Abstract
Secretion of the Haemophilus influenzae HMW1 adhesin occurs via the two-partner secretion pathway and requires the HMW1B outer membrane translocator. HMW1B has been subjected to extensive biochemical studies to date. However, direct examination of the structure of HMW1B has been lacking, leaving fundamental questions about the oligomeric state, the membrane-embedded β-barrel domain, the approximate size of the β-barrel pore, and the mechanism of translocator activity. In the current study, examination of purified HMW1B by size exclusion chromatography and negative staining electron microscopy revealed that the predominant species was a dimer. In the presence of lipid, purified HMW1B formed two-dimensional crystalline sheets. Examination of these crystals by cryo-electron microscopy allowed determination of a projection structure of HMW1B to 10 Å resolution. The native HMW1B structure is a dimer of β-barrels, with each β-barrel measuring 40 Å by 50 Å in the two orthogonal directions and appearing largely occluded, leaving only a narrow pore. These observations suggest that HMW1B undergoes a large conformational change during translocation of the 125-kDa HMW1 adhesin.
Gram-negative bacteria have evolved a number of sophisticated pathways for secreting proteins (19). Among the simplest of these pathways is the two-partner secretion (TPS) system, which consists of an exoprotein termed TpsA and a pore-forming outer membrane (OM) translocator protein termed TpsB (16). Based on experimental data and genome sequence analysis, over 100 TPS members have been identified (16). The TpsB proteins are large (60 to 80 kDa), pore-forming, outer membrane proteins that are predicted to form β-barrels and that translocate the cognate TpsA protein from the periplasm to the bacterial surface (16).
Among the predicted TpsB proteins, the transmembrane topologies of the Serratia marcescens ShlB protein, the Bordetella pertussis FhaC protein, and the Haemophilus influenzae HMW1B protein have been studied. ShlB is involved in secretion and activation of the ShlA hemolysin, a pore-forming cytotoxin. Based on sequence alignments, secondary structure predictions, and characterization of M2 epitope insertions, ShlB is predicted to have 20 β strands with a periplasmic N terminus and two large surface loops (18). FhaC is involved in secretion of filamentous hemagglutinin (FHA), a multifunctional adhesin. Initial work with FhaC suggested that this protein contains 19 transmembrane β strands with a surface-exposed N terminus and forms two large loops on the bacterial surface (12, 15, 22). However, more recent studies suggest that FhaC has a maximum of 16 transmembrane β strands and contains two functional domains, including an N-terminal domain that modulates pore properties and may participate in the recognition of FHA and a 350-amino-acid C-terminal domain that forms the pore (22). HMW1B facilitates surface localization of the HMW1 adhesin, a 125-kDa protein that forms fibers on the bacterial surface and mediates H. influenzae interaction with respiratory epithelium (26). In recent studies, we found that HMW1B has pore-forming activity (28) and contains three domains, namely, a surface-localized N-terminal domain, an internal periplasmic domain, and a C-terminal pore-forming membrane anchor. Based on epitope tagging and cysteine substitution mutagenesis, we concluded that the C-terminal membrane anchor contains 10 β strands (27).
In previous work using single-particle electron microscopy, we observed that HMW1B containing a HAT tag at the N terminus formed predominantly tetramer-like particles in dodecyl-maltoside (DDM) detergent (28). Because the cognate HMW1 adhesin is generally present on the bacterial surface in pairs (6) and because modification such as tagging or fusion can change the oligomerization properties of a protein (21), we elected to reassess the oligomeric state of HMW1B using purified untagged protein. As a first step, the HMW1B gene was amplified by PCR from the plasmid pHMW1-15 and was cloned into pTrc99A, yielding pTrc:HMW1B. Subsequently, cultures of Escherichia coli DH5α harboring pTrc:HMW1B were incubated overnight and then harvested and resuspended in sample buffer (20 mM HEPES [pH 7.4], 150 mM NaCl supplemented with Complete Mini protease inhibitor mixture tablets [Roche]). Bacteria were disrupted by using a French press (Thermo Electron Corp., Milford, MA), and OM proteins were recovered on the basis of sodium N-lauroyl sarcosinate (Sarkosyl) insolubility (7). OM proteins were solubilized with 1% Elugent (Calbiochem) in sample buffer by being rocked overnight at 4°C. The insoluble OM fraction was pelleted by centrifugation at 40,000 × g for 45 min at 4°C. The soluble OM fraction was dialyzed with 0.5% Elugent, 20 mM HEPES (pH 7.4), and 10 mM NaCl and then loaded onto Source 15S and Source 15Q columns (GE Healthcare) in tandem. The sample flowthrough was collected, dialyzed with 0.5% Elugent, 50 mM sodium phosphate (pH 7.0), and 1.2 M ammonium sulfate and applied to a Source 15 Phe column (GE Healthcare) preequilibrated in the same buffer. HMW1B was eluted with a 10-column-volume gradient from 100% 0.5% Elugent, 50 mm sodium phosphate (pH 7.0), 1.2 M ammonium sulfate to 50% 0.5% Elugent, 50 mM sodium phosphate (pH 7.0). Fractions containing HMW1B were pooled and concentrated by using an Amicon Ultra 10 concentrator (Millipore). The concentrated fraction was then further purified by size exclusion chromatography by using a HiPrep 16/60 Sephacryl S-200 HR column (GE Healthcare) equilibrated in 0.5% Elugent, 50 mM sodium phosphate (pH 7.0), 150 mM NaCl. The final HMW1B-containing fractions were concentrated by using an Amicon Ultra 10 concentrator (Fig. 1A).
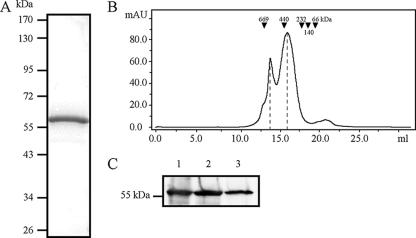
FIG. 1.
Examination of wild-type untagged HMW1B by size exclusion chromatography suggests a dimer in DDM solution. (A) Coomassie blue-stained SDS-PAGE gel of purified HMW1B. (B) Size exclusion chromatographic profile of purified HMW1B. The horizontal axis shows the elution volume, and the vertical axis shows absorbance at 280 nm. Positions for molecular size standards are indicated. The peaks at 16.6 and 14.4 ml correspond to molecular masses of∼300 kDa (dimers) and ∼600 kDa (dimers of dimers), respectively. (C) Western immunoblot of samples from peak 1 and peak 2. Lane 1 is purified HMW1B, lane 2 is a fraction from the 16.6-ml peak, and lane 3 is a fraction from the 14.4-ml peak. The blot was prepared with a guinea pig polyclonal antiserum raised against HMW1B.
Assessment of purified HMW1B on a Superose-6 size exclusion column using DDM in the running buffer revealed two peaks (Fig. 1B). The smaller peak (peak 1) at the elution volume of 14.4 ml had a nominal molecular mass of ∼600 kDa, and the larger peak (peak 2) at the elution volume of 16.6 ml had a nominal molecular mass of ∼300 kDa. The identities of peaks 1 and 2 were confirmed to be HMW1B by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western analysis (Fig. 1C). In the presence of detergent, the apparent mass of a membrane protein can be ∼200 kDa greater than the actual mass, reflecting the surrounding detergent micelle in aqueous solution (21, 30). Taking the effect of detergent into account, we concluded that peak 2 corresponds to a dimer and that peak 1 corresponds to a dimer of dimers or a tetramer.
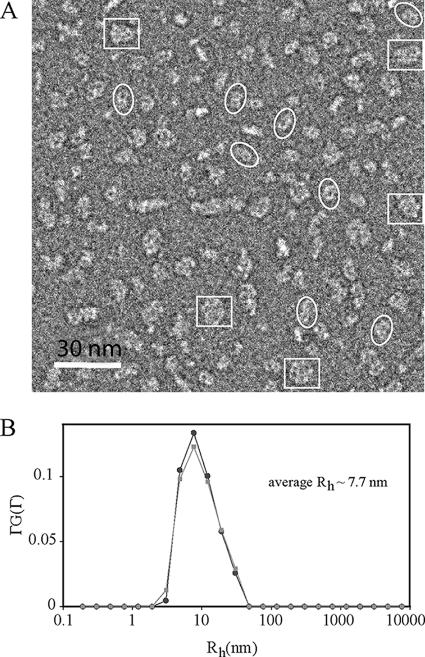
To extend our observations, we examined the HMW1B preparation by negative staining electron microscopy, as described previously (21). As shown in Fig. 2A, we observed two predominant species of particles. The first species was a double ring approximately 100 Å long and 50 Å wide (Fig. 2A, white ellipses). These particles apparently correspond to HMW1B dimers. The second species was a four-ring cluster approximately 100 Å in both dimensions (Fig. 2A, white rectangles). These particles apparently represent an association of two dimers (a dimer of the first species). We further characterized purified HMW1B by dynamic light scattering (DLS), which measures the hydrodynamic radius, Rh, and the size distribution of molecules in solution. DLS measurements were performed using a commercial Brookhaven Instrument light-scattering spectrometer with a BI-9000AT digital correlator. The concentration of HMW1B was ∼1.2 mg/ml. At this protein concentration, light scattering from the empty detergent micelles is insignificant, comprising less than 1% of the total scattering as estimated from control detergent buffer. The CONTIN method (24) was used to analyze the DLS data and calculate the Rh of the particles from the characteristic line width, Γ. The DLS measurements provided information about the particle size distribution in solution from a plot of ΓG(Γ) versus Rh. Data were collected at 25°C, and a viscosity (η) of 1.2 mPas was used. A typical CONTIN analysis from the DLS measurements of purified HMW1B showed a peak with an averaged Rh value of 7.7 ± 0.2 nm and a relatively broad sized distribution (Fig. 2B), suggesting that the majority of the HMW1B preparation is in a dimeric state. The average Rh value measuring 2 nm larger than expected for a dimer and the broad peak probably indicate that the HMW1B solution contains a mixture of detergent micelles and protein-detergent-lipid ternary micelles of different sizes.
FIG. 2.
Analysis of wild-type untagged HMW1B by electron microscopy and dynamic light scattering suggests a dimer in DDM solution. (A) An electron micrograph of HMW1B particles negatively stained with phosphotungstic acid. Electron microscopy was performed with a Jeol-1200EX microscope, and images were recorded on a Gatan 791 charge-coupled device camera. White ellipses mark examples of dimers, and white rectangles mark examples of dimers of dimers. (B) Size distribution of HWM1B in detergent micelles as derived from the CONTIN analysis of the dynamic light scattering measurement. The two curves represent two separate measurements.
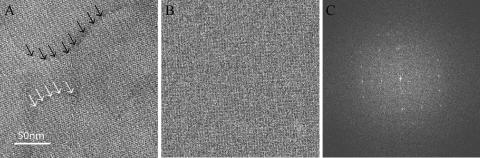
In order to further define the structure of the HMW1B dimer, we reconstituted purified HMW1B in lipid bilayers at low lipid-to-protein ratios (LPR), aiming to generate two-dimensional (2D) crystals. HMW1B was adjusted to a concentration of 2 mg/ml and mixed with detergent-solubilized E. coli polar lipid extract (Avanti Polar Lipids, Inc.) at LPR that ranged from 0.5 to 2.0. The detergent in the mixture was removed by dialysis for 5 days in 20 mM HEPES (pH 6.5), 100 mM NaCl, 5 mM MgCl2, 0.02% NaN3, and 0.1 mM TCEP (17). Following dialysis, examination by electron microscopy revealed micrometer-sized crystalline sheets in preparations with an LPR of 1:2 (wt/wt). While some of these sheets were single layers, many were relatively thick and contained two to five layers. Figure 3A shows a three-layered sheet and demonstrates lattice lines that are continuous as they cross the thickness steps (noted by white arrows and black arrows), indicating that this sheet is an in-register stack of three single-layered 2D crystals, similar in nature to type I three-dimensional membrane protein crystals (23). Figure 3B shows an image of a multilayered sheet preserved in glucose and, again, demonstrates lattice lines that are continuous across the entire crystalline area. Computational Fourier transform of the image in Fig. 3B revealed sharp reflection spots, further supporting the conclusion that the thicker crystalline sheets are in-register stacked 2D crystals (Fig. 3C).
FIG. 3.
Electron microscopy of HMW1B reconstituted in lipid bilayers reveals 2D crystalline sheets. (A) An electron micrograph of an HMW1B crystalline sheet negatively stained with phosphotungstic acid. Electron microscopy was performed with a Jeol-1200EX microscope, and images were recorded on a Gatan 791 charge-coupled device (CCD) camera. The multilayered nature of this sheet is evident from the thickness steps. One such step is indicated by the black arrows, and a second such step is indicated by the white arrows. The lattice lines are continuous as viewed at a glazing angle, even when crossing the thickness steps, indicating in-register stacking between individual layers in the sheet. (B) A cryo-electron micrograph of a single crystalline sheet embedded in glucose. Cryo-electron microscopy was performed with a Jeol-2010F FasTEM microscope, and images were recorded on a Tietz 2K CCD camera at a magnification of ×40,000. (C) Fourier transform of the image in panel B shows periodic and sharp reflections.
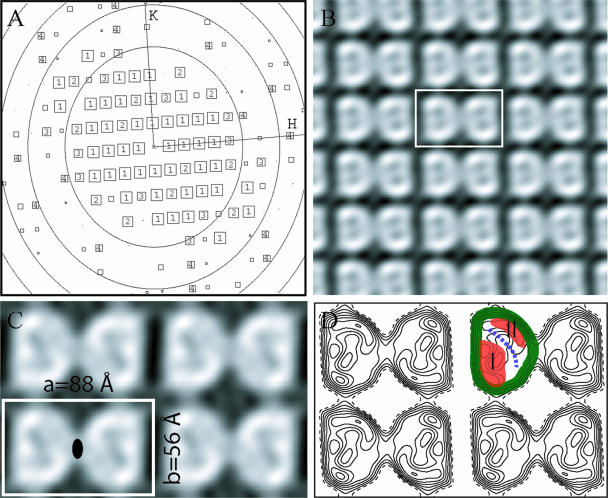
Using the images of 2D crystals, MRC image 2000 software, and a standard image processing procedure, we were able to compute a projection structure of HMW1B (3, 9). We began by digitally correcting electron micrographs for the microscope contrast transfer effect (Fig. 4A), then improved the lattice order by image unbending (14), and then calculated a projection structure for each image. Using this approach, we found that the projection structures calculated from single-layered crystals were virtually the same as the projection structures calculated from the larger and more abundant multilayered sheets. Figure 4B shows the structure calculated from one crystal without applying symmetry. The structural factors from eight images with essentially the same individual projection structures were brought to a common phase origin and were averaged to produce an improved projection structure at ∼10 Å, as shown in Fig. 4C. Figure 4D is a contour plot presentation of the structure shown in Fig. 3C. The crystals had a space group of p2, with the unit cell dimensions a = 88 ± 2 Å, b = 56 ± 2 Å, and γ = 91° ± 2°. The detected twofold symmetry was applied when the averaged structure was calculated (Fig. 4C and 4D). There is a dimer in each unit cell, and each monomer measures ∼40 Å by ∼50 Å in the two orthogonal directions, dimensions that are appropriate for a β-barrel structure with a molecular mass of ∼60 kDa.
FIG. 4.
HMW1B is a dimer when reconstituted in lipid bilayers. (A) A representation of the quality of amplitudes of structure factors derived from the Fourier transform of an image after correction of lattice distortions. The number in each square is the intensity quotient (IQ), i.e., the strength of the spot (13), which ranges between 1 (strongest) and 9 (weakest). The reflections with IQ more than 8 are not shown. The concentric oval rings represent the zero values of the contrast transfer function. The edge of the plot corresponds to a resolution of 8 Å. H and K are reciprocal lattice indices. (B) A gray scale plot of the projection structure of HMW1B calculated from structure factors obtained from one 2D crystal image. The unit cell is marked by a white rectangle. No symmetry was applied, but the twofold symmetry relating the two monomers in a unit cell is clear. (C) The projection structure of HMW1B with p2 symmetry enforced. The cell dimensions are labeled. The filled oval indicates the twofold symmetry axis. (D) Contour plot of the projection structure shown in panel C. The highest density band, corresponding to the projection of the wall of the β-barrel, is colored green. Two lower density regions are orange and are labeled I and II, limiting the low-density pore region, which is indicated by a blue dashed line, to ∼10 Å.
The 2D projection structure of a β-barrel structure is informative because the projection direction is along the axis of the β-barrel, i.e., perpendicular to the lipid membrane. In this projection, the wall of the β-barrel forms an easily recognizable high-density ring, as shown in Fig. 4D by a green band traced along one HMW1B monomer ring. Both the gray scale plot (Fig. 4C) and the contour plot (Fig. 4D) reveal extra densities inside the β-barrel (Fig. 4D, highlighted in orange and labeled I and II). In addition, there is a narrow low-density region that likely represents a channel inside the β-barrel (Fig. 4D, as marked by a blue dashed curved line). Although the β-barrel encloses a large space (30 Å), the channel is only ∼10 Å wide, reflecting the density areas I and II.
In this study, we have shown that the HMW1B outer membrane translocator protein forms a dimeric complex both in solution and in lipid bilayers. Based on the 10-Å projection structure, there is a low-density area consistent with a narrow channel at the center of each subunit in the dimer. This information suggests that HMW1B is a twin-pore complex, analogous to a variety of other protein translocating channels, including the PapC outer membrane usher involved in the assembly of E. coli P pili, the Sec complex responsible for transport of proteins across the bacterial inner membrane and the eukaryotic endoplasmic reticulum, the TIM22 protein insertion complex in the mitochondrial inner membrane, and the TOM protein import complex in the mitochondrial outer membrane, among others (1, 2, 4, 20, 21, 25).
In earlier work, we examined whole bacteria expressing the H. influenzae HMW1 adhesin by using quick-freeze deep-etch transmission electron microscopy and observed that HMW1 forms thin hairlike fibers that are typically present on the bacterial surface in pairs (6). Additional analysis demonstrated that HMW1 is anchored to the bacterial surface via a noncovalent interaction with HMW1B, resulting in a correlation between the quantity of surface-associated HMW1 and the level of HMW1B in the outer membrane (6). Interestingly, HMW1 anchoring to the bacterial surface requires a disulfide bond between two conserved cysteine residues present in the C-terminal 20 amino acids of the protein, a region that appears to be embedded in the pore in HMW1B (6). These observations are consistent with our finding in the current study that HMW1B forms a dimeric complex.
The observation that HMW1B is a dimer contrasts with our earlier analysis suggesting that HAT-tagged HMW1B forms a tetrameric complex. In considering this discrepancy, it is known that an affinity tag can modify the oligomerization behavior of a protein. For example, the PapC usher formed mainly hexamers when a six-His affinity tag was present at the C terminus (29) but formed a dimeric structure in the absence of the His tag, both in detergent and when reconstituted in lipid bilayers (21). We now believe that the tetramer-like particle that we observed when HAT-tagged HMW1B was examined by single-particle electron microscopy was an artifact due to the HAT tagging.
The pore inside the HMW1B β-barrel is partially occluded by density regions on opposite sides of the β-barrel and is thus relatively narrow. These two density regions likely represent the internal periplasmic domain that is postulated to interact with the HMW1 secretion domain and the loops (periplasmic and/or surface) that connect the transmembrane β strands (27). Accordingly, the HMW1B architecture appears to resemble TonB-dependent outer membrane transporters, which are characterized by an N-terminal globular domain that plugs the β-barrel (31). Based on the crystal structures of FepA, BtuB, FecA, and FhuA and molecular dynamic simulation results, one proposal is that the globular plug domain of TonB-dependent outer membrane transporters retracts from the β-barrel for the passage of solute molecules of up to 600 Da in mass (5, 8, 10, 11). With this information in mind, we propose that HMW1B undergoes a major conformational change during translocation of HMW1, perhaps activated by the interaction between the HMW1B periplasmic domain and the HMW1 secretion domain. In particular, the periplasmic domain and some loops in HMW1B may move away from the pore, allowing HMW1 to slide through the open β-barrel. Consistent with this idea, Guedin et al. have noted that the FhaC translocator undergoes a conformational change during FHA secretion (12).
In considering the advantage of a twin-pore complex for HMW1B, it is unclear whether dimerization is essential for HMW1B translocation of HMW1. One possibility is that the formation of a dimer results in a more stable HMW1B structure in the outer membrane, thus increasing the efficiency of translocator activity. Independent of translocator activity per se, it is also possible that the presentation of two physically linked HMW1 fibers facilitates HMW1-mediated adherence to respiratory epithelium, perhaps potentiating a bivalent interaction with the host cell surface. Along these lines, identification of the HMW1 receptor may provide insights into the selective pressures that resulted in the evolution of the HMW1B dimeric structure.
Acknowledgments
This work was supported partially by BNL LDRD grant 06-60 and NIH grant GM75985 to H.L. and by NIH grant DC02873 to J.W.S.G.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Ahting, U., M. Thieffry, H. Engelhardt, R. Hegerl, W. Neupert, and S. Nussberger. 2001. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J. Cell Biol. 153:1151-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahting, U., C. Thun, R. Hegerl, D. Typke, F. E. Nargang, W. Neupert, and S. Nussberger. 1999. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amos, L. A., R. Henderson, and P. N. Unwin. 1982. Three-dimensional structure determination by electron microscopy of two-dimensional crystals. Prog. Biophys. Mol. Biol. 39:183-231. [DOI] [PubMed] [Google Scholar]
- 4.Breyton, C., W. Haase, T. A. Rapoport, W. Kuhlbrandt, and I. Collinson. 2002. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418:662-665. [DOI] [PubMed] [Google Scholar]
- 5.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 6.Buscher, A. Z., S. Grass, J. Heuser, R. Roth, and J. W. St. Geme III. 2006. Surface anchoring of a bacterial adhesin secreted by the two-partner secretion pathway. Mol. Microbiol. 61:470-483. [DOI] [PubMed] [Google Scholar]
- 7.Carlone, G. M., M. L. Thomas, H. S. Rumschlag, and F. O. Sottnek. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J. Clin. Microbiol. 24:330-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10:394-401. [DOI] [PubMed] [Google Scholar]
- 9.Crowther, R. A., R. Henderson, and J. M. Smith. 1996. MRC image processing programs. J. Struct. Biol. 116:9-16. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 12.Guedin, S., E. Willery, J. Tommassen, E. Fort, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2000. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 275:30202-30210. [DOI] [PubMed] [Google Scholar]
- 13.Henderson, R., J. M. Baldwin, T. A. Ceska, F. Zemlin, E. Beckmann, and K. H. Downing. 1990. An atomic model for the structure of bacteriorhodopsin. Biochem. Soc. Trans. 18:844. [DOI] [PubMed] [Google Scholar]
- 14.Henderson, R., J. M. Baldwin, T. A. Ceska, F. Zemlin, E. Beckmann, and K. H. Downing. 1990. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 213:899-929. [DOI] [PubMed] [Google Scholar]
- 15.Jacob-Dubuisson, F., C. El-Hamel, N. Saint, S. Guedin, E. Willery, G. Molle, and C. Locht. 1999. Channel formation by FhaC, the outer membrane protein involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 274:37731-37735. [DOI] [PubMed] [Google Scholar]
- 16.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 17.Jap, B. K., M. Zulauf, T. Scheybani, A. Hefti, W. Baumeister, U. Aebi, and A. Engel. 1992. 2D crystallization: from art to science. Ultramicroscopy 46:45-84. [DOI] [PubMed] [Google Scholar]
- 18.Konninger, U. W., S. Hobbie, R. Benz, and V. Braun. 1999. The haemolysin-secreting ShlB protein of the outer membrane of Serratia marcescens: determination of surface-exposed residues and formation of ion-permeable pores by ShlB mutants in artificial lipid bilayer membranes. Mol. Microbiol. 32:1212-1225. [DOI] [PubMed] [Google Scholar]
- 19.Kostakioti, M., C. L. Newman, D. G. Thanassi, and C. Stathopoulos. 2005. Mechanisms of protein export across the bacterial outer membrane. J. Bacteriol. 187:4306-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkele, K. P., S. Heins, M. Dembowski, F. E. Nargang, R. Benz, M. Thieffry, J. Walz, R. Lill, S. Nussberger, and W. Neupert. 1998. The preprotein translocation channel of the outer membrane of mitochondria. Cell 93:1009-1019. [DOI] [PubMed] [Google Scholar]
- 21.Li, H., L. Qian, Z. Chen, D. Thibault, G. Liu, T. Liu, and D. G. Thanassi. 2004. The outer membrane usher forms a twin-pore secretion complex. J. Mol. Biol. 344:1397-1407. [DOI] [PubMed] [Google Scholar]
- 22.Meli, A. C., H. Hodak, B. Clantin, C. Locht, G. Molle, F. Jacob-Dubuisson, and N. Saint. 2006. Channel properties of TpsB transporter FhaC point to two functional domains with a C-terminal protein-conducting pore. J. Biol. Chem. 281:158-166. [DOI] [PubMed] [Google Scholar]
- 23.Michel, H. 1983. Crystallization of membrane proteins. Trends Biochem. Sci. 8:56-59. [Google Scholar]
- 24.Provencher, S. W. 1976. A Fourier method for the analysis of exponential decay curves. Biophys. J. 16:27-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehling, P., K. Model, K. Brandner, P. Kovermann, A. Sickmann, H. E. Meyer, W. Kuhlbrandt, R. Wagner, K. N. Truscott, and N. Pfanner. 2003. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299:1747-1751. [DOI] [PubMed] [Google Scholar]
- 26.St. Geme, J. W., III, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 27.Surana, N. K., A. Z. Buscher, G. G. Hardy, S. Grass, T. Kehl-Fie, and J. W. St. Geme III. 2006. Translocator proteins in the two-partner secretion family have multiple domains. J. Biol. Chem. 281:18051-18058. [DOI] [PubMed] [Google Scholar]
- 28.Surana, N. K., S. Grass, G. G. Hardy, H. Li, D. G. Thanassi, and J. W. St. Geme III. 2004. Evidence for conservation of architecture and physical properties of Omp85-like proteins throughout evolution. Proc. Natl. Acad. Sci. USA 101:14497-14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanassi, D. G., E. T. Saulino, M. J. Lombardo, R. Roth, J. Heuser, and S. J. Hultgren. 1998. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc. Natl. Acad. Sci. USA 95:3146-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei, Y., H. Li, and D. Fu. 2004. Oligomeric state of the Escherichia coli metal transporter YiiP. J. Biol. Chem. 279:39251-39259. [DOI] [PubMed] [Google Scholar]
- 31.Wiener, M. C. 2005. TonB-dependent outer membrane transport: going for Baroque? Curr. Opin. Struct. Biol. 15:394-400. [DOI] [PubMed] [Google Scholar]