbcD2 (Table 2) and cloned into pLL38. To clone the sbcDC genes for complementation, a 4.8-kb PCR fragment was amplified from Newman chromosome by use of primers sbcFP and sbcRP and cloned into the BamHI-PstI sites of pZC3618 to form pZC3624. The amplified fragment, which matched completely with that of strain COL, was verified by sequencing.
TABLE 2.
Primers used in this research
| Primer | Sequence |
|---|---|
| sbcD1 | AATTGAATTCGGACTTCTCGATTTGAAGTC |
| sbcD2 | AATGCCCGGGCCTCTCTTTACCATCGTGAT |
| sbcD3 | ATTAGGATCCAGCTAAAGGGTATAGACGTG |
| sbcD4 | AATGCTGCAGCGGCTGTTTACCATCAGCGA |
| sbcC1 | AATTGAGCTCAGGATTAATGAATGAACCA |
| sbcC2 | ATTAGGATCCGCCATAAAAGGTCAATTTGTTGA |
| sbcC3 | AATTGGATCCGAAATGGAAATAGCTAGGTTAG |
| sbcC4 | ATTCGAATTCTCAATTCGCTCACATGTGAA |
| sbcFP1 | CGGGATCCGAAGGTGTCTGCGTGCTC |
| sbcFP | GGATCCTGGAGAATTAGGCGGCATGTTCTT |
| sbcRP | CCGCGGGCTAAGGTTGTTCTATACATTCCA |
| RNAIII 1 | AGGAAGGAGTGATTTCAATG |
| RNAIII 2 | ACTCATCCCTTCTTCATTAC |
| mgrA38 | CATATGTCTGATCAACATAATTTAAAAG |
| mgrA39 | GGATCCGTTAATTATTTTTCCTTTGTTTC |
| arlR1 | CATATGACGCAAATTTTAATAG |
| arlR2 | GGATCCTCATCGTATCACATAC |
| SGcap8A1 | ACTAAGGGTGACAATCCTCAG |
| SGcap8A2 | AAGTCCTTTGACACCTCATCTA |
Allele replacement of sbcD, sbcC, and sbcDC in Newman.
To construct the ΔsbcD, ΔsbcC, and ΔsbcDC mutants from Newman, two sets of primers (Table 2) were used to amplify the upstream and downstream fragments (about 0.5 kb) of each target gene. The primer pairs were as follows: sbcD1/sbcD2 and sbcD3/sbcD4 for ΔsbcD deletion, sbcC1/sbcC2 and sbcC3/sbcC4 for ΔsbcC deletion, and sbcD1/sbcD2 and sbcC3/sbcC4 for ΔsbcDC deletion. The amplified fragments were sequence verified and cloned into pLL28 such that the upstream and downstream fragments flanked the cat gene and had the same orientation as in the chromosome. The resultant plasmids were used for allele replacement as described previously (17). The mutations were verified by PCR.
RNA isolation, purification, and transcriptional analyses.
Total RNAs were isolated as described previously (21). Quantification of cap5 mRNA by real-time RT-PCR using SGcap8A1 and SGcap8A2 primers (Table 2) was performed as described previously (19). Northern hybridization was carried out as described in instructions for a Roche digoxigenin high prime DNA labeling and detection starter kit II (Roche Applied Science, Indianapolis, IN). Briefly, total RNAs were resolved in a formaldehyde agarose gel and capillary transferred to a positively charged nylon membrane (Immobilon NY+; Millipore Corp.). RNAs were immobilized by UV cross-linking at 25,000 μJ/cm2 at 254 nm using an HL-2000 Hybrilinker (UVP, Inc., Upland, CA). The DNA probes were generated by PCR and labeled using a PCR-based digoxigenin probe synthesis kit (Roche Applied Science).
UV survival test.
Cultures of Newman and the ΔsbcDC mutant at 4 h and 24 h were diluted 10−3 to 10−6 with Trypticase soy broth (TSB), and 100 μl of the diluted cells was plated on Trypticase soy agar plates supplied with appropriate antibiotics. Plates with the cover open were irradiated under UV light at 4,000 μJ/cm2 using an HL-2000 Hybrilinker and then incubated in the dark at 37°C overnight. Control plates were prepared similarly but without irradiation. Survival rates for each strain were calculated by dividing the CFU of the UV-treated plates with those of the control plates.
SOS induction.
Overnight cultures were inoculated at a 1:100 dilution into 10 ml TSB and incubated for 2 h at 37°C with aeration. Mitomycin C or ciprofloxacin was added directly to the growing cultures at a subinhibitory level of 1 μg/ml or 2 μg/ml, respectively. At this concentration, the growth rates of the cultures were not affected. The cultures were further incubated until specific time points for analyses.
Other tests.
CP5 capsule was quantified as previously described (20). β-Lactamase (BlaZ) was assayed by the nitrocefin method as previously described (21). XylE activity was assayed as described by Zukowski et al. (31), with the following modifications. Overnight Staphylococcus aureus culture was diluted into 8 ml prewarmed TSB with antibiotic in 50-ml conical tubes to an optical density at 660 nm (OD660) of 0.05 and incubated at 37°C with shaking at 225 rpm. At various time points, OD660 was measured and 1.5 ml culture was collected by centrifugation. The pellet was washed once and resuspended in 500 μl 20 mM phosphate buffer (pH 7.5). The cells were mixed with 10 μl 55 mg/ml catechol and incubated at 30°C for 10 min. The supernatant after centrifugation was measured at A375. Promoter activity was expressed as the ratio of A375 to OD660 of the culture.
Statistical analysis.
Data from reporter gene fusion analyses were analyzed by the GraphPad Prism program (San Diego, CA) using a paired Student t test for comparing two samples. P values of <0.05 were considered statistically significant.
RESULTS
The sbcDC locus negatively affects capsule production.
Previously, we screened a transposon Tn551 library of strain COL with the plasmid pCL8704 containing a Pcap8::xylE fusion to identify genes affecting capsule gene expression. Seven genes, including two adjacent sbcD and sbcC genes, were identified by the genetic screen. Interestingly, among 17 unique Tn551 insertions, 6 were found in the sbcD coding region, whereas only 1 site was found in each of the other genes (19). These mutants showed increased capsule promoter activity, indicating that the sbcCD genes negatively regulate capsule. Backcross experiments showed that the transposon insertions were responsible for the phenotype. To confirm that the two genes are involved in cap5 gene regulation, we first constructed a deletion-insertion mutation in each gene or in both genes in the chromosome by allele replacement in strain Newman as described in Materials and Methods. All three resultant mutants had no detectable differences in growth rate compared to that of the wild type (not shown).
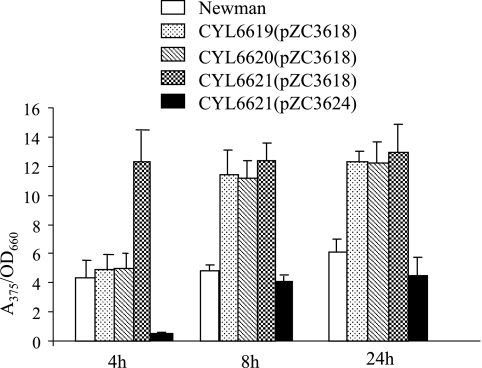
The mutants and the wild-type strain Newman containing the pZC3618 plasmid carrying the Pcap5::xylE fusion were tested for their effects on capsule promoter activity by XylE assay. As shown in Fig. 1, all of the mutations resulted in increased capsule promoter activities at 8 h and 24 h time points (all P values were <0.0028). At the 4 h time point, only the mutant with both the sbcD and sbcC genes deleted, but not the mutants with either gene deleted, resulted in increased capsule promoter activity (P = 0.0085). It is not clear why the results were different at the 4 h time point. To confirm further that the sbcDC genes are responsible for the repression of capsule promoter, we performed complementation tests by cloning a 4.8-kb fragment containing the sbcDC genes amplified by PCR from strain Newman in pZC3618. The resultant plasmid, pZC3624, was transferred to the mutants for complementation, and the resulting strains were assayed for XylE activities. The results in Fig. 1 show that the fragment was able to complement all mutations at all time points tested (all P values were <0.0019). In fact, the complementation resulted in reduced promoter activities in all mutants compared to that of the wild type, especially at 4 h (P = 0.0027). This could be explained by the effect of overproduction of SbcDC due to the multiple-copy plasmid used in the experiments. The much-reduced level of cap5 promoter activities at 4 h suggests that sbcDC acts early in the growth phase.
FIG. 1.
XylE activities of Pcap5::xylE transcriptional fusion. Strains CYL6619, CYL6620, and CYL6621 are ΔsbcC, ΔsbcD, and ΔsbcDC deletion mutants, respectively. The complementation of CYL6619 and CYL6620 by pZC3624 (not shown) resulted in values similar to those obtained by complementation of CYL6621(pZC3624). The XylE activities are expressed as the ratio of A375 of the enzymatic reaction to the OD660 of the culture. The error bars indicate standard deviations of at least three independent experiments.
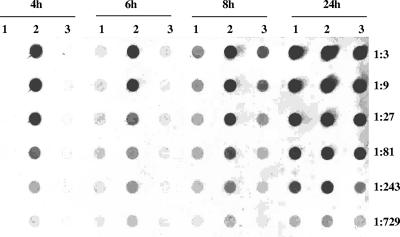
To determine whether the sbcDC effect on the capsule promoter activity reflects the effect on capsule production, the levels of CP5 production from Newman, the ΔsbcDC mutant, and the complementation strain were assayed by immunoblotting. As shown in Fig. 2, the mutation resulted in a substantial increase in CP5 production and the mutant phenotypes were restored by the complementation. The effect was much more profound at early time points again, suggesting that sbcDC exerts its effect early in the growth phase. Together, these results showed that sbcDC negatively affected CP5 production by affecting the cap5 promoter activity.
FIG. 2.
CP5 assays of (lanes 1) Newman(pZC3618), (lanes 2) CYL6621(pZC3618), and (lanes 3) CYL6621(pZC3624) at the time points shown at top. Threefold serial dilutions of the samples were analyzed by Western dot blotting.
The sbcDC locus affects CP5 production mainly at the transcriptional level.
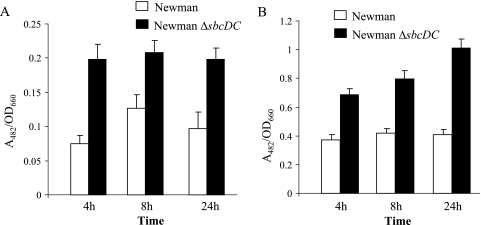
The above-mentioned results suggest that sbcDC genes affect the transcription of the cap5 genes. However, it is possible that they are also involved in translational regulation. To test this possibility, we analyzed the effect of sbcDC on translational and transcriptional Pcap5::blaZ fusions in Newman and the ΔsbcDC mutant. As shown in Fig. 3, the effects of ΔsbcDC mutation by transcriptional fusion assays are similar to those of the translational fusion assays, suggesting that the regulation is most likely at the transcriptional level (all P values were <0.0025).
FIG. 3.
Promoter activities of Pcap5::blaZ transcriptional fusion in plasmid pAM3176 (A) and translational fusion in plasmid pAM3175 (B) in strains Newman and CYL6621 (Newman ΔsbcDC) were analyzed at 4, 8, and 24 h. The BlaZ activities are expressed as the ratio of A482 of the enzymatic reaction to the OD660 of the culture. The error bars indicate standard deviations of at least three independent experiments.
The sbcDC genes form an operon transcribed early in growth phase.
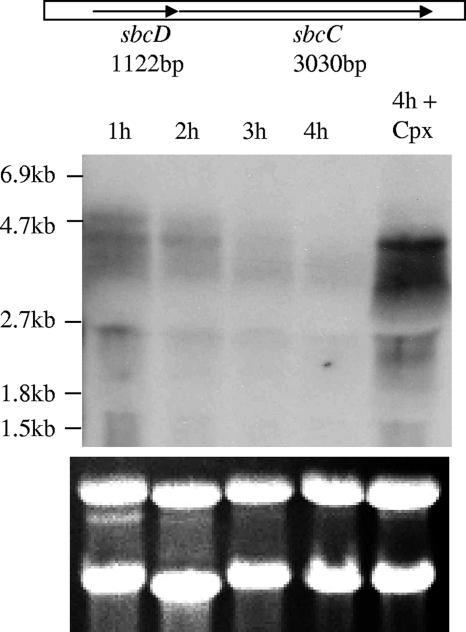
The sequence information indicates that there are only 4 bp in the intergenic region between the sbcD and sbcC genes, suggesting that the two genes may form an operon (Fig. 4). To test this possibility, we performed Northern hybridization. We found a major band at about 4.3 kb and a faint band at around 4.8 kb (Fig. 4, leftmost lane). The size of both bands is large enough to encompass both genes, suggesting that the two genes form an operon. There were two other much smaller bands and some smearing detected, which could be the degradation products. Using individual gene probes, we also identified the larger bands (data not shown), which further supports the operonic structure. The Northern results in Fig. 4 also showed that sbcDC transcripts were expressed early, at 1 h, and were quickly degraded. Indeed, use of a Psbc::xylE fusion containing 603 bp upstream of the sbcD start codon revealed that the XylE activities were very high at early time points but reduced drastically thereafter (data not shown), confirming the Northern blot data.
FIG. 4.
Northern blot of Newman RNA samples isolated at various time points, probed with a DNA fragment containing both sbcDC genes. The positions of the RNA markers (Roche Applied Science) are shown on the left. The last lane indicates an RNA sample obtained from Newman incubated for 2 h in TSB at 37°C and treated with ciprofloxacin (Cpx) for an additional 2 h. The closely linked sbcDC genes are shown above the blot. rRNA as a loading control is shown below the blot.
sbcDC is involved in UV resistance.
The sbcD and sbcC genes in E. coli have been shown to encode an exonuclease involved in DNA recombination and repair upon DNA damage (10). However, the S. aureus sbcDC genes have not been studied. To investigate whether the sbcDC locus is involved in DNA recombination and repair of UV lesions, we compared the survival rates upon UV irradiation between strain Newman and the isogenic sbcDC mutant. We found that there was no difference between wild-type Newman and the sbcDC mutant when the cells were collected at 4 h. However, when the cells were collected at 24 h, the survival rate of the sbcDC mutant was 50.68% (standard deviation of 8.55% for five independent experiments) that of Newman, indicating that sbcDC genes are involved in UV resistance when the cultures are in stationary phase. It is not clear why there was no difference at the 4 h time point. Perhaps at log phase but not at stationary phase there is a redundant system for complementing sbcDC mutation. In Pseudomonas putida, a mutT mutation has been shown to cause a much stronger mutator phenotype in starved bacteria than in actively growing bacteria, suggesting that there is a backup system complementing the lost MutT function in the growing cells (27).
The SOS system affects type 5 capsule through sbcDC.
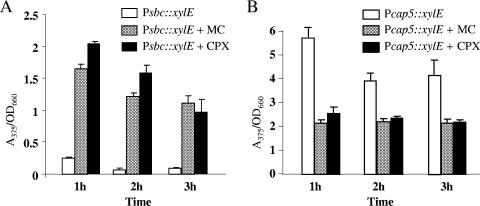
The involvement of sbcDC genes in UV resistance suggests that the genes could be part of the SOS regulon. Indeed, it has been reported that the sbcD open reading frame is preceded by a LexA box (6). Recently, microarray studies have also shown that the SOS response induces sbcD and sbcC genes (1, 6). To further test that the sbcDC genes are part of the SOS regulon, Newman cultures were treated with ciprofloxacin or mitomycin C, conditions that have been shown to induce an SOS response in S. aureus. The expression of sbcDC was analyzed by XylE reporter fusion assays. As shown in Fig. 5A, both chemicals substantially induced sbcDC promoter activity, indicating that the sbcDC genes are regulated by the SOS response. To confirm these results, we performed Northern hybridization using a DNA fragment within the sbcDC locus as the probe. The results in Fig. 4 showed that sbcDC mRNA was highly induced (Fig. 4, compare the two rightmost lanes at the 4 h time point), which is consistent with the results of the XylE reporter assay (all P values were <0.0031) (Fig. 5A). To test whether SOS induction also affects CP5 production, Newman containing the Pcap5::xylE fusion plasmid was treated with ciprofloxacin or mitomycin C and assayed for the reporter activities. The induction of the SOS response reduced the Pcap5 activities to about 50% at 1, 2, and 3 h time points (all P values were <0.0023) (Fig. 5B). Taken together, our results support the notion that sbcDC and its target cap5 genes are part of the SOS regulon.
FIG. 5.
Effects of mitomycin C (MC) and ciprofloxacin (CPX) on PsbcDC::xylE activities in fusion plasmid pZC3419 (A) and on Pcap5::xylE activities in fusion plasmid pZC3618 (B). The XylE activities are expressed as the ratio of A375 of the enzymatic reaction to the OD660 of the culture. The error bars indicate standard deviations of three independent experiments.
sbcDC downregulates cap5 genes via arl and mgr.
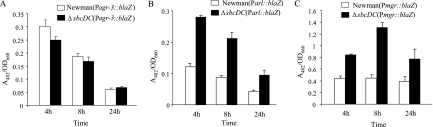
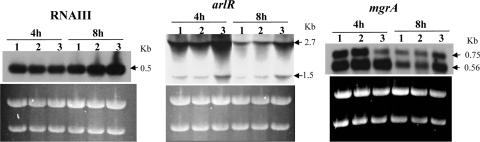
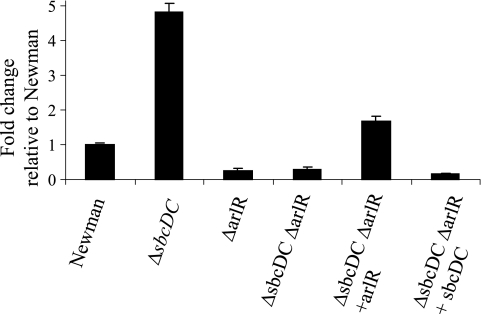
The similarity to other exonucleases and the ability to enhance cell survival of UV irradiation indicate that the sbcDC genes are most likely involved in recombination repair. Their role in capsule gene regulation is, however, not apparent. Since their role in direct gene regulation is not obvious, we considered the possibility of indirect regulation through other regulators. Several regulators have been shown to be involved in capsule gene regulation in our laboratory; these include arlRS, agr, and mgrA (19-21). To test this possibility, we compared the BlaZ reporter activities between Newman and the ΔsbcDC deletion mutant by using transcriptional fusion plasmids containing fusions Parl::BlaZ, Pagr-3::BlaZ, and Pmgr::BlaZ. As shown in Fig. 6, the promoter activities of arlRS and mgrA were increased in the ΔsbcDC mutation (all P values were <0.0075); however, the P3 promoter of agr (from which RNAIII is transcribed) was not affected. To further confirm the fusion results, we employed Northern blotting. The results shown in Fig. 7 confirmed that sbcDC repressed arlRS and mgrA but not RNAIII. It should be noted here that we have also performed reciprocal experiments in which PsbcDC::blaZ fusions were compared in wild-type and agr, mgrA, and arlR mutant strains. We found that none of these regulators affected PsbcDC activity (data not shown). These results suggest that sbcDC acts upstream of arl and mgr in capsule regulation. We have shown previously that arl activates the cap5 genes mostly through mgrA-dependent pathways (19). The above-mentioned results therefore suggest that sbcDC affects capsule through the arl-mgr pathway. To test this hypothesis, we performed genetic epistasis assays in which ArlR or SbcDC was expressed from a multiple-copy plasmid to complement the arlR sbcDC double mutant. The cap5A gene was assayed by real-time RT-PCR. As shown in Fig. 8, we found that the double mutant could be complemented by the plasmid containing the arlR gene (P = 0.0094) but not by the plasmid containing sbcDC. In addition, the double Δarl ΔsbcDC mutant had the same phenotype as the Δarl single mutant (P = 0.065) but a different phenotype than the ΔsbcDC mutant (P = 0.0007). Taken together, these epistasis results suggest that arl acts downstream of sbcDC in the pathway.
FIG. 6.
Promoter activities of the blaZ reporter in Newman and ΔsbcDC mutant CYL6621 containing Pagr-3::blaZ fusion plasmid pTL3564 (A), Parl::blaZ fusion plasmid pTL3562 (B), or Pmgr::blaZ fusion plasmid pTL3580 (C). The BlaZ activities are expressed as the ratio of A482 of the enzymatic reaction to the OD660 of the culture. The error bars indicate standard deviations of at least three independent experiments.
FIG. 7.
Northern blots probed with DNA fragment containing RNAIII, arlR, or mgrA. RNA samples were isolated at 4 h and 8 h from (lanes 1) Newman(pTL3618), (lanes 2) ΔsbcDC mutant complemented strain CYL6621(pZC3624), and (lanes 3) ΔsbcDC mutant CYL6621(pZC3618). rRNA as a loading control is shown below each blot.
FIG. 8.
Epistasis assay using relative real-time RT-PCR analysis of the cap5 mRNA levels in the various strains indicated below the chart (Newman, ΔsbcDC mutant CYL6621, ΔarlR mutant CYL1164, ΔsbcDC ΔarlR mutant CYL6851, ΔsbcDC ΔarlR mutant CYL6851 complemented with arlR in pTL3287, and ΔsbcDC ΔarlR mutant CYL6851 complemented with sbcDC in pZC3624). The changes (n-fold) are expressed relative to the level for Newman, which was arbitrarily set at 1. Error bars indicate standard deviations from at least three independent experiments.
DISCUSSION
Previously, we have shown that sbcD and sbcC were two of seven genes involved in type 5 capsule gene regulation identified by a genetic screen. In this report, we confirmed that both genes were indeed involved in CP5 regulation. We provided the evidence first by using reporter gene fusion analyses to show that mutations in the two genes caused increased cap5 promoter activity and that the regulation was most likely at the transcriptional level. The downregulation of CP5 by sbcDC was then confirmed by assaying capsule production using immunoblotting and analyzing the cap5 mRNA using real-time RT-PCR. Additionally, we showed that the mutant phenotypes could be complemented by providing, in trans, a DNA fragment containing only the sbcC and sbcD open reading frames.
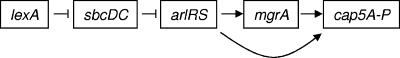
We have shown previously that staphylococcal capsule is regulated positively by agr, mgr, and arl, in which arl acts upstream of mgr in strain Newman but can also regulate capsule independently from mgr (19-21). We now show that sbcDC regulated cap5 by repressing arl and thus mgr but did not affect agr. This conclusion was based on genetic epistasis assays by assessing single and double mutant phenotypes of arlR and sbcDC and by complementation of the double mutant with each gene. Since we have shown previously that arl acts mostly through mgrA to regulate capsule (19), we propose that the signal transduction is transmitted through sbcDC, arl, and mgr (in that order) and finally to cap5. Based on these data, we propose the putative pathway shown in Fig. 9. In this model, activated RecA induced by the SOS response causes LexA to autocleave itself, thereby derepressing the SOS regulon including SbcDC. The increased SbcDC results in reduced ArlRS, MgrA, and CP5. Our results that the cap5 promoter activity was repressed by a subinhibitory concentration of ciprofloxacin or mitomycin C (Fig. 5B) strongly support this hypothesis. It should be noted that the pathway is not complete and is likely to involve additional regulators, particularly downstream of sbcDC. In fact, our preliminary studies indicated that ArlR did not bind directly to the mgrA promoter and MgrA did not bind to the cap5 promoter, suggesting that additional intermediary genes may be involved in the pathway. Thus, the signaling pathway is rather lengthy even without additional players. The lengthy pathway could allow different inputs to the pathway that may reflect various needs for fine tuning capsule production in various lifestyles of S. aureus.
FIG. 9.
Proposed regulatory pathway for cap5 regulation by sbcDC. Arrows indicate positive regulation, and blocked arrows indicate negative regulation.
In E. coli, SbcD and SbcC form a complex that cleaves DNA hairpins (7, 9). It has been proposed that the protein complex could weakly bind double-stranded DNA nonspecifically and migrates to the blocked DNA end for cleavage during DNA repair (8). In Bacillus subtilis, SbcC is involved in DNA interstrand cross-link repair (22). The functions of SbcD and SbcC in other bacteria, including S. aureus, have not been studied. Based on homology, it is most likely that the two S. aureus proteins perform similar DNA repair functions. Our UV survival test supports this notion, though the effect was not observed when the experiments were performed using 4-h cultures. The involvement in DNA damage repair is also consistent with our Northern and reporter gene fusion results showing that the sbcDC genes were induced by ciprofloxacin and mitomycin C, indicating that the sbcDC genes are part of the SOS regulon, which includes genes mostly involved in DNA repair in E. coli (30). Two recent microarray studies (1, 6) also showed that the sbcDC genes were induced in the SOS response in S. aureus. While the involvement of sbcDC in DNA damage repair is expected, our finding that SbcDC acted as a repressor in capsule regulation is surprising. SbcDC has not been implicated as a transcriptional regulator in any organism. Indeed, it is difficult to envision how a DNA damage repair exonuclease can function as a transcriptional regulator. Since SbcDC can bind DNA nonspecifically, perhaps the DNA binding capability endows the protein complex to regulate gene expression. However, we showed that sbcDC repressed arl, mgr, and cap5 but had no effect on agr, suggesting that the regulation is specific and that the nonspecific DNA binding property may not play a role in regulation. Thus, it is more likely that the SbcDC complex exerts its regulatory function not by direct specific DNA binding on the cap5 promoter but by a posttranscriptional mechanism through other regulators. Our results showed that arl and mgr were such intermediary regulators in which arl functions upstream of mgr (Fig. 9). It has been reported previously that DNA supercoiling is involved in arl regulation of protein A (11). Since SbcDC cleaves DNA, which could result in relaxation of DNA supercoils, it is possible that a change in supercoiling could be the mechanism involved in the transduction of signal from sbcDC to arl. However, our results also indicate that SbcDC affects arl mRNA level and promoter activity, suggesting the effect is at the transcriptional level. Further studies are needed to determine whether supercoiling plays a role in the regulation and, if not, what other intermediary regulators are involved.
A recent microarray study by Cirz et al. (6) revealed that agrB and saeRS were downregulated upon the SOS response in S. aureus. The agr locus has been shown to activate the cap5 genes (21), whereas the sae locus has been shown to repress the cap5 genes (29; unpublished data). Since our results outlined above (Fig. 7) showed that RNAIII was not affected by sbcDC, the microarray results of Cirz et al. would suggest that additional pathways may be involved in capsule regulation upon the SOS response. This is not surprising since virulence gene regulation in S. aureus has been shown to be extremely complicated (4, 5, 23). However, it should be noted that strain 8325 was used in the microarray study. This strain is known to be defective in the sigB locus, which has been shown to affect capsule production and other regulatory genes (2, 15, 29). Thus, the microarray results by Cirz et al. may not be applicable to other strains, especially strains with an intact sigB locus, such as Newman used in our study. The different strains used in different laboratories may also explain why arl, mgr, and cap5 genes were not detected in the two microarray studies (1, 6) that profiled SOS response genes (note that the other study by Anderson et al. [1] used the UAMS1 strain). However, the differences in growth conditions may also contribute to the discrepancies.
DNA repair function could be considered a stress response for bacteria. Capsule has been shown to be an important virulence factor for S. aureus, whose expression has been shown to be influenced by various environmental cues, including stress conditions (16, 24). However, regulation of capsule in response to DNA-damaging agents has not been reported. What is the biological significance for the capsule being repressed by the SOS induction? One plausible hypothesis is that capsule biosynthesis is an energy-demanding process. The cells may sense the need for saving energy in order to perform DNA repairs upon the SOS response. However, fibronectin binding protein gene fnbB has been shown to be repressed directly by LexA and is induced by the SOS response in the presence of ciprofloxacin (3). Since reduction of capsule would unmask the cell surface proteins, it is most likely that the fnbB and cap5 genes are regulated in a coordinate fashion upon the SOS response, resulting in an increased capability of adherence for the bacteria. It is not apparent how this increase of adherence property would benefit the bacteria after DNA damage. However, it has been shown that S. aureus can survive intracellularly (reference 28 and references therein). Perhaps by increasing adherence capability the bacteria would promote internalization by the host cells, thereby avoiding DNA-damaging agents.
It is interesting to note that, without SOS induction, the sbcDC genes were expressed very early in the growth cycle but that the expression declined quickly thereafter. The early expression was detected by Northern blotting and manifested by several of our assays, showing that the mutations had the most profound effects at the early growth phase. Although SbcDC is involved in DNA repair after DNA damage, in E. coli or B. subtilis the enzyme is also involved in repairing replication errors (7, 22). Thus, it is likely that the increased expression in the early growth phase could be due to a high DNA replication activity at this growth phase. The finding that sbcDC is expressed early is also consistent with our previous results showing that cap5 and cap8 genes are expressed highly only after the exponential growth phase (19, 21). This temporal regulation of the cap genes is likely due to activation by the cell density-dependent agr system. However, since sbcDC is expressed early and it represses the cap genes, it is tempting to speculate that sbcDC plays a role in the repression of the cap genes at the early growth phase.
Acknowledgments
This work was supported by grant AI37027 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 17 August 2007.
REFERENCES
- 1.Anderson, K. L., C. Roberts, T. Disz, V. Vonstein, K. Hwang, R. Overbeek, P. D. Olson, S. J. Projan, and P. M. Dunman. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188:6739-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischoff, M., P. Dunman, J. Kormanec, C. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. Projan. 2004. Microarray-based analysis of Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisognano, C., W. L. Kelley, T. Estoppey, P. Francois, J. Schrenzel, D. Li, D. P. Lew, D. C. Hooper, A. L. Cheung, and P. Vaudaux. 2004. A RecA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 279:9064-9071. [DOI] [PubMed] [Google Scholar]
- 4.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Cirz, R. T., M. B. Jones, N. A. Gingles, T. D. Minogue, B. Jarrahi, S. N. Peterson, and F. E. Romesberg. 2007. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 189:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connelly, J. C., E. S. de Leau, and D. R. F. Leach. 1999. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 27:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connelly, J. C., E. S. de Leau, and D. R. Leach. 2003. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair 2:795-807. [DOI] [PubMed] [Google Scholar]
- 9.Connelly, J. C., L. A. Kirkham, and D. R. F. Leach. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 95:7969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connelly, J. C., and D. R. F. Leach. 2002. Tethering on the brink: the evolutionarily conserved Mre 11-Rad 50 complex. Trends Biochem. Sci. 27:410-418. [DOI] [PubMed] [Google Scholar]
- 11.Fournier, B., and A. Klier. 2004. Protein A gene expression is regulated by DNA supercoiling which is modified by the ArlS-ArlR two-component system of Staphylococcus aureus. Microbiology 150:3807-3819. [DOI] [PubMed] [Google Scholar]
- 12.Herbert, S., S. W. Newell, C. Y. Lee, K. P. Wieland, B. Dassy, J. M. Fournier, C. Wolz, and G. Doring. 2001. Regulation of Staphylococcus aureus type 5 and type 8 capsular polysaccharides by CO2. J. Bacteriol. 183:4609-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373-376. [Google Scholar]
- 14.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 15.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, C. Y., and J. C. Lee. 2006. Staphylococcal capsules, p. 456-463. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. American Society for Microbiology, Washington, DC.
- 17.Lin, W. S., T. Cunneen, and C. Y. Lee. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 176:7005-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 19.Luong, T. T., and C. Y. Lee. 2006. The arl locus regulates Staphylococcus aureus type 5 capsule via an mgr-dependent pathway. Microbiology 152:3123-3131. [DOI] [PubMed] [Google Scholar]
- 20.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luong, T. T., S. Sau, M. Gomez, J. C. Lee, and C. Y. Lee. 2002. Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect. Immun. 70:444-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mascarenhas, J., H. Sanchez, S. Tadesse, D. Kidane, M. Krisnamurthy, J. C. Alonso, and P. L. Graumann. 2006. Bacillus subtilis SbcC protein plays an important role in DNA inter-strand cross-link repair. BMC Mol. Biol. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novick, R. P. 2006. Staphylococcal pathogenesis and pathogenicity factors: genetics and regulation, p. 496-516. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 24.O'Riordan, K., and J. C. Lee. 2004. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17:218-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouyang, S., S. Sau, and C. Y. Lee. 1999. Promoter analysis of the cap8 operon involved in type 8 capsular polysaccharide production in Staphylococcus aureus. J. Bacteriol. 181:2492-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sau, S., N. Bhasin, E. R. Wann, J. C. Lee, T. J. Foster, and C. Y. Lee. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143:2395-2405. [DOI] [PubMed] [Google Scholar]
- 27.Saumaa, S., A. Tover, M. Tark, R. Tegova, and M. Kivisaar. 2007. Oxidative DNA damage defense systems in avoidance of stationary-phase mutagenesis in Pseudomonas putida. J. Bacteriol. 189:5504-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shompole, S., K. T. Henon, L. E. Liou, K. Dziewanowska, G. A. Bohach, and K. W. Bayles. 2003. Biphasic intracellular expression of Staphylococcus aureus virulence factors and evidence for Agr-mediated diffusion sensing. Mol. Microbiol. 49:919-927. [DOI] [PubMed] [Google Scholar]
- 29.Steinhuber, A., C. Goerke, M. G. Bayer, G. Döring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker, G. C. 1996. The SOS response of Escherichia coli, p. 1400-1416. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 31.Zukowski, M. M., D. F. Gaffney, D. Speck, M. Kauffmann, A. Findeli, A. Wisecup, and J. P. Lecocq. 1983. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc. Natl. Acad. Sci. USA 80:1101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]