Abstract
MreB is an actin homolog required for the morphogenesis of most rod-shaped bacteria and for other functions, including chromosome segregation. In Caulobacter crescentus and Escherichia coli, the protein seems to play a role in the segregation of sister origins, but its role in Bacillus subtilis chromosome segregation is less clear. To help clarify its role in segregation, we have here studied the protein in Vibrio cholerae, whose chromosome I segregates like the one in C. crescentus and whose chromosome II like the one in E. coli or B. subtilis. The properties of Vibrio MreB were similar to those of its homologs in other bacteria in that it formed dynamic helical filaments, was essential for viability, and was inhibited by the drug A22. Wild-type (WT) cells exposed to A22 became spherical and larger. The nucleoids enlarged correspondingly, and the origin positions for both the chromosomes no longer followed any fixed pattern. However, the sister origins separated, unlike the situation in other bacteria. In mutants isolated as A22 resistant, the nucleoids in some cases appeared compacted even when the cell shape was nearly normal. In these cells, the origins of chromosome I were at the distal edges of the nucleoid but not all the way to the poles where they normally reside. The sister origins of chromosome II also separated less. Thus, it appears that the inhibition or alteration of Vibrio MreB can affect both the nucleoid morphology and origin localization.
Evidence for the evolutionary connection between bacteria and eukaryotes was strengthened by the discovery of homologs of all three types of cytoskeletal proteins of eukaryotes in bacteria (19, 45). The tubulin homolog, FtsZ, a highly conserved bacterial protein, has been known for some time and is essential for cytokinesis (3). The actin homologs MreB and Mbl, recognized first in Bacillus subtilis, are also conserved proteins in rod-shaped bacteria (7, 26). Their similarity to F-actin became clear when the structure of MreB crystals was solved (40, 50). More recently, a homolog of the third major class of cytoskeletal proteins, the intermediate filament proteins, has also been found in Caulobacter crescentus (1). These proteins are considered cytoskeletal elements because they form extended filaments beneath the cytoplasmic membrane and their inactivation by mutation or antibiotics changes the cell shape (4, 21, 45). It is believed that MreB filaments distribute the murein biosynthetic enzymes along the length of the cells and thereby help to maintain their rod shape (7, 14). Among the three cytoskeletal proteins, MreB interests us because of its suggested role in chromosome segregation.
Bacterial chromosomes are highly organized structures, and sister chromosomes segregate to opposite cell halves by active processes (22, 25, 32, 36, 37, 48, 51, 53). However, the equivalent of the microtubules of the eukaryotic mitotic apparatus has not been found in bacteria, which made MreB an attractive candidate for a player in chromosome segregation. In eukaryotes, the microtubules pull sister chromosomes apart by depolymerization. MreB filaments, like microtubules, are also dynamic structures (9, 26, 28). The participation of an actin homolog in bacterial chromosome segregation became clear in studies of the ParM protein of plasmid R1 in E. coli (34). ParM forms filaments that segregate R1 sisters to opposite cell poles in a process that is dependent upon a kinetochore-like structure on the plasmids. Mechanistically, the R1 system remains the best-understood partitioning system in bacteria (18).
The homology with ParM also inspired studies of MreB in the segregation of the E. coli chromosome (29, 30). E. coli tolerates deletion of the mreB gene but with a greatly disturbed cell shape and chromosome segregation pattern (30, 52). The segregation defect was seen even when the cell shape change was minimal. This was achieved by overproducing mutant forms of MreB in WT cells, which elongated the cells without distorting their rod shape. In these cells, loci near the origin and terminus of replication were mislocalized and the nucleoids did not separate, suggesting a direct role of MreB in chromosome segregation. This became more evident in a subsequent study in C. crescentus, which identified a step in the segregation process in which MreB seemed to participate (21). The authors made use of a drug, A22, which specifically and rapidly inactivated the protein. These studies showed that MreB interacts directly or via some other proteins with an origin-proximal DNA sequence (the putative centromere) and separates the sister origins to opposite poles. The protein did not play any role in the separation of the rest of the chromosome. However, in B. subtilis, although an earlier study demonstrated origin segregation defects and the formation of anucleate cells upon the depletion of MreB or another MreB-like protein, Mbl, a later study implied that these defects could be due to cell shape change (8, 17, 46). When the cell shape of ΔmreB cells was maintained by using high concentrations of magnesium and the osmoprotectant sucrose, the chromosome segregation appeared normal (17). Because B. subtilis has, in addition to Mbl, another actin homolog, MreBH, it has been suggested that their functions could be somewhat redundant in chromosome segregation (29, 49). Nevertheless, these studies called into question the results on segregation in other bacteria, which could also be due to an indirect effect of changes in cell shape.
Here we have studied MreB of Vibrio cholerae to determine whether the protein plays a role in the segregation of one or both of its two chromosomes (chrI and chrII). The two chromosomes follow different replication and segregation patterns (13, 16). The origin of chrI (oriI) duplicates at a pole. One of the sister origins stays at the pole of birth, and the other migrates to the opposite pole by an active mechanism requiring the plasmid-type ParABS system (15, 41). This segregation pattern mimics that of C. crescentus (51). The origin of chrII (oriII) duplicates at the cell center, and the sister origins migrate close to cell quarter positions as in E. coli and B. subtilis (16, 55). ChrII also encodes its own Par proteins, but how they function is not known. Although both the chromosomes encode Par proteins, these might not be the only players in the partition process. In plasmids, it is clear that ParB binds to the centromere, and ParA is believed to bear the burden of segregating the sister copies to opposite cell halves (31). In the absence of ParA, the oriI of V. cholerae no longer localizes to the pole, but the localization is not random either, suggesting the participation of other active mechanisms in the segregation process that might include MreB (15, 41). It is also not known whether the ParABS system suffices for the segregation of the centromeric region or whether it also requires help from global mechanisms, such as the DNA condensation, transcription, and transertion that are believed to segregate the bulk of the chromosomal DNA, the nucleoid (49).
We show that the inhibition of MreB or mutations in its coding sequence can affect nucleoid morphology and mislocalize the origins. Particularly revealing was the defect in the positioning of oriI, which no longer localized to the poles in some of the MreB mutants that were nearly normal in cell shape. An authentic polar protein, IcsA, known to be dependent on MreB for its localization, was still polar in the mutants (20, 38, 44). These studies thus support the view that altering MreB can affect chromosome segregation without gross changes in cell morphology.
MATERIALS AND METHODS
Strains, plasmids, and growth medium.
The bacterial strains and plasmids used in this study are listed in Table 1. The primers used in strain and plasmid constructions are listed in Table S1 in the supplemental material. Bacteria were grown in Luria broth (LB).
TABLE 1.
Bacterial strains and plasmids used in the present study
| Strain or plasmid | Relevant description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α(λpir) | Supplies R6K pi protein | M. Waldor |
| SM10(λpir) | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu(λpir) pro endA hsdA hsdR supF | 11 |
| DH5α-T1R | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK−) phoA supE44 thi-1 gyrA96 relA1 tonA | Invitrogen |
| V. cholerae | ||
| CVC116 | N16961 | 23 |
| CVC209 | Strr derivative of N16961 | M. Waldor |
| CVC305 | CVC209 with parS-Kn at 40 kb in chrII | This study |
| CVC1302 | CVC209 with parS-Kn at −90 kb in chrI | This study |
| Plasmids | ||
| pALA2705 | gfp-parB under pTrc promoter, Apr | 35 |
| pDS132 | R6K γori mobRP4 cat sacB Cmr; suicide vector for conjugal transfer and integration | 39 |
| pEM7-Zeo | Source of Zeo cassette | Invitrogen |
| pMAC362 | Para-icsAΔ507-620::gfp | 6 |
| pPS2 | Coordinates 39216 to 41197 of chrII cloned in pDS132 | This study |
| pPS37 | Coordinates 2869408 to 70626 (pullulanase gene) cloned in pDS132 | This study |
| pPS44 | P1parS cloned in pNEB193, Apr | This study |
| pPS46 | parS-Kn cloned at NheI site in pPS37 | This study |
| pPS49 | parS-Kn cloned at NsiI site in pPS2 | This study |
| pPS68 | Modified pALA2705, Apr | This study |
| pPS69 | pPS68 carrying mCherry-mreB gfp-parB Apr | This study |
| pPS75 | Same as pPS96 except for a mutation in mreB (same as in M5) | This study |
| pPS89 | Source of parS-Kn (bounded by iterons) cassette, Apr Knr | This study |
| pPS91 | pPS69 with gfp-parB deleted, Apr | This study |
| pPS96 | mreB under pBAD promoter, Apr | This study |
| pPS106 | Same as pPS96 except for a mutation in mreB (same as in M4) | This study |
| pRE11 | Source of parS (bounded by iterons) cassette, Apr | 10 |
| pRSETb | Source of mCherry | 42 |
| pRFB122 | Source of Knr cassette | 12 |
Chromosomal integration of P1parS.
The P1parS of plasmid pRE11 was used where the locus is bounded by iterons. First, the locus was linked to a kanamycin resistance gene cassette (Kn), obtained from pRFB122 by digestion with HindIII. pRE11 was linearized with StyI and ligated to the Kn cassette after both were blunted with Klenow fragment. The resultant plasmid, pPS89, was used as a source of the parS-Kn cassette.
The parS-Kn cassette was inserted near oriI as follows. PCR primers PS65 and PS66 containing SphI and PstI sites were used to amplify the pullulanase gene (VC2700) from chrI. The PCR product was ligated to SphI- and PstI-digested pDS132 to obtain pPS37. To clone the parS-Kn cassette, the plasmid was digested with NheI, blunted with Klenow fragment, and ligated to the parS-Kn cassette, obtained from pPS89 by digestion with SmaI and ScaI. The resultant plasmid, pPS46, was transferred by conjugation to V. cholerae CVC209, and integrants of parS-Kn without the plasmid backbone were obtained (47). The strains were confirmed by PCR using primers PS70 and PS37, internal to the pullulanase and Kn genes, respectively. One such strain, V. cholerae CVC1302, was used to follow the dynamics of oriI.
The parS-Kn cassette was inserted near oriII in the intergenic region between VCA32 and VCA33, both coding for hypothetical proteins. Approximately 1 kb on either side of the point of insertion was amplified using PCR primers PS20 and PS21 and cloned in the TOPO vector (Invitrogen, Carlsbad, CA). The plasmid was digested with BamHI, and the ends blunted with Klenow fragment followed by digestion with XhoI. One of the digestion products (2 kb) was purified and cloned into pDS132, which was predigested with SacI, blunted with Klenow fragment, and further digested with SalI. The plasmid so constructed was named pPS2. The plasmid was digested with NsiI, blunted with T4 DNA polymerase, and ligated to the parS-Kn cassette, obtained from pPS89 by digestion with SmaI and ScaI. The resultant plasmid, pPS49, was used to transfer the parS-Kn cassette to the V. cholerae CVC209 chromosome. The integration was verified by PCR using primers PS26 and PS37, internal to the VCA33 and Kn genes, respectively. One such strain, V. cholerae CVC305, was used to follow the dynamics of oriII.
Construction of mCherry-MreB.
The mreB gene was amplified from the chromosome using primers PS88 and PS89. The PCR product was digested with AatII and SacII. The mCherry fragment was obtained from pRSETb by PCR using primers PS86 and PS87, containing XhoI and AatII sites, respectively. These fragments were cloned under an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter, pTrc, present in pALA2705 (33). To be able to clone the fragments in the order XhoI-AatII and AatII-SacII between the pTrc and Shine-Dalgarno sequences of the gfp-parB gene, the vector was first modified by using two primer pairs to introduce the XhoI and SacII sites; the first pair, PS82 and PS83, had enzyme sites for EcoRV and XhoI, respectively, and the second pair, PS84 and PS85, had sites for XhoI and SacII (internal) and SacI, respectively. Ligation of the PCR products, after digestion of their ends with cognate enzymes, with the vector backbone (obtained by digesting pALA2705 with EcoRV and SacI and gel purifying the large fragment) generated pPS68, which is identical to pALA2705 except for the two new sites (for XhoI and SacII). pPS68 was digested with XhoI and SacII and ligated to the mCherry and mreB fragments to generate pPS69 (see Fig. S1 in the supplemental material). The order of the elements downstream of pTrc is XhoI-mCherry-AatII-mreB-SacII-gfp-parB-SmaI. The gfp-parB gene was removed from pPS69 by digestion with SacII, end blunting with Klenow fragment, and further digestion with SmaI. The large fragment was gel eluted and self-ligated to get the plasmid pPS91.
Construction of an inducible source of MreB.
The mreB gene of V. cholerae was amplified using PCR primers PS132 and PS133. The PCR product was digested with NheI and SmaI and ligated to plasmid pBAD24, digested with NheI and SmaI, to get the plasmid pPS96. The same primers were used to amplify the mreB genes for mutants M4 and M5, and the products were cloned in pBAD24 as above to get plasmids pPS106 and pPS75, respectively.
Deletion of mreB.
A PCR-based one-step method was developed to delete the mreB gene of V. cholerae and replace it with a zeocin (Zeo) drug cassette. The method is analogous to the Red recombineering system (54) but uses the native recombination system of the host. Briefly, PCR primers were designed to amplify the drug cassette such that they have about 50 bp of homology with the chromosomal DNA at the 5′ end and about 18 to 20 bp of homology with the drug cassette at the 3′ end. These primers (PS232 and PS233) were then used to amplify the Zeo gene from the plasmid pEM7-Zeo. The PCR product was digested with DpnI (to remove the template plasmid) and gel purified. About one μg of the PCR product was electroporated into about 4 × 107 cells containing an mreB-complementing plasmid, pPS96, where the gene was transcribed from an arabinose-inducible promoter. The cells were grown for 2 to 3 h in LB at 30°C to allow time for recombination and drug gene expression (the use of 30°C was incidental). The cells were spread on LB plates containing 100 μg/ml ampicillin for selecting pPS96, 25 μg/ml Zeo for selecting ΔmreB mutants, and 0.2% arabinose for expressing MreB from pPS96. The colonies were subsequently checked for the deletion with PCR using primers PS236, specific for the Zeo gene, and PS127, specific for a chromosomal sequence near the mreB gene. All 40 colonies tested showed a PCR product of the expected length.
Selection of A22-resistant mutants.
We selected A22-resistant mutants by plating 2 × 108 cells on LB plates containing A22 at 10 μg/ml. After the plates were incubated overnight at 37°C, only a few colonies appeared. The colonies were purified in the presence of the drug, and their mreB genes were sequenced. First, the genes were amplified by PCR using primers PS124 and PS127 and the proofreading enzyme platinum Taq polymerase (Invitrogen, Carlsbad, CA). The PCR product was then sequenced either directly or after being cloned in the pCR2.1-TOPO vector (Invitrogen).
Fluorescence microscopy.
Exponentially growing cells were concentrated by centrifugation using a microfuge at 2,500 rpm (600 × g) for 5 min and washed once with 1× PBS containing 1 mM EDTA. Approximately 2.5 μl of cells was placed on a slide and overlaid with a coverslip treated with poly-l-lysine (Sigma-Aldrich, St. Louis, MO) before microscopy (12). The staining of nucleoids was done by incubation with DAPI (4′,6′-diamidino-2-phenylindole) at 50 μg/ml for 10 min at room temperature before sampling the cells on the microscope slide.
Time-lapse microscopy was done using a grooved microscope slide (homemade) that was overlaid with an agarose pad. The slide with the pad was placed on the microscope stage, heated to 37°C. The pad was equilibrated for about an hour with LB containing A22 (10 μg/ml) by continually running the medium through the grooves by gravity flow. About one μl of log-phase cells was placed on the pad and overlaid with an untreated coverslip, and imaging was done every 2 (see Fig. 7) or 10 (see Fig. 4) min.
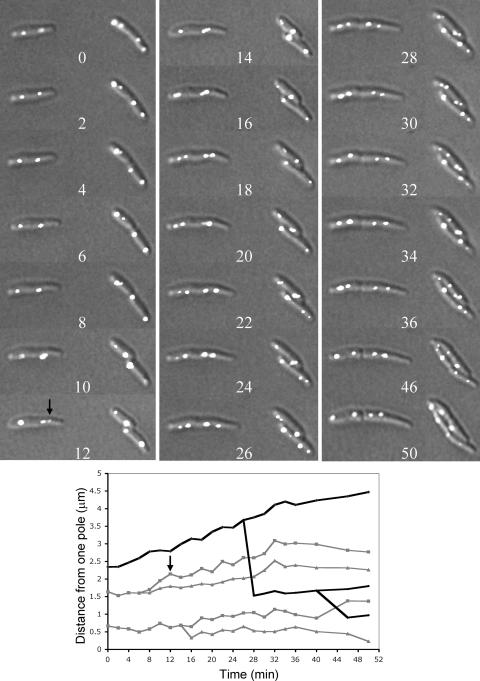
FIG. 7.
Time-lapse microscopy showing positions of oriI at 2-min intervals in A22-resistant cells with mutant M4. In the plot below the micrographs, positions of origins (gray lines) and cell poles (black lines) are shown for the cell lying horizontally. The arrow (12-min time point) marks an origin of that cell which never approached a cell pole in subsequent panels. The branch points of the gray line indicate the times at which the origin foci split. The branch points of the black line mark the time of appearance of invagination of the cell envelope. The data for the 40-min point only appears on the plot and is omitted from the micrographs to save space.
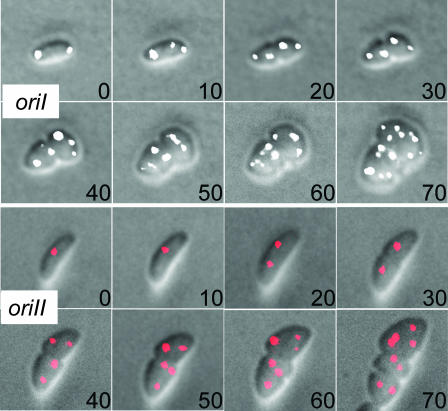
FIG. 4.
Time-lapse microscopy showing cell shape change and origin segregation upon exposure to A22. The cells were grown on an agarose pad by continually feeding them from underneath with fresh medium (LB with 10 μg/ml A22) using a homemade flow cell. The cells were exposed to the drug at time 0 and imaged every 10 min. The oriI foci are shown in white, and oriII foci in red.
Deconvolution microscopy, fluorescence recovery after photobleaching (FRAP) analysis, and image analysis.
Bacterial cells were mounted on LabTek II coverglass chambers (Nalge Nunc International) by placing 2.5 μl of cell suspension under a 1-cm2 slab of 1% agarose containing LB. Using a Nikon TE300 inverted microscope, images were obtained with the imaging system controlled by Metamorph software (Molecular Devices). Images were deconvolved with SoftWoRx software (Applied Precision, Inc.).
Time-lapse single-focal-plane images were obtained by using an automated Olympus IX81 inverted microscope with the imaging system controlled by Metamorph software. Montages and movies of time-lapse images were also obtained with Metamorph software.
FRAP experiments were performed on an LSM510 confocal microscope (Carl Zeiss, Inc). Cells were imaged with a 543-nm laser line from a HeNe laser with acousto-optical tunable filter (AOTF) transmission set to 10%. FRAP bleaching was done with a 488-nm laser line from a 40-mW argon laser, with laser power set to 20%. A single iteration was used for a laser pulse which lasted 23 ms. Fluorescence recovery was monitored every 660 ms for 65 s. The average intensity curves from bleached and nonbleached protein spots were obtained from the FRAP data sets, and the background was subtracted from those curves. To correct for the bleaching due to imaging, the curves for the bleached spots were normalized to the curves of the nonbleached spots. No less than 10 cells were tested in each experiment.
RESULTS
mreB is an essential gene.
To determine if MreB is essential for V. cholerae growth in LB, we attempted to delete the gene and replace it with a Zeo drug cassette. The deletion/substitution was attempted by transformation with a linear DNA. Although Zeor colonies were obtained, they were tiny and failed to grow after restreaking in the presence of the drug. When the deletion was attempted in cells that also carried a plasmid-borne mreB gene (as in pPS96) transcribed from an arabinose-inducible promoter, at least 100 colonies were obtained per μg of electroporated DNA. The colonies were of normal size and morphology, and the cells appeared normal under the microscope. Depletion of MreB by withdrawing arabinose from the medium resulted in spherical cells that continued to enlarge and ultimately lysed. MreB thus appears essential for the viability of V. cholerae. We also succeeded in deleting mreB from cells transformed with a plasmid (pPS91) supplying the mCherry-MreB fusion protein. However, ΔmreB cells with pPS91 were smaller in size than when they carried pPS96, indicating that the mCherry-MreB protein fusion is partially functional or that the fusion protein made from pPS91 was less than optimal in concentration (see Fig. S2 in the supplemental material). MreB is also essential for B. subtilis and C. crescentus, but not for E. coli, although the cell plating efficiency can be significantly lower in ΔmreB derivatives (21, 26, 30).
MreB forms dynamic helical filaments.
We used an mCherry-MreB fusion protein in the WT (mreB+) background to locate the intracellular position of the protein by microscopy. The protein was expressed from the pTrc promoter, which was induced with 100 μm IPTG for 1.5 h before microscopy. The level of induced protein apparently is much less than the physiological level, as the fusion protein band could not be seen by Western analysis (see Fig. S2 in the supplemental material), and the increase in fluorescence upon induction was marginal. The protein appeared mostly as a bright focus either at the cell center or at the poles but could also be more spread out (Fig. 1A). However, as imaged by deconvolution microscopy, it appeared to form a continuous spiral spanning the entire cell length (Fig. 1B; see also the movie in Fig. S3 in the supplemental material). The mean pitch from well-resolved regions of the spirals was about 0.3 ± 0.1 μm (mean ± standard deviation), similar to the values found in E. coli and B. subtilis (26, 30, 44).
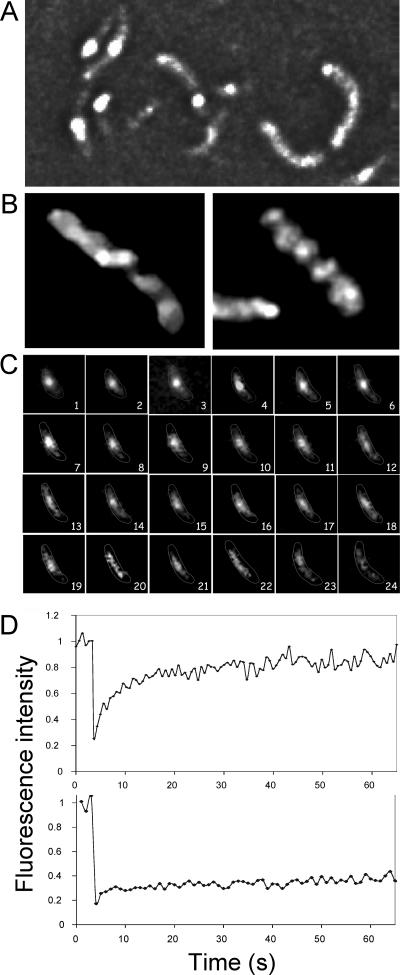
FIG. 1.
mCherry-MreB is a dynamic protein. (A) Localization of mCherry-MreB by fluorescence microscopy in log-phase (unfixed) cells. The protein can be distributed over the entire cell length but is often seen as concentrated in a few spots. (B) Deconvolution microscopy of fixed cells showing that the protein forms a helical filament. (C) Time-lapse microscopy at 1-min intervals showing that the protein concentration along the cell length changes with time. (D, top panel) FRAP. Cells with at least two adjacent fluorescent spots as seen in panel A were chosen, one of which was bleached. The average intensity curve of the bleached spot was normalized to the average intensity curve of the nonbleached spot; both spots were imaged simultaneously. Individual curves were then averaged to obtain the mean curve. (D, bottom panel) FRAP of cells with only one fluorescent spot, which was bleached entirely as described above. To normalize this curve, time-lapse images of unbleached cells were used first to get the average intensity curves of individual cells, which were then averaged, and the resultant mean curve was used for normalization.
To follow MreB localization during the entire cell cycle, time-lapse microscopy was done at the ambient temperature at 1-min intervals without changing the focal plane (Fig. 1C). In the example shown, a bright focus is seen in the cell center at the 1-min time point. At some later times, the focus appeared more diffuse and extended (for example, at 13 min). Although the time-averaged position of MreB was preferentially at the mid cell, the filamentous nature of the protein was obvious with the loss of fluorescence at later time points. These results suggest that the MreB spiral is dynamic.
To further test whether the MreB spiral is dynamic, FRAP analysis was done. Log-phase cells were treated with 10 μg/ml cephalexin for 60 min when the generation time was 20 min. The drug induced cell filamentation, which made bleaching a part of the cell easier. In these cells, mCherry-MreB was seen primarily as bright spots, as in Fig. 1A, and filaments were selected where at least two spots were adjacent to each other. One spot was bleached and its recovery was measured. The unbleached spot was used for normalization purposes. The results from 10 such cells from different experiments were averaged and plotted (Fig. 1D, top panel). About 50% of the fluorescence intensity lost due to bleaching recovered in about 11 s. In other bacteria, depending upon the system, MreB structures were shown to move in a time scale of seconds to minutes (5, 9, 28). When small cells were bleached entirely with exposures identical to those used for filamentous cells, no recovery could be seen during the 65-s interval of the experiment (Fig. 1D, bottom panel). This indicates that the bleaching step inactivated the chromophores in the exposed areas and that the observed recovery is from mCherry-MreB molecules that existed in the unbleached areas rather than from newly synthesized molecules. The FRAP results confirm that the macromolecular structure of mCherry-MreB is dynamic.
A22 affects cell shape and targets MreB.
We first confirmed that MreB is sensitive to the drug A22 and that the inhibition of the protein alters the shape of V. cholerae cells, as has been shown in C. crescentus and in E. coli (21, 29). Increasing concentrations of A22 were used for a fixed period of 1.5 h. Even at the lowest concentration (0.05 μg/ml), the drug was effective in distorting the cell shape (data not shown). At 10 μg/ml A22, all cells were nearly spherical. Removal of the drug after 1.5 h allowed the cells to gradually resume the rod shape, indicating that the A22 effect can be reversible. The cell shape change was followed by time-lapse microscopy.
The effect of the drug on cell growth was also measured. When an overnight culture was diluted to an optical density at 600 nm of 0.005 in LB containing the drug at 10 μg/ml, there was no significant increase in the optical-density value beyond 0.2, while the value in the control culture without the drug reached 1.2. Thus, A22 appears to be an effective inhibitor of V. cholerae growth.
To test whether A22 targets MreB, we selected A22-resistant mutants on LB plates containing the drug at 10 μg/ml. The mutants appeared at a frequency of 10−6 or lower. Sequencing of the mreB gene of the mutants revealed a base change within the gene in all 30 colonies tested (Fig. 2 and Table 2; see also Fig. S4 in the supplemental material). The mutations were mostly but not always in the ATP binding pocket (Fig. 2). Some of the mutations were found repeatedly, and they were at positions also found in A22-resistant mutants of C. crescentus, implying the importance of these positions for the A22-resistant phenotype (21). These results show that MreB is the preferred target of A22, if not the only target.
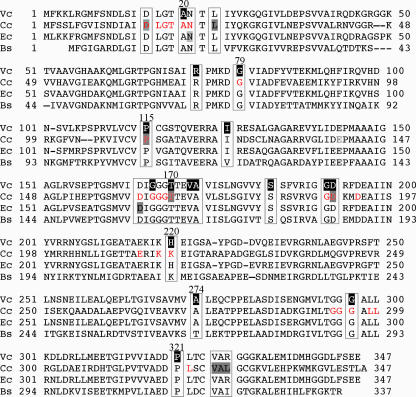
FIG. 2.
Alignment of the amino acid sequence of MreB protein of V. cholerae (Vc) with sequences of its homologs in C. crescentus (Cc), E. coli (Ec), and B. subtilis (Bs). The amino acids altered in A22-resistant mutants of V. cholerae, C. crescentus, and E. coli are shown by making the background black (for V. cholerae) or gray (for C. crescentus and E. coli). The positions of alterations are also boxed for easier identification. The amino acids in red (shown for C. crescentus only) comprise the ATP binding pocket of MreB (21, 50). The positions of alterations may also be seen on a three-dimensional model of MreB in Fig. S4 in the supplemental material.
TABLE 2.
Properties of A22-resistant MreB mutant cellsa
| Strain | Chromosome containing parS | Mutation
|
Length (μm) ± SD
|
|||
|---|---|---|---|---|---|---|
| Name (times found) | Base change | Amino acid change | Cell | Nucleoid | ||
| CVC1302 | chrI | WT | None | None | 1.31 ± 0.28 | 1.25 ± 0.28 |
| CVC305 | chrII | WT | None | None | 1.49 ± 0.26 | 1.50 ± 0.25 |
| CVC1303 | chrI | M1 (2) | G→A | A20T | 0.97 ± 0.18 | 0.88 ± 0.17 |
| CVC1304 | chrII | M2 (2) | C→T | R74C | ND | ND |
| CVC1305 | chrII | M3 | G→A | R74H | 0.95 ± 0.16 | 0.92 ± 0.17 |
| CVC1306 | chrI | M4 (3) | G→A | G79S | 1.67 ± 0.36 | 0.78 ± 0.24 |
| CVC1307 | chrII | M4 (1) | G→A | G79S | 1.56 ± 0.35 | 0.75 ± 0.18 |
| CVC1308 | chrI | M5 (2) | C→T | P115S | 1.34 ± 0.27 | 0.66 ± 0.19 |
| CVC1309 | chrII | M5 (1) | C→T | P115S | 1.31 ± 0.26 | 0.57 ± 0.11 |
| CVC1310 | chrI | M6 (3) | A→C | I126L | ND | ND |
| CVC1311 | chrII | M7 (2) | T→G | I126S | 1.35 ± 0.25 | 1.31 ± 0.25 |
| CVC1312 | chrII | M8 (2) | G→A | G167S | 1.66 ± 0.36 | 0.75 ± 0.28 |
| CVC1313 | chrI | M9 | C→T | T170M | 1.31 ± 0.24 | 0.71 ± 0.15 |
| CVC1314 | None | M10 | T→G | V173G | ND | ND |
| CVC1315 | chrII | M11 | C→A | A174E | ND | ND |
| CVC1316 | chrI | M12 | C→A | S184Y | 0.97 ± 0.17 | 0.49 ± 0.11 |
| CVC1317 | chrI | M13 | G→T | G191C | ND | ND |
| CVC1318 | chrII | M14 | C→G | D192E | ND | ND |
| CVC1319 | None | M15 | A→C | H220P | ND | ND |
| CVC1320 | chrI | M16 | G→A | A274T | ND | ND |
| CVC1321 | chrII | M17 | G→A | G297N | ND | ND |
| CVC1322 | chrII | M18 (2)b | C→T | P321L | ND | ND |
In the mutants shown in bold, the nucleoids were significantly compacted. ND, not determined.
One of the isolates had a second (synonymous) change (G→A, L322L).
We also tested the effect of A22 on DNA replication, since chromosome segregation is intimately tied to it. Replication was checked by replication runout using flow cytometry (27, 47). It appeared that A22 blocks neither replication initiation nor elongation (data not shown). This was also confirmed by counting replication origins by time-lapse microscopy (see Fig. 4).
Origin localization is disturbed upon change of cell shape.
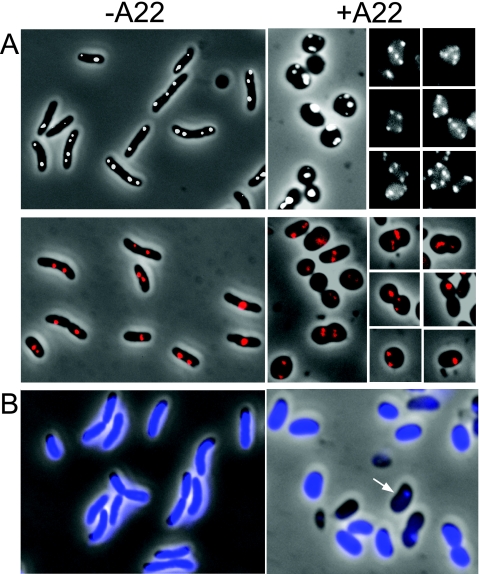
To find if MreB has a role in chromosome segregation, we used two V. cholerae strains, CVC1302 and CVC305, marked with P1parS sites near oriI and oriII, respectively. Before use, the cells were transformed with pPS68 to supply green fluorescent protein (GFP)-ParB so that its binding to parS would fluorescently tag the origin-proximal DNA and allow its mapping. We treated early-log-phase cells with A22 at 10 μg/ml and allowed the culture to grow for about two to three generations before sampling the cells for fluorescence microscopy. The drug treatment made the cells mostly spherical and the positions of the origin foci quite variable for both the chromosomes. The focus positions were not always random, as they were sometimes in a straight line or symmetrically disposed (selected examples are shown in Fig. 3). This suggests that the segregation system may still be partly functional in the presence of A22. However, because of the change in cell shape, it could not be concluded to what extent the origin positions were altered and whether the changes were a direct effect of MreB inhibition or an indirect effect of cell-shape change. The distribution of the numbers of origin foci per cell showed that, upon the drug treatment, the number of cells with multiple foci increased (Table 3). The drug apparently delayed cell division but allowed the replication cycle to continue (also evident in Fig. 4). The cells were filled with the nucleoids, but there was also an increase in the number of anucleate cells from 3 to 24% in the presence of the drug. Most likely, the nucleoid-free cells were initially born without chrII that caused degradation of chrI also due to activation of some toxins of the postsegregational killing systems encoded in chrII that degraded chrI also (55). The formation of anucleate cells has also been reported in E. coli ΔmreB cells (30, 44) and in MreB-depleted B. subtilis cells even when cell shape change was unnoticeable (46).
FIG. 3.
Effect of A22 on cell shape, origin positions, and nucleoid segregation, as determined by fluorescence microscopy. (A) Origins were visualized by tagging with the P1parS-GFP-ParB system. The oriI foci are shown in white, and oriII foci in red. In panels showing cells treated with A22 (+A22), positions of origin foci were variable but not necessarily random, as they sometimes appeared in straight lines or symmetrically disposed with respect to each other (selected examples). −A22, not treated with A22. (B) Formation of anucleate cells upon exposure to A22. The cells were analyzed after staining with DAPI. Cells with DNA are seen as blue, and cells without DNA as black (arrow).
TABLE 3.
Distribution of ori foci in cells grown in LB with and without A22
| Chromosome | Presence of A22 (10 μg/ml)a | % of cells with indicated no. of origin foci
|
||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | ||
| chrI | − | 5 | 17 | 63 | 0 | 15 |
| + | 18 | 26 | 20 | 16 | 20 | |
| chrII | − | 6 | 66 | 27 | 1 | 0 |
| + | 26 | 27 | 30 | 11 | 6 | |
+, present; −, absent.
The effect of A22 on chromosome segregation was also followed by time-lapse microscopy using a flow cell. An exponentially growing culture was grown in the absence of A22, placed over an agarose pad preequilibrated with LB containing 10 μg/ml of A22, and continually fed from underneath with fresh drug-containing medium during the course of the experiment. Distortion of the cell shape and a tendency to round up became apparent by 20 min (i.e., within a generation's time), which made the localization of new poles ambiguous (Fig. 4). The lack of well-defined reference points, such as the poles, also precluded definitive statements on origin localization. By 40 min, the positions of both oriI and oriII in the two sister cells of the same age bore little similarity to each other. It appears that origin mislocalization is simultaneous with cell-shape change, leaving open the possibility that the effect of MreB inhibition on the positioning of both the Vibrio origins could be indirect, due to cell-shape change. Alternatively, MreB could be affecting chromosome segregation that in turn changes the cell shape.
The results of time-lapse microscopy were informative in other respects. They showed that origin numbers continue to increase even without cell division in some cases (apparent in Fig. 4, 70-min panels). This indicates that replication initiation was not disturbed in the distorted cells and that there was no strong tendency for the origins to cohere, as by 70 min, we could discern up to 16 foci for oriI and 8 foci of oriII (Fig. 4). We have shown elsewhere that V. cholerae maintains two to four copies of oriI, as opposed to one to two copies of oriII, when grown in LB (P. Srivastava and D. K. Chattoraj, submitted for publication). The results were thus expected if both the origins underwent three doublings during the 70-min period of the experiment. We conclude that the cell shape-determining activity of MreB is not required for the separation of sister origins.
Chromosome segregation defect in MreB mutants.
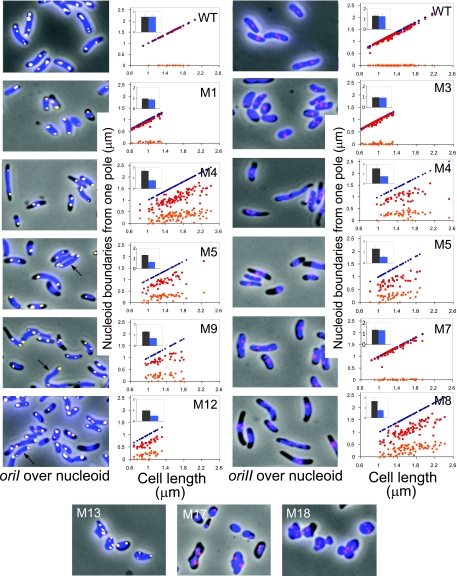
In the A22-resistant mutants studied here, the mutated mreB gene was present in its natural chromosomal position and context. The mutants were characterized for cell and nucleoid morphology and positioning of the two chromosomal origins. Out of 18 different mutants characterized, only cells with mutant M7 appeared normal (Table 2; see also Fig. S4 in the supplemental material). Thus, it is possible to acquire A22 resistance with a single amino acid change without any apparent change in cell or nucleoid morphology. This was also true for the MreB221 mutant of E. coli (29). In the remaining mutants, cell shape was variously affected (Fig. 5). Changes at the C-terminal end of the protein (mutants M13 to M18) affected cell shape the most. For the lack of easily recognizable poles, these mutants were not characterized any further. Several mutants were still rod-shaped or nearly so, although their average width and length could be different from those of the WT cells. The cell lengths of cells with M1, M3, and M12 were significantly shorter (average lengths were about 1 μm compared to about 1.3 to 1.5 μm for the WT; Fig. 5, insets, and Table 2). In several mutants, the nucleoids did not span the entire cell length and were mostly away from both the poles (cells with M4, M5, M8, M9, and M12 (Fig. 5). The average distance from a pole to the nearer nucleoid boundary was 0.19, 0.18, and 0.08 in fractional cell length at one end and 0.34, 0.39, and 0.47 at the other for cells with M4, M5, and M8, respectively. The nucleoids were thus mostly asymmetrically located in the cell. In these mutants, one of the nucleoid boundaries was only at the pole in about 7% of the cells (about 250 cells were measured).
FIG. 5.
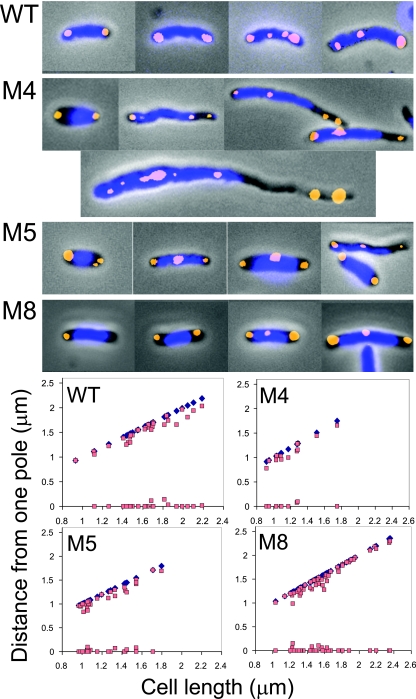
Mapping of nucleoid boundaries with respect to cell poles in A22-resistant cells with mutants of MreB. The nucleoids were stained with DAPI and are shown in blue. The pictures also show oriI (yellow spots) and oriII (red spots), and plots of their positions are shown in Fig. 6. The mutants were named M1 to M18 (Table 2). In the plots shown here, the positions of nucleoid boundaries are shown in red and orange symbols, and the distal cell poles as dark blue symbols. The proximal cell pole is placed on the abscissa, and it is chosen as the pole to which a nuclear boundary was closer. The plots were not made for cells with M13, M17, and M18 (bottom panels) because their poles could not be identified unambiguously. The insets show the average sizes of cells (black bars) and nucleoids (blue bars). Strains with M5, M9, and M12 mutants had mixtures of cells with either compacted nucleoids or normal nucleoids (arrow) that spanned the entire cell volume as in WT cells. The plots for these mutants include only the compact nucleoids. Cells with other mutants were more homogeneous in terms of nucleoid compaction and were not sorted for plotting purposes.
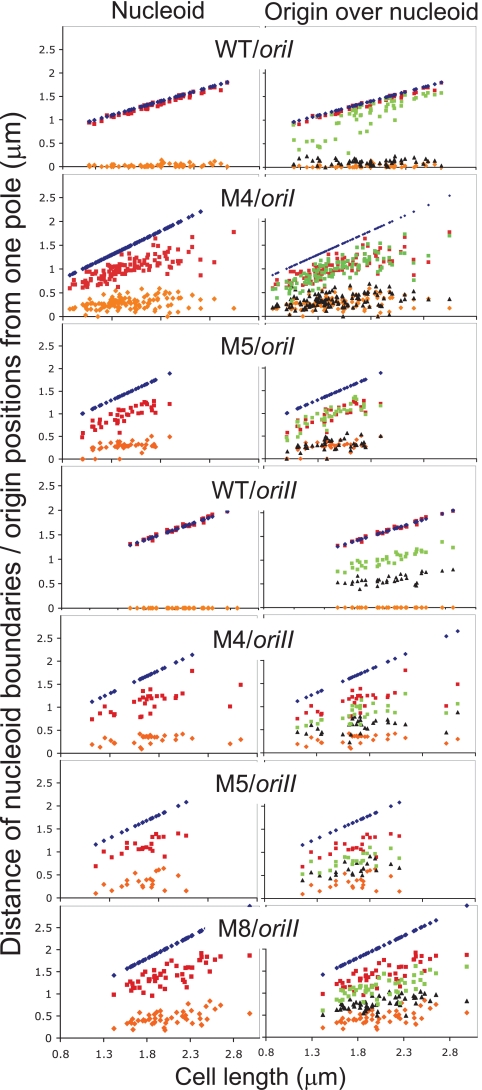
M4 and M5 were chosen for further study because the same mutation was independently found in both oriI- and oriII-marked cells, allowing the study of the effect of the same MreB change on the two origins. In some cases, M8 was also studied, because the position of the amino acid change was close to position 165 of E. coli MreB, where changes made the protein dysfunctional (Fig. 2) (28). Simultaneous localization of oriI and the nucleoid showed that the origin was also no longer at the poles but resided mostly at the poleward edges of the nucleoid (Fig. 5 and 6). In two-focus cells, the mean separation between the oriI foci (in fractional cell length) was reduced from 0.7 in WT cells to about 0.4 in cells with mutants M4 and M5 (Table 4). The distance between the nucleoid boundaries along the long axis of the cells was reduced from 0.95 to about 0.5, respectively. The reduction in oriI separation thus appears correlated with the degree of nucleoid compaction. The position of oriII was mostly at the center of the nucleoid and did not appear to be grossly mislocalized, although the separation between the sister origins was significantly less than in the WT (Fig. 6; Table 4). We conclude that both the origins do not separate optimally in cells with MreB mutants where the nucleoids appear compacted.
FIG. 6.
Mapping of origin foci and nucleoid boundaries with respect to cell poles in WT cells and cells with MreB mutants M4, M5, and M8. Other details are as described in the Fig. 5 legend. Only the cells with two origin foci (green and black) were included in the plot.
TABLE 4.
Origin separation and nucleoid compaction in cells with MreB mutantsa
| Strain | oriI separation | Nucleoid size | oriII separation | Nucleoid size |
|---|---|---|---|---|
| WT | 0.70 ± 0.21 | 0.95 ± 0.04 | 0.26 ± 0.07 | 0.97 ± 0.04 |
| M4 | 0.42 ± 0.16 | 0.50 ± 0.02 | 0.15 ± 0.07 | 0.48 ± 0.08 |
| M5 | 0.44 ± 0.10 | 0.48 ± 0.07 | 0.12 ± 0.05 | 0.48 ± 0.08 |
The values are fractional cell lengths ± standard deviations.
To test whether oriI could be found at the poles at least temporarily, time-lapse microscopy was done using M4. However, imaging at 2-min intervals gave no indication of oriI reaching the pole transiently (Fig. 7). The fact that oriI was still localized at the poleward edges of the nucleoid and not positioned randomly (Fig. 5) suggests that the active partitioning of oriI was essentially functional in this mutant.
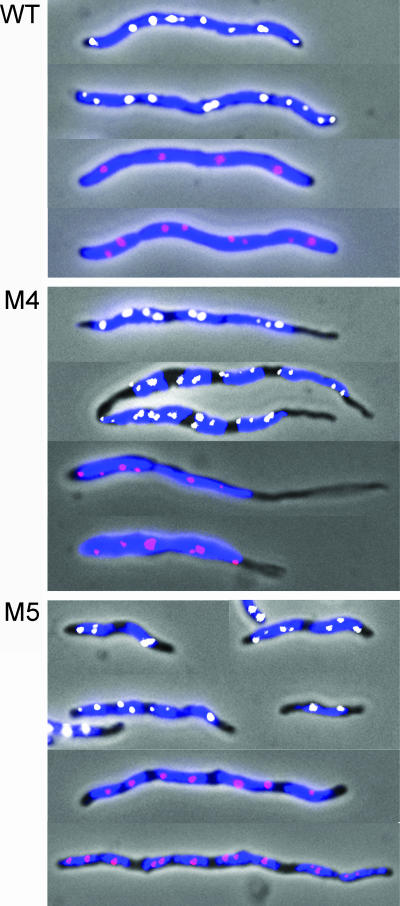
To get more insight on oriI localization and nucleoid condensation, the mutants were studied after treatment with cephalexin for about two generations (total, 40 min). In E. coli, the drug blocks cell division, which elongates cell length without blocking other cell cycle events (30). Our expectation was that chromosome segregation defects would be more pronounced in elongated cells. The cells elongated as expected and were filled with the nucleoids in the case of the WT strain (Fig. 8). The nucleoid separation thus appeared normal. However, in the mutants, instead of forming a continuous body, the nucleoids separated into discrete units, supporting the view that they are more compacted. Both the origins were also found along the entire filament and within the nucleoids in all cases. The poles were still devoid of DNA, as in cells not treated with cephalexin. The results thus support the view that the extension to the poles of both nucleoids and oriI is particularly affected in the mutants.
FIG. 8.
Origin and nucleoid positions in cephalexin-treated WT cells and cells with MreB mutants M4 and M5. The nucleoids are shown in blue, oriI foci in white, and oriII foci in red. Note that although individual nucleoids are more compact in the mutants (evident from the presence of nucleoid-free zones), they nonetheless segregate and span the entire cell length except for the polar regions. Two to four oriI foci and one to two oriII foci were expected per nucleoid.
Secondary mutations are not responsible for MreB mutant phenotypes.
We found that M4 became drug sensitive when streaked a few times in the absence of A22. The mutant phenotypes, however, were the same when M4 cells were grown for about 10 generations in the absence of the drug before microscopy. Although other mutants were not checked similarly, to prevent the possible accumulation of revertants, the results reported here were obtained in the presence of A22 for all the mutants.
To further rule out the possibility that secondary mutations could be contributing to the phenotypes reported here, we took two different approaches. First, we deleted the mreB gene anew in the presence of complementing plasmids carrying either the WT mreB gene (pPS96) or the mutant genes for M4 or M5 (pPS106 and pPS75, respectively). The genes were expressed by induction with arabinose. The efficiency of deletion was comparable in the three cases: after transformation with linear DNA and overnight growth, about 10 to 50 colonies of heterogeneous sizes appeared, irrespective of the type of complementing plasmid. As before, only the big colony formers were found to have the mreB gene deletion. The similar numbers and growth of the colonies in the three cases suggest that additional mutations are not required for the growth of the two mutants tested. Cultures grown from purified colonies (i.e., after two cycles of overnight growth, ∼40 generations) showed nucleoid condensation only when the two mutant MreBs were present (data not shown).
In the second approach, WT (mreB+) cells were transformed with the above three plasmids (pPS96, pPS106, and pPS75) and maintained in the presence of glucose (0.2%). For microscopy, single colonies from LB-glucose plates were inoculated into LB-arabinose (0.2%) medium and the culture was grown to log phase (optical density at 600 nm, ∼ 0.2). In such cultures, nucleoid condensation (for M4 and M5) and cells with tapered ends (for M4) were conspicuous in the majority of the cells (data not shown). These results suggest that the mutant mreB genes are directly responsible for the mutant phenotypes. We note that A22 was not used in these experiments. Thus, although the mutations were originally selected in the presence of A22, its continued presence appears unnecessary for the phenotypes of M4 and M5.
Polar localization of IcsA-GFP in mreB mutants.
Since oriI did not locate to the pole, we wanted to know if the pole is the normal localization of known polar proteins. Towards this end, we examined the localization of IcsA-GFP, which has been shown to depend on MreB to localize to the poles in different bacteria, including V. cholerae (6, 24). In our MreB mutants characterized for oriI localization, the fusion protein could be found at polar zones devoid of nucleoids, suggesting that polar localization is not generally affected (Fig. 9). It is also known that IcsA localizes to the pole in anucleate cells, indicating that its localization is independent of chromosome segregation (24). Thus, the role of MreB in nucleoid and origin segregation need not be identical to its role in the positioning of polar proteins.
FIG. 9.
Localization of a polar protein, IcsA, in cells with MreB mutants M4, M5, and M8. The nucleoids are shown in blue, and the GFP-fused IcsA foci in brown. In some examples, spontaneously elongated (not cephalexin treated) cells were selected to show that IcsA could be polar even where nucleoids were far removed from the pole. Plots show positions of IcsA (brown squares) in cells with two foci in an otherwise unselected field of cells. The abscissa positions the poles chosen for distance measurements. The distal poles are shown in blue diamonds.
DISCUSSION
MreB is a well-conserved actin-like protein in rod-shaped bacteria that not only plays a crucial role in determining cell shape but also has important roles in cell division, cell polarity, chromosome segregation, and the organization of membranous organelles (45). Here we have studied the dynamics of the protein in live cells and have determined the consequences of inhibition or changes in the amino acids of the protein in the segregation of the two V. cholerae chromosomes. The V. cholerae MreB formed dynamic filaments whose characteristics were similar to those of its homologs in other bacteria, particularly B. subtilis and C. crescentus, that have been studied more extensively. This is expected, since V. cholerae MreB is 55% and 61% identical to its homologs in B. subtilis and C. crescentus, respectively (Fig. 2). The protein is also 89% identical to E. coli MreB.
The V. cholerae mreB was found to be an essential gene. This necessitated studies of the protein by short-term inhibition with the drug A22 or by using missense mutants isolated as resistant to A22. Some of these mutants retained their rod shape enough that the poles could be unambiguously recognized. The poles were also normal for the localization of a polar protein, IcsA. In some of these cells, the nucleoid often appeared more compacted and the origin of chrI was no longer at the cell pole, its normal address during most of the cell cycle. The separation of the sister origins of chrII was also reduced significantly. The extent of reduction in origin separation for both the chromosomes, which are partitioned by two different Par systems, correlated with the degree of nucleoid compaction (Table 4). This suggests that the nucleoid compaction could have caused the defects. However, as discussed below, it is equally possible that the origin segregation defects, particularly of chrI, made the nucleoids more compact.
The partitioning of chromosomal origins is likely to involve several steps. It is believed that sister chromosome cohesion that must be overcome to separate the origins occurs in bacteria as in eukaryotes. The origins then need to be moved to their subcellular addresses. Finally, after arrival, the origins need to be retained there during most of the cell cycle. In the MreB mutants studied here, the oriIs were well separated, poleward directed, and made it all the way to the boundaries of the condensed nucleoid. It appears that only the later stages of origin segregation were affected. It is possible that MreB could be required only for anchoring oriI to the pole. The origin anchor could also be required to extend the nucleoid to the pole and control its volume. On the other hand, the polar anchoring of oriI may not be defective per se but the origins were not polar because the increased force of nucleoid condensation either did not allow the origin to extend to the pole or pulled it away from the pole. Time-lapse microscopy, however, did not indicate that the origins first went to the pole but then retracted back to the nucleoid (Fig. 7). The causal relationship of nucleoid compaction and origin segregation defects thus remains to be understood.
In V. cholerae, polymers of ParA of chrI cover a significant fraction of the cell length and appear to move the origins to the pole as do microtubules in eukaryotic anaphase. It is possible that ParA polymers span the gap between the pole and oriI and provide the polar directionality to the origin positions (15). The localization of ParA polymers in the MreB mutants could provide further insights on how oriI achieves polar directionality without ever reaching the pole.
From the studies on MreB, some inferences can be drawn on the relationship between the segregation of the origin and the bulk chromosomal DNA (nucleoid). The inhibition of MreB by A22 in C. crescentus only affected the origin separation; when added after the origins had separated, the drug had no effect on the segregation of the rest of the chromosome (21). The results indicate that the requirements for segregation of the origin-proximal DNA and the nucleoid are not the same. The segregation of the two could also be uncoupled genetically. In E. coli rpoC(Ts) and rpoD(Ts) mutants, origin segregation, but not nucleoid segregation, was normal at the nonpermissive temperature, indicating that only the latter requires help from transcription (29). In elongated (cephalexin treated) E. coli, A22 affected the separation of both the sister origins and the nucleoid, but apparently in opposite ways (29). The sister origins stayed cohered, but the nucleoids were enlarged, in that they appeared as a continuous body filling up the entire cell volume rather than separating into discrete bodies. In V. cholerae, as in E. coli, A22 did not restrain the nucleoid from covering the entire cell volume (Fig. 3B). However, in contrast to E. coli and C. crescentus, the sister origins did separate in V. cholerae (19, 28). Overall, these results suggest that the origin-proximal (“centromeric”) DNA is treated differently than the bulk of the nucleoid for segregation purposes.
In contrast to A22-treated WT cells, the nucleoids appeared more compact in the presence of MreB mutants in both E. coli and V. cholerae (Fig. 5) (29). Thus, MreB might play a role in controlling the nucleoid volume. When MreB filaments depolymerize upon exposure to A22, the nucleoids apparently expand unhindered, but they could not get to their normal volume in the mutants, possibly due to resistance from dysfunctional filaments. In either case, when the nucleoid morphology changed, so did the origin segregation. The apparent coupling of the two processes both in E. coli and in V. cholerae raises the possibility that the bulk chromosome segregation contributes to the origin segregation.
Although a lingering concern in studies of MreB relating to chromosome segregation has been that the role of the protein could be indirect, through its role in cell shape, the opposite possibility of chromosome segregation causing the cell shape change cannot be ruled out either. For example, the nucleoid expansion could be making the cells spherical (Fig. 3B) or nucleoid compaction could be making the cells smaller (Fig. 5, M1). In several A22-resistant mutants, the cell diameter was not uniform across the cell length and the nucleoid-free regions were often thinner, as if the nucleoid forced the diameter to increase (Fig. 8 and 9). The nucleoid normally is a highly compact structure (43), and the possibility remains that its relaxation might impact the cell shape, in addition to the role it plays to block cytokinesis (2).
Supplementary Material
Acknowledgments
We thank Maria Sandkvist for the generous supply of MreB antibody and Richard Fekete for designing the flow cell for optical microscopy. We also thank our lab members for much help and Michael Yarmolinsky, Rotem Edgar, Michael Lichten, Zemer Gitai, and the anonymous reviewers for many thoughtful comments.
The Intramural Research Program of the Center for Cancer Research, NCI, NIH, supported this work.
Footnotes
Published ahead of print on 17 August 2007.
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Ausmees, N., J. R. Kuhn, and C. Jacobs-Wagner. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705-713. [DOI] [PubMed] [Google Scholar]
- 2.Bernhardt, T. G., and P. A. de Boer. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi, E. F., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 4.Briegel, A., D. P. Dias, Z. Li, R. B. Jensen, A. S. Frangakis, and G. J. Jensen. 2006. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol. Microbiol. 62:5-14. [DOI] [PubMed] [Google Scholar]
- 5.Carballido-Lopez, R., and J. Errington. 2003. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev. Cell 4:19-28. [DOI] [PubMed] [Google Scholar]
- 6.Charles, M., M. Perez, J. H. Kobil, and M. B. Goldberg. 2001. Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio. Proc. Natl. Acad. Sci. USA 98:9871-9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 8.Defeu Soufo, H. J., and P. L. Graumann. 2005. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Defeu Soufo, H. J., and P. L. Graumann. 2004. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 5:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar, R., D. K. Chattoraj, and M. Yarmolinsky. 2001. Pairing of P1 plasmid partition sites by ParB. Mol. Microbiol. 42:1363-1370. [DOI] [PubMed] [Google Scholar]
- 11.Egan, E. S., and M. K. Waldor. 2003. Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114:521-530. [DOI] [PubMed] [Google Scholar]
- 12.Fekete, R. A., and D. K. Chattoraj. 2005. A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol. Microbiol. 55:175-183. [DOI] [PubMed] [Google Scholar]
- 13.Fiebig, A., K. Keren, and J. A. Theriot. 2006. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol. Microbiol. 60:1164-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 15.Fogel, M. A., and M. K. Waldor. 2006. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 20:3269-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogel, M. A., and M. K. Waldor. 2005. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol. Microbiol. 55:125-136. [DOI] [PubMed] [Google Scholar]
- 17.Formstone, A., and J. Errington. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol. Microbiol. 55:1646-1657. [DOI] [PubMed] [Google Scholar]
- 18.Garner, E. C., C. S. Campbell, D. B. Weibel, and R. D. Mullins. 2007. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315:1270-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gitai, Z. 2005. The new bacterial cell biology: moving parts and subcellular architecture. Cell 120:577-586. [DOI] [PubMed] [Google Scholar]
- 20.Gitai, Z., N. Dye, and L. Shapiro. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA 101:8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 22.Gordon, G. S., D. Sitnikov, C. D. Webb, A. Teleman, A. Straight, R. Losick, A. W. Murray, and A. Wright. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 23.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janakiraman, A., and M. B. Goldberg. 2004. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc. Natl. Acad. Sci. USA 101:835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen, R. B., and L. Shapiro. 1999. Chromosome segregation during the prokaryotic cell division cycle. Curr. Opin. Cell Biol. 11:726-731. [DOI] [PubMed] [Google Scholar]
- 26.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 27.Kim, M. S., S. H. Bae, S. H. Yun, H. J. Lee, S. C. Ji, J. H. Lee, P. Srivastava, S. H. Lee, H. Chae, Y. Lee, B. S. Choi, D. K. Chattoraj, and H. M. Lim. 2005. Cnu, a novel oriC-binding protein of Escherichia coli. J. Bacteriol. 187:6998-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S. Y., Z. Gitai, A. Kinkhabwala, L. Shapiro, and W. E. Moerner. 2006. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 103:10929-10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruse, T., B. Blagoev, A. Lobner-Olesen, M. Wachi, K. Sasaki, N. Iwai, M. Mann, and K. Gerdes. 2006. Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli. Genes Dev. 20:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruse, T., J. Moller-Jensen, A. Lobner-Olesen, and K. Gerdes. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemonnier, M., J. Y. Bouet, V. Libante, and D. Lane. 2000. Disruption of the F plasmid partition complex in vivo by partition protein SopA. Mol. Microbiol. 38:493-505. [DOI] [PubMed] [Google Scholar]
- 32.Lewis, P. J., and J. Errington. 1997. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein. Mol. Microbiol. 25:945-954. [DOI] [PubMed] [Google Scholar]
- 33.Li, Y., K. Sergueev, and S. Austin. 2002. The segregation of the Escherichia coli origin and terminus of replication. Mol. Microbiol. 46:985-996. [DOI] [PubMed] [Google Scholar]
- 34.Moller-Jensen, J., R. B. Jensen, J. Lowe, and K. Gerdes. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen, H. J., Y. Li, B. Youngren, F. G. Hansen, and S. Austin. 2006. Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 61:383-393. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen, H. J., J. R. Ottesen, B. Youngren, S. J. Austin, and F. G. Hansen. 2006. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 62:331-338. [DOI] [PubMed] [Google Scholar]
- 37.Niki, H., and S. Hiraga. 1998. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 12:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsen, T., A. W. Yan, G. Gale, and M. B. Goldberg. 2005. Presence of multiple sites containing polar material in spherical Escherichia coli cells that lack MreB. J. Bacteriol. 187:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Philippe, N., J. P. Alcaraz, E. Coursange, J. Geiselmann, and D. Schneider. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246-255. [DOI] [PubMed] [Google Scholar]
- 40.Roeben, A., C. Kofler, I. Nagy, S. Nickell, F. U. Hartl, and A. Bracher. 2006. Crystal structure of an archaeal actin homolog. J. Mol. Biol. 358:145-156. [DOI] [PubMed] [Google Scholar]
- 41.Saint-Dic, D., B. P. Frushour, J. H. Kehrl, and L. S. Kahng. 2006. A parA homolog selectively influences positioning of the large chromosome origin in Vibrio cholerae. J. Bacteriol. 188:5626-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 43.Sherratt, D. J. 2003. Bacterial chromosome dynamics. Science 301:780-785. [DOI] [PubMed] [Google Scholar]
- 44.Shih, Y. L., I. Kawagishi, and L. Rothfield. 2005. The MreB and Min cytoskeletal-like systems play independent roles in prokaryotic polar differentiation. Mol. Microbiol. 58:917-928. [DOI] [PubMed] [Google Scholar]
- 45.Shih, Y. L., and L. Rothfield. 2006. The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 70:729-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soufo, H. J., and P. L. Graumann. 2003. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr. Biol. 13:1916-1920. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava, P., R. A. Fekete, and D. K. Chattoraj. 2006. Segregation of the replication terminus of the two Vibrio cholerae chromosomes. J. Bacteriol. 188:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teleman, A. A., P. L. Graumann, D. C. Lin, A. D. Grossman, and R. Losick. 1998. Chromosome arrangement within a bacterium. Curr. Biol. 8:1102-1109. [DOI] [PubMed] [Google Scholar]
- 49.Thanbichler, M., and L. Shapiro. 2006. Chromosome organization and segregation in bacteria. J. Struct. Biol. 156:292-303. [DOI] [PubMed] [Google Scholar]
- 50.van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 51.Viollier, P. H., M. Thanbichler, P. T. McGrath, L. West, M. Meewan, H. H. McAdams, and L. Shapiro. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. USA 101:9257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wachi, M., M. Doi, S. Tamaki, W. Park, S. Nakajima-Iijima, and M. Matsuhashi. 1987. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 169:4935-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, X., X. Liu, C. Possoz, and D. J. Sherratt. 2006. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 20:1727-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaichi, Y., M. A. Fogel, and M. K. Waldor. 2007. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 104:630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.