Abstract
Infection of Escherichia coli by Shiga toxin-encoding bacteriophages (Stx phages) was the pivotal event in the evolution of the deadly Shiga toxin-encoding E. coli (STEC), of which serotype O157:H7 is the most notorious. The number of different bacterial species and strains reported to produce Shiga toxin is now more than 500, since the first reported STEC infection outbreak in 1982. Clearly, Stx phages are spreading rapidly, but the underlying mechanism for this dissemination has not been explained. Here we show that an essential and highly conserved gene product, YaeT, which has an essential role in the insertion of proteins in the gram-negative bacterial outer membrane, is the surface molecule recognized by the majority (ca. 70%) of Stx phages via conserved tail spike proteins associated with a short-tailed morphology. The yaeT gene was initially identified through complementation, and its role was confirmed in phage binding assays with and without anti-YaeT antiserum. Heterologous cloning of E. coli yaeT to enable Stx phage adsorption to Erwinia carotovora and the phage adsorption patterns of bacterial species possessing natural yaeT variants further supported this conclusion. The use of an essential and highly conserved protein by the majority of Stx phages is a strategy that has enabled and promoted the rapid spread of shigatoxigenic potential throughout multiple E. coli serogroups and related bacterial species. Infection of commensal bacteria in the mammalian gut has been shown to amplify Shiga toxin production in vivo, and the data from this study provide a platform for the development of a therapeutic strategy to limit this YaeT-mediated infection of the commensal flora.
Shiga toxin-producing Escherichia coli (STEC) strains, the most notorious serotype of which is O157:H7, possess a variety of factors that contribute to their pathogenic profiles. The emergence of these strains as serious food-borne pathogens of humans has been multifaceted (55), but the pivotal event in their emergence was undoubtedly acquisition of the ability to produce Shiga toxin, which is also referred to as Shiga-like toxin or verotoxin (19), following infection by a bacteriophage carrying the stx genes (36). Stx phages are lambdoid phages that carry two small genes comprising the Shiga toxin operon that encodes a typical AB5 toxin. Essentially, the only trait that all Stx phages share is carriage of an stx operon, which can encode one of two distinct toxin types, Stx1 or Stx2; each type is comprised of multiple variants (3). Stx phages have different morphologies (4, 34, 38, 39), and the exact identities of the genetic modules that make up their mosaic genomes vary, although the general organization of these modules is conserved. Recombination events within bacterial lysogens are thought to promote bacteriophage mosaicism, which is found among all lambdoid phages (3). Determination of the means by which Stx phages recognize and infect their bacterial hosts has been confounded by this vast heterogeneity.
STEC strains can possess more than one stx operon and can be infected with multiple Stx phages (4, 39). Even a single E. coli cell can be infected more than once by the same Stx phage or related Stx phages (4). However, no phage that carries more than one stx operon has ever been identified. Thus, a multiply Stx phage-infected E. coli strain can produce more than one type of Shiga toxin and/or high levels of Shiga toxin, and both of these properties are associated with a greater potential to cause serious life-threatening disease (4). A multiply infected host cell also serves as a reservoir for recombination events in situ, potentiating the creation of novel Stx phages, which may ultimately expand their host range and thus result in the emergence of new toxigenic bacterial strains (4). This could partially explain the heterogeneity of Stx phages and the rapid evolution and spread of STEC and STEC-like disease that has been remarkable since the first discovery in the early 1980s (40).
It was initially proposed that only rough strains of E. coli, lacking an intact cell envelope and unable to colonize the gastrointestinal tract, could be infected by Stx phages (50). For this reason, the crucial role of Stx phages in the continuing evolution and spread of STEC and STEC-like disease was largely ignored. However, it has been established more recently that smooth, wild-type strains of E. coli can be infected by Stx phages (22, 44) and that ab initio Stx phage infection of E. coli can actually take place in the animal gut (14, 15). Originally, it was hypothesized that Stx phages recognized some epitope on the lipopolysaccharide (LPS) of rough E. coli strains that was masked in smooth, wild-type strains possessing an intact cell envelope, a concept for which there is a precedent (50). Since it has been shown that the epitope recognized by Stx phages is not masked in smooth strains (22, 44), there must be some other target for adsorption to the host surface. Stx phages are heterogeneous, and many different bacterial species, even members of different bacterial genera belonging to and not belonging to the Enterobacteriaceae, have been reported to produce Shiga toxin (4, 18, 22). Therefore, either multiple host proteins are recognized by different Stx phages or the phages must adsorb to a highly conserved gene product on the bacterial surface. Alternatively, both mechanisms might control recognition of potential host cells.
Here, the host recognition marker that φ24B, an Stx phage studied in some detail previously (4, 22, 23, 42), uses to initiate infection is described. We chose to measure phage-host cell recognition directly through adsorption assays rather than relying upon the ability of the Stx phage to produce plaques on indicator cell lawns. This is important when the ability of isogenic mutant host cells or cells of other bacterial strains to support phage binding is examined because the metabolic state of the host cell, the presence of restriction and modification systems, and other factors can significantly affect the ability to detect plaques on bacterial lawns (12, 24, 28, 47, 49).
MATERIALS AND METHODS
Bacterial strains, plasmids, and bacteriophages.
E. coli K-12 strains MC1061 and DM1187 are susceptible to a variety of Stx phages (4, 22) and were used as hosts for all phage infections, as indicated below. Other bacterial strains, plasmids, and bacteriophages used in this study are described in Table 1. Unless indicated otherwise, cultures were grown in Luria-Bertani Miller broth (1% tryptone, 0.5% yeast extract, 1% NaCl [VWR)]) or on plates prepared by addition of 1.5% (wt/vol) agar (Difco). When bacteriophages were used, the growth medium was supplemented with 0.01 M CaCl2 (LB-Ca). The antibiotics used were kanamycin (50 μg ml−1; Merck), ampicillin (100 μg ml−1; Sigma), and chloramphenicol (30 μg ml−1; Sigma).
TABLE 1.
Strains, plasmids, and bacteriophages used in this study
| Strain(s), plasmid, or bacteriophage | Relevant characteristics | Reference(s) or source |
|---|---|---|
| E. coli strains | ||
| MC1061 | 8 | |
| DM1187Rif | recA441 lexA51 sfiA, rifampin resistant | 22 |
| MC1061-MRL1 | Phage resistant | This study |
| MC1061-MRL2 | Phage resistant | This study |
| MC1061-MRL3 | Phage resistant | This study |
| SG13009 | 17 | |
| M15 | QIAGEN | |
| S. flexneri PT2a and PT6 | 22 | |
| S. sonnei PT36 | 22 | |
| P. luminescens subsp. laumondii TTO1 | Genome sequencing completed at the Institut Pasteur | Institute Pasteur, Paris France |
| S. enterica serovar Choleraesuis SC-B67 | Genome sequencing completed at the Chang Gung Genomic Medical Center, Chang Gung Memorial Hospital | Cheng-Hsun Chiu, Chang Gung Children's Hospital, Chang Gung University College of Medicine, Taoyuan, Taiwan |
| E. carotovora subsp. atroseptica SCRI1043 | Genome sequencing completed at the Sanger Institute | ATCC BAA-672 |
| C. rodentium ATCC 51459 | ATCC 51459 | |
| Plasmids | ||
| pUC19 | Cloning vector, Ampr | 57 |
| pUC18 | Cloning vector, Ampr | 57 |
| pUCΦR1 | pUC19 with a 4.5-kb BamHI chromosome fragment | This study |
| pUCΦR1A | pUC18 with a 2.5-kb BamHI-KpnI fragment | This study |
| pUCΦR1B | pUC19 with a 2.0-kb KpnI-BamHI fragment | This study |
| pUCΦR1C | pUC18 with a 1.9-kb XhoI fragment | This study |
| pUCΦR1D | pUC19 with a 2.8-kb BamHI-PstI frgament | This study |
| pQE32 | Cloning vector, places six His residues at the amino terminus of a recombinant protein, Ampr | QIAGEN |
| pSCHyaeT | pQE32 possessing yaeT minus the leader peptide coding region | This study |
| pREP4 | Constitutively produces the lac repressor protein encoded by lacI | QIAGEN |
| Bacteriophages | ||
| φE86654-VT1 | Wild-type Stx1-encoding phage | 4 |
| φE86654-VT2 | Wild-type Stx2-encoding phage | 4 |
| φE85539-VT2a | Wild-type Stx2-encoding phage | 4 |
| φE83819-VT1 | Wild-type Stx1-encoding phage | 4 |
| φD155-VT1 | Wild-type Stx1-encoding phage | 4 |
| φ24B::kan | Mutant φE86654-VT2, VT operon interrupted with aph-3, lysogens are Kanr | 22, 42 |
| φ24B::cat | Mutant φE86654-VT2, VT operon interrupted with cat, lysogens are Catr | 4 |
Plaque assay.
E. coli MC1061 was cultured to an optical density at 600 nm (OD600) of 0.5 in LB-Ca at 37°C with shaking. Aliquots (200 μl) were then incubated at 37°C with 200 μl of serially diluted phage suspensions. After 30 min, 5 ml of VT top agar (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 1% [wt/vol] NaCl, 0.4% [wt/vol] Difco agar, 0.01 M CaCl2) was added to each infection mixture, which was then mixed and poured onto LB plates. The plastic petri plates were vented and incubated overnight at 37°C to allow plaques to develop.
Identification of an Stx phage-resistant mutant.
We used a strategy similar to that of Kilno and Rothman-Denes (26) in which proteins involved in infection with bacteriophage N4 were identified. A 100-ml culture of E. coli strain MC1061 was infected at a multiplicity of infection of 0.1 with phage φE86654-VT2 in LB-Ca. The infection was allowed to proceed overnight at 37°C, and then the culture was serially diluted before plating onto LB agar. The majority of cells were lysed during this initial infection step, and while most of the survivors were lysogens, a few cells were found to have lost their susceptibility to Stx phages. This was confirmed experimentally using a phage cocktail consisting of φE86654-VT1, φE86654-VT2, φE85539-VT2a, φE83819-VT1, and φD155-VT1 (4) and parameters identical to those used for the initial infection step. Any colonies were examined for their ability to adsorb phage in order to identify potential phage-resistant mutants.
Phage adsorption assay.
All bacterial strains were grown to an OD600 of 0.5 in LB-Ca with shaking at 37°C. Approximately 107 PFU ml−1 phage was added to each culture or LB-Ca alone. The phage were allowed to adsorb to the cell surfaces for 15 min at 37°C. Subsequently, the infection mixtures were centrifuged in a microcentrifuge for 5 min at 5,000 rpm to remove cells and adherent phage. The supernatant was harvested and titrated in a classical plaque assay using the wild-type MC1061 strain as the host. The data from the wild-type strain infection served as the control. The level of phage adsorption was determined as follows: percentage of phage adsorption = [(PFU from LB-Ca control − PFU from sample infection)/PFU from wild-type MC1061 infection] × 100.
Identification of yaeT.
A genetic library from E. coli MC1061 was created following the ligation of partially Sau3AI-digested chromosomal DNA (2- to 5-kb fragments) into BamHI-restricted, dephosphorylated expression vectors pUC18 and pUC19 (Amersham-Pharmacia). This library was transformed into MC1061-MRL1. Transformants were isolated on LB agar containing 100 μg ml−1 ampicillin and then used to inoculate LB-Ca. A phage, φ24B::kan (22), was added to this culture at a concentration of 2.5 × 109 PFU ml−1, and the infection mixture was incubated at 37°C with shaking (200 rpm). After 5 h, 1-ml portions of the infection mixture were plated onto LB agar containing 50 μg ml−1 kanamycin and 100 μg ml−1 ampicillin. No colonies were isolated. However, when 1 ml of the infection mixture was subcultured into 10 ml LB-Ca and further incubated overnight, 30 colonies ml−1 were obtained on the selective medium.
DNA preparations.
Plasmid DNA preparations were made routinely using a QIAprep Spin miniprep kit according to the manufacturer's recommendations (QIAGEN). Chromosomal preparations were made using 2 ml of an overnight culture, the cells of which were harvested by centrifugation and suspended in 600 μl of lysing buffer (0.5% [wt/vol] sodium dodecyl sulfate [SDS], 0.1 mg proteinase K [Sigma], 10 mM Tris, 0.5 mM EDTA; pH 8.0) for 1 h at 37°C. The DNA was further purified by cetyltrimethylammonium bromide treatment (100 μl of 5 M NaCl, 80 μl of 10% [wt/vol]) cetyltrimethylammonium bromide in 0.7 M NaCl) followed by extraction with an equal volume of chloroform-isoamyl alcohol (24:1) and a second extraction with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1). The DNA was precipitated with 0.6 volume of isopropanol, and RNA was removed by treatment with 50 ng of RNase (Sigma) followed by a second phenol-chloroform-isoamyl extraction and ethanol precipitation (5).
SDS-PAGE and Western blot analysis.
Three identical SDS-polyacrylamide gel electrophoresis (PAGE) gels were prepared by the method of Laemmli (27), using a 10% separating gel and a 5% stacking gel. One gel was stained with Coomassie blue R250, and the other two were transferred to nitrocellulose membranes (Bio-Rad) using a semidry transfer apparatus according to the manufacturer's (Bio-Rad) instructions. The blots were incubated in blocking buffer consisting of 3.0% (wt/vol) bovine serum albumin in Tris-buffered saline (10 mM Tris-HCl [pH 7.5], 150 mM NaCl) for 1 h at room temperature, which was followed by incubation with the primary antibody, either anti-RGS-His (1:1,000; Clontech) or anti-YaeT (1:50,000; this study) in the same buffer for 1 h at room temperature. The blots were washed twice for 10 min in 20 mM Tris-HCl (pH 7.5)-500 mM NaCl-0.05% Tween 20-0.2% Triton X-100; then one of the blots was incubated in blocking buffer containing anti-mouse alkaline phosphatase conjugate (1:10,000; Sigma), and the other blot was incubated in blocking buffer containing anti-rabbit alkaline phosphatase conjugate (1:10,000; Sigma). The blots were developed using Sigma Fast 5-bromo-4-chloro-3-indolylphosphate (BCIP)—nitroblue tetrazolium tablets according to the manufacturer's instructions.
Creation of H-YaeT.
The yaeT gene from pUCφR1D was amplified using primers BamH I YaeT5′ (5′-GTA TAC GGT GCT GAA GGG ATC CTA GTG AAA G-3′) and yaeT 3′PstI (5′-CTT AGC TTG CAT GCC TGC-3′). These primers were used to amplify the portion of the yaeT open reading frame that corresponded to the mature, processed protein product. This was achieved by using Platinum Pfx DNA polymerase (Invitrogen Life Technologies) according to the manufacturer's instructions and the following parameters: denaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 68°C for 1.5 min. Primer BamH I YaeT5′ has nucleotide substitutions at residues 21 and 24, which result in addition of a BamHI restriction endonuclease site. Cleavage of the amplification product with BamHI endonuclease removed all sequences encoding the leader peptide of YaeT, and subsequent cleavage with the restriction endonuclease PstI removed all traces of the pUC19 vector. The BamHI-PstI fragment was then cloned into similarly digested pQE32 (QIAGEN) to create plasmid pSCHyaeT, which codes for expression of histidine-tagged recombinant YaeT (H-YaeT). This clone was identified in E. coli strain SG13009 carrying plasmid pREP4, which prevents expression from the lacZ promoter in the absence of an inducer molecule, by growth on LB agar plates containing ampicillin and kanamycin. Its identity was confirmed by Western blot analysis using the primary antibody RGS-His (QIAGEN) to identify a ∼90-kDa histidine-tagged protein. H-YaeT differs from the wild-type processed YaeT in that the sequence of the former begins with MRGSHHHHHHGILVKDIHFE, while the sequence of the latter begins with AEGFVVKDIHFE.
Purification of H-YaeT protein.
H-YaeT was purified using affinity Ni-nitrilotriacetic acid chromatography (QIAGEN) under denaturing conditions, as specified by the manufacturer. Briefly, E. coli strain SG13009 carrying both pSCHyaeT and pRep4 was grown in 200 ml LB containing both ampicillin and kanamycin to an OD600 of 0.6. Production of H-YaeT was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cells were incubated for a further 4 h. The cells were harvested by centrifugation at 4,000 × g for 15 min and suspended in 20 ml lysing buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea; pH 8). Cells were then lysed using a probe sonicator set to an amplitude of 22 μm using four 15-s bursts with intervening 30-s intervals at 0°C. The resultant lysates were centrifuged at 4,000 × g for 25 min, and the supernatant was collected. The lysates were then incubated with 5 ml of the Ni-nitrilotriacetic acid matrix for 1 h at room temperature with shaking. The resulting slurry was then poured into a chromatography column, and the flowthrough was collected for analysis. The column was washed with 8 ml of wash buffer (100 mM Na2PO4, 10 mM Tris-HCl, 8 M urea; pH 6.5), His-YaeT was eluted with elution buffer (100 mM Na2PO4, 10 mM Tris-HCl, 8 M urea; pH 5.9), and fractions were collected at 2-min intervals. Samples were subsequently analyzed by SDS-PAGE and Western blotting.
Protein assay.
Protein concentrations were determined using the bicinchoninic acid assay. Briefly, twofold dilutions of fraction V purified bovine serum albumin (Sigma Chemical Co.) were prepared as standards. These samples, along with dilutions of the Ni+-purified H-YaeT, were incubated with a bicinchoninic acid-CuSO4 solution (50:1, vol/vol; Sigma Chemical Co.) and incubated for 1 h at 37°C. Concentrations of protein were determined spectrophotometrically at 562 nm.
Antibody production.
Approximately 5 mg of H-YaeT was purified by nickel affinity chromatography in the denaturing elution buffer. The resulting preparation was sent to Biogenesis (Poole, England), where it was used to inoculate two New Zealand White rabbits using the custom antibody production service of this company.
RESULTS
In order to investigate the initial steps in the infection of E. coli by Stx phage φ24B, a strategy similar to that used to identify the host ligand for bacteriophage N4 was adopted (26). Spontaneous Stx phage-resistant mutants were identified following infection of an E. coli K-12 strain with a cocktail of Stx phages induced from clinical isolates (4). Each Stx phage was independently capable of infecting the host strain. Three resistant mutants that failed to produce phage particles following UV induction were obtained, and one strain (MRL1), which was later determined to be incapable of supporting the adsorption of φ24B, was used to screen an expression library created from the Stx phage-susceptible E. coli K-12 strain MC1061. Four clones that conferred phage adsorption potential as well as phage susceptibility to MRL1 were identified, and all four shared a region of identity containing two open reading frames (Fig. 1). Following independent subcloning of the two open reading frames, it was determined that the gene conferring the ability to support phage adsorption, as well as the phage-susceptible phenotype, was yaeT (11, 16, 30, 54), also referred to as yzzN (2) and ecfK (10), while the second gene, skp, encoding a periplasmic molecular chaperone, did not confer phage susceptibility. The yaeT gene has recently been identified elsewhere as an essential gene in E. coli (16) involved in populating the LPS outer membrane with proteins (11, 43, 54, 56).
FIG. 1.
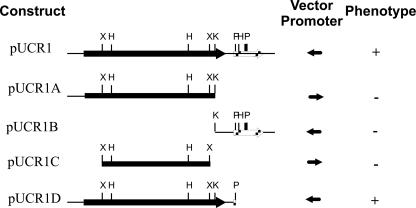
Subcloning of the open reading frame associated with adsorption of φ24B to MRL1. Construct pUCR1 was one of the four original clones that complemented the φ24B phage adsorption defect. All four clones shared the two open reading frames present in pUCR1; the solid box represents yaeT, and the striped box represents skp. When fragments from pUCR1 were subcloned into pUC19 or pUC18 (indicated by the vector promoter direction: leftward, pUC19; rightward, pUC18), only the complete yaeT gene found in pUCR1 and pUCR1D complemented the MRL1 adsorption defect. Restriction sites are as follows: X, XhoI; H, HincII; K, KpnI; P, PstI.
BLASTP analyses of YaeT identified a homologue in all gram-negative bacterial genome sequences (Fig. 2), predictably in view of its proposed role in biogenesis of the LPS membrane. YaeT has an especially high level of sequence conservation among most members of the Enterobacteriaceae (Fig. 3), yet the orthologue in Erwinia carotovora subsp. atroseptica fell outside the main clade (Fig. 2). The degree of conservation among members of the Enterobacteriaceae is particularly apposite considering that many of its members have been reported to produce Shiga toxin, implying that at some time they were infected by an Stx phage (22, 37, 45, 48). It would have been informative to have sequenced the yaeT gene possessed by the MRL1 strain, but unfortunately this strain was lost. However, complementation of MRL mutants identified only YaeT, a predicted outer membrane protein, as the potential phage adsorption target. A series of experiments designed to ablate expression of yaeT in the wild-type E. coli K-12 strain MC1061 were initiated, but these experiments did not produce knockout mutants and predated the discovery that yaeT was an essential gene (16). Although a yaeT deletion mutant has been described previously (10), it was created only after the gene was first duplicated in cells by complementation (D. L. Smith and D. Missiakas, personal communication). Conditional mutants or complementation strategies used to study sigma E-regulated genes (10) were also unsuitable as even short-term expression of yaeT would be sufficient to populate the cell surface with YaeT, which would subsequently be able to support phage adsorption. Consequently, the initial mutation and complementation experiments with the MRL mutants served to identify YaeT as a potential receptor for the short-tailed Stx phage, and the experiments described below were designed to provide the formal proof. These experiments centered on the use of an in vivo binding assay based on measurement of phage adsorption in the presence and absence of specific YaeT antiserum. This approach was supported by experiments in which the relationship between phage binding and yaeT expression and copy number were studied, together with heterologous cloning of E. coli yaeT to confer phage adsorption ability on E. carotovora. The data were correlated with predicted phage adsorption profiles for other species with natural variations in the yaeT sequence.
FIG. 2.
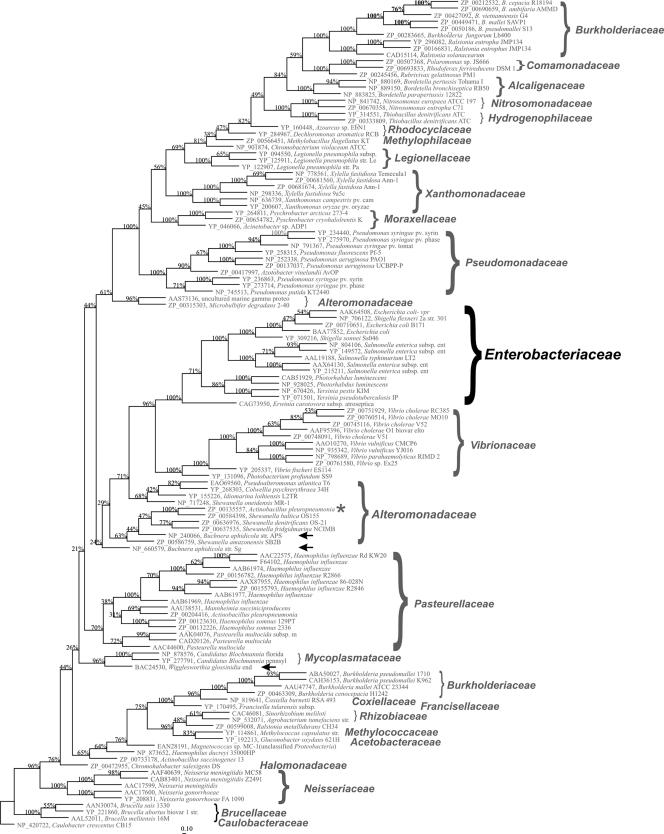
Phylogenetic tree of YaeT orthologues for most of the gram-negative bacteria for which there are complete genome sequences. Homologues of YaeT were identified by BLASTP analyses and used to identify all homologues of YaeT possessing at least 30% amino acid identity. The sequences were aligned and then subjected to a maximum parsimony analysis using ARB (29). Branches were validated through 100 bootstrap analyses. Scale bar = 1% amino acid changes. The Enterobacteriaceae is indicated by larger type, and a few unusual members of this family are indicated by arrows. The asterisk indicates a bacterial species that clusters with an unrelated bacterial family.
FIG. 3.
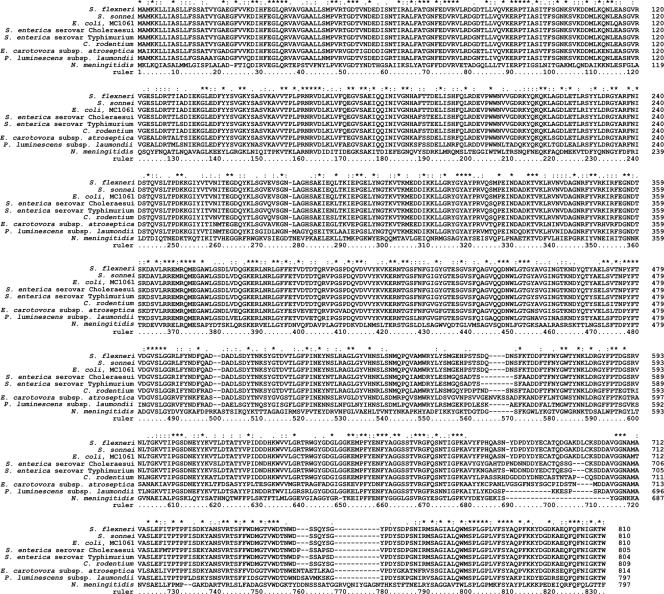
Multiple alignment of YaeT from members of the Enterobacteriaceae. Clustal X (46) was used to align YaeT sequences from eight members of the Enterobacteriaceae against the orthologue from Neisseria meningitidis, for which a structure has been proposed (51). The alignments were used to identify regions possessing the greatest similarities and disparities; the latter are mostly associated with the carboxyl terminus and correspond to predicted extracellular loops. The conservation of residues is indicated above the alignments as follows: asterisk, complete identity; colon, conservation of a strong group (46); period, conservation of a weak group (46).
The amino terminus of YaeT was fused to a histidine tag (H-YaeT) to enable easy purification of recombinant H-YaeT, and monospecific rabbit antiserum was raised against H-YaeT (Fig. 4). This antiserum was capable of recognizing both recombinant and native YaeT in Western blots at a 1:50,000 dilution (Fig. 3B), and it could also be used to block phage adsorption in a dose-dependent fashion (Fig. 5). Preimmune sera from the same rabbit, which recognized many E. coli proteins in Western blots at a lower dilution (1:10,000) (data not shown), had no effect in the adsorption assay. Additionally, pyaeT was fused to a promotorless lacZ gene in order to confirm that growth conditions shown to affect yaeT expression (10) affected its expression in our strain; phage adsorption levels correlated to the expression levels of pyaeT, and factors such as growth at 42°C caused a twofold increase in expression from pyaeT and also doubled the number of phages that could become adsorbed to a single cell (data not shown). E. coli clones carrying yaeT on a high-copy-number vector (pUC19 or TOPO vectors [Invitrogen]) also exhibited the ability to adsorb significantly increased numbers of phage (data not shown). All of this evidence indicated that YaeT was acting as the cell surface ligand for phage adsorption, but independent confirmation was still required.
FIG. 4.
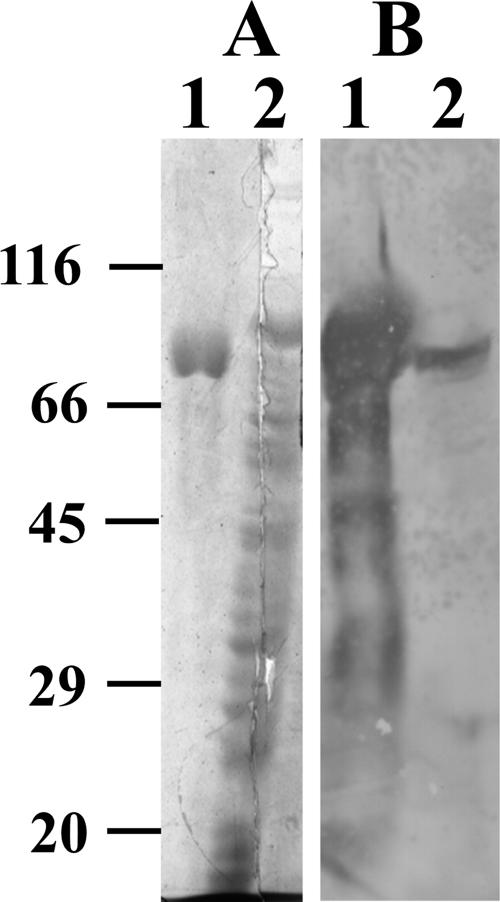
SDS-PAGE and Western blot identification of purified histidine-tagged recombinant YaeT. (A) Coomassie blue-stained SDS-PAGE gel. (B) Western blot analysis using rabbit anti-H-YaeT (1:50,000). Lane 1, nickel affinity-purified H-YaeT; lane 2, whole-cell lysates of E. coli strain MC1061.
FIG. 5.
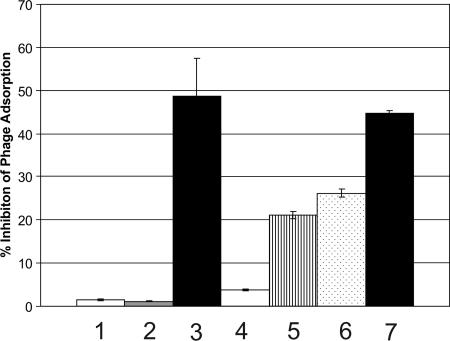
Inhibition of φ24B adsorption using rabbit anti-H-YaeT antiserum. A total of 5 × 107 MC1061 cells were incubated with no serum (bar 1), rabbit preimmune serum (1:5 dilution) (bar 2), or rabbit anti-H-YaeT serum (1:5 dilution) (bar 3) prior to infection with φ24B (5 × 108 PFU). The error bars indicate the standard errors of the means (n = 15). In a separate experiment, using the same numbers of cells and phage, various dilutions of the anti-H-YaeT serum were added to the cells prior to infection with φ24B (bar 4, none; bar 5, 1:600 dilution; bar 6, 1:60 dilution; bar 7, 1:6 dilution) The error bars indicate the standard errors of the means (n = 5).
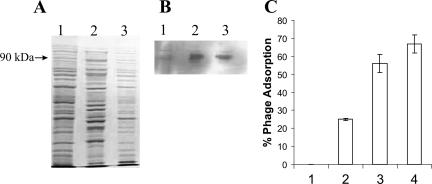
There have been several previous reports of the functional expression of outer membrane proteins that have been exchanged between various members of the Enterobacteriaceae, including E. coli and E. carotovora (1, 9, 25). The latter species is an unusual member of the Enterobacteriaceae in that it is a plant pathogen and is not known to colonize animals. Use of this species as a recombinant host for studying Stx phage adsorption in the laboratory also guards against the potential creation of a new human or animal pathogen. Our adsorption assays demonstrated that phage φ24B was not capable of adsorption to the surface of E. carotovora strain SCRI1043. So, in essence, E. carotovora was a potential adsorption-deficient mutant that still possessed a functional orthologue of YaeT. The E. coli yaeT gene was introduced into E. carotovora carried on the low-copy-number vector pKT230 (6) possessing the RSF1010 broad-host-range replicon, which is known to function in E. carotovora (52). Western blot analysis using the antiserum raised against recombinant E. coli H-YaeT indicated that the E. coli YaeT was expressed in the E. carotovora clone carrying pKT230 yaeT (Fig. 6). Although the anti-H-YaeT serum was able to recognize the endogenous Erwinia YaeT orthologue, its reactivity with E. coli YaeT was much greater (Fig. 6B). The ability of φ24B to adsorb to E. carotovora containing pKT230 with or without the cloned yaeT gene was determined (Fig. 6C). Adsorption could be observed in a dose-dependent fashion only in the E. carotovora strain carrying the E. coli yaeT gene, and this adsorption was specifically inhibited with the anti-H-YaeT sera, thus providing formal proof that YaeT serves as the adsorption target for φ24B infection of E. coli.
FIG. 6.
Complementation of a natural yaeT mutant confers the ability to support φ24B adsorption. (A) SDS-PAGE analysis of total cell protein from E. carotovora subsp. atroseptica (lane 2), E. coli strain MC1061 (lane 2), and E. carotovora subsp. atroseptica carrying the construct pKT230yaeT (lane 3). The region corresponding to 90 kDa is indicated on the left. (B) Western blot analysis of total cell proteins from panel A. (C) Phage binding to YaeT on the surface of E. carotovora carrying pKT230yaeT. Data were calculated for plasmid-free and pKT230-containing strains challenged with 3.4 × 107 phage particles. The x axis indicates different total numbers of E. carotovora cells, as follows: bar 1, 1.5 × 108 cells bearing pKT230; and bars 2, 3, and 4, 1.5 × 108, 3 × 108, and 5 × 108 cells carrying pKT230yaeT, respectively. The error bars indicate the standard errors of the means (n = 5).
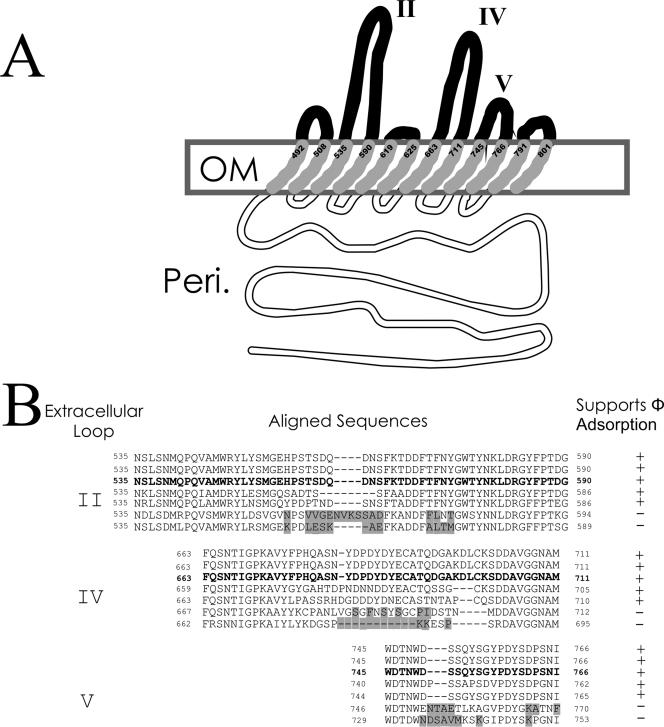
Mutational studies of essential gene products present challenges, especially if the only available measure of their activity is the ability to support the binding of a ligand once the essential gene product has been appropriately folded and inserted into the LPS outer membrane. In order to better understand the adsorption of φ24B to YaeT, the natural sequence variation in yaeT orthologues across members of the Enterobacteriaceae was utilized. Most of the sequence diversity can be found in the carboxy-terminal portion of the YaeT protein (Fig. 3). The model produced by Voulhoux et al. (51) predicted the conformation of Omp85 (a YaeT orthologue) in the Neisseria LPS membrane and is used here to propose a model for the E. coli YaeT molecule (Fig. 7). The majority of unconserved residues in the YaeT orthologues across the Enterobacteriaceae are associated with regions that are predicted to be extracellular loops and specifically the middle sequences of those loops (Fig. 7). In order to investigate which orthologues of YaeT could support phage adsorption and thus identify important sequences for φ24B adsorption, assays were carried out with Shigella flexneri strains PT2a and PT6, Shigella sonnei strain PT36, E. coli strain MC1061, Salmonella enterica serovar Choleraesuis strain SC-B67, Citrobacter rodentium strain ICC168, E. carotovora subsp. atroseptica strain SCRI1043, and Photorhabdus luminescens subsp. laumondii strain TTO1 (Fig. 6). Only E. carotovora subsp. atroseptica and P. luminescens subsp. laumondii were unable to support φ24B adsorption. The greatest degree of sequence diversity is found in the central sections of extracellular loops II, IV, and V, and thus predictions concerning the important residues in φ24B adsorption can be made (Fig. 7), although additional experiments are necessary to prove these predictions. The anti-H-YaeT serum was capable of blocking φ24B adsorption to all of the susceptible strains (data not shown).
FIG. 7.
Relationship between φ24B phage adsorption and yaeT sequence variation with the predicted protein conformation in Enterobacteriaceae strains. (A) Proposed configuration of YaeT in the E. coli LPS membrane. The black loops are the predicted extracellular loops; the gray segments are predicted outer membrane (OM)-spanning regions; and the open segments are predicted periplasmic (Peri.) regions of YaeT. The extracellular loops are numbered from the amino terminus. (B) Identification of amino acid substitutions that correlate with the loss of Stx phage adsorption ability. The highlighted sequences are the changes predicted to be responsible for the loss of the ability to support phage adsorption. The sequences are sequences of, from top to bottom, S. flexneri, S. sonnei, E. coli strain MC1061, S. enterica serovar Choleraesuis strain SC-B67, C. rodentium strain ICC168, E. carotovora subsp. atroseptica strain SCRI1043, and P. luminescens subsp. laumondii strain TTO1.
DISCUSSION
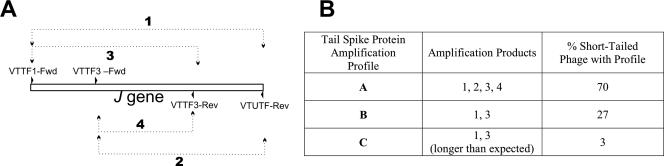
It is widely accepted that lambdoid bacteriophages are genetic mosaics, and the Stx phages are no exception. In fact, the only single attribute known to be shared by all Stx phages is the carriage of the genes encoding some variant of Shiga toxin (3). Thus, the impact of the identification of a ubiquitous host receptor for the short-tailed Stx phage φ24B is dependent on the relative frequency of this phage type among circulating populations of Stx phages. A total of 76 Stx phages were induced from a collection of 460 E. coli O157:H7 strains that were obtained from a large farming area over a 5-year period in Cheshire, United Kingdom (41). Most (70%) of the induced Stx phages are Podoviridae (short tail, icosahedral head), like 933W (35) and φ24B (4). The predominance of short-tailed Podoviridae among Stx phages isolated directly from the environment has also been reported previously (13, 33). Examination of the tail spike sequences from the induced Stx phages, as well as 933W, φ24B, and short-tailed phages in the DNA sequence databases (GenBank accession numbers BAB87868, NP_612899, NP_050557, NP_286993), NP_309255, and NP_859099), confirmed that all of these phages possess tail spike proteins with identical or highly conserved sequences (Fig. 8). Through examination of the short-tailed Stx phage genomes that are available, it is clear that they possess only the tail spike protein for host recognition, as does bacteriophage lambda for recognition of its potential host cells (7, 31). The ability of all of the short-tailed phages in our collection to adsorb to the surface of E. coli K-12 strain MC1061 was demonstrated, as was inhibition of binding in competition assays with anti-H-YaeT (data not shown). Thus, YaeT is likely to be an Stx phage recognition site of considerable importance to the epidemiology of Shiga toxin dispersal among populations of E. coli and possibly gram-negative bacteria in general.
FIG. 8.
Variations in tail spike proteins that recognize YaeT as an adsorption target. Amplification primers were designed against the gene sequence encoding the tail spike protein (the J gene orthologue) of φ24B and 933W (VTTF1-Fwd, GTTGTTGTTTCGGGGACG; VTUTF-Rev, TCATTCTCCTGTTCTGCC; VTTF3-Fwd, TGCAGAGGAAAGCTCGAC; VTTF3-Rev, GCAGCCTCTTCTGCCTTT). The primers were used in combinations of VTTF1-Fwd with VTUTF-Rev or VTTF3-Rev and of VTTF3-Fwd with VTUTF-Rev or VTTF3-Rev (combinations 1, 3, 2, and 4, respectively). All variations of tail spike proteins identified by this means were capable of adsorption to E. coli MC1061, and this adsorption could be blocked with the anti-YaeT sera. Three different amplification profiles were found for the short-tailed phages listed in the table in panel B. The majority (70%) of phages possessed amplification profile A, which results from the production of all four amplification products of the anticipated size. Profile C was found only rarely and is characterized by the failure of the VTUTF-Rev primer to produce any products in combination with either 5′ primer, while the products of VTTF3-Rev were ∼800 bp larger than expected.
There have been previous attempts to identify the host ligand recognized by Stx phages (21, 32, 53), using long-tailed or short-tailed phages. It is clear that long-tailed Stx phages, such as H19-B (35), PP01 (32), and P27 (38), recognize other host-encoded surface molecules, and there are data that support the use of surface ligands, such as OmpC (52). The data on short-tailed phage adsorption sites have been more difficult to interpret (35). The approach reported here directly determines adsorption of the phage to the host surface, in contrast to other studies that have examined the ability of the phage to infect a host cell, propagate, and infect surrounding cells to a level sufficient to produce plaques on bacterial lawns. Many contributing factors can affect the overall ability of a phage to elicit a productive infection and form plaques, and measuring phage adsorption is the most direct and appropriate technique for identifying phage-host binding ligands.
We propose that the recognition of YaeT and its orthologues among the members of the Enterobacteriaceae has contributed to the rapid evolution of STEC and STEC-like pathogens. Although there is considerable genetic heterogeneity among Stx phages, our data suggest that the large majority share an identical tail spike protein and thus the ability to recognize susceptible host cells in the same fashion, via YaeT. The use of a highly conserved essential gene product as the receptor for adsorption by a phage is likely to ensure that host bacterial cells will always be available in the gut or other environments that support Enterobacteriaceae populations. The function of YaeT and its conserved structure ensure that it will always be present on the surface of potential host cells and that susceptible bacterial populations will always be present in the gut environment. This differs fundamentally from the other phage adsorption targets, which do not provide an essential function to the cell. Mutations or conditions that abrogate production of the phage adsorption target are very unlikely to occur, so spread of the bacteriophage should be enhanced according to our current understanding of herd immunity concepts (20). Together then, the ability of Stx phage to convert its host cell into a deadly human pathogen and its ability to infect related strains or species by utilizing a conserved surface receptor underlie the rapid evolution, emergence, and expansion of STEC and STEC-like pathogens, which have echoed throughout the scientific literature since the first E. coli O157:H7 outbreak in 1982. These abilities also support the view that STEC and STEC-like pathogens will continue to evolve as the short-tailed Stx phages direct conversion of additional susceptible bacterial strains. For example, we have determined that pathogenic strains of S. enterica can support the adsorption of the short-tailed Stx phages (Fig. 7), with corresponding inhibition by anti-H-YaeT (data not shown), and this was predicted by the conservation of the yaeT allele with that of E. coli (Fig. 3 and 7). Salmonellae have yet to be linked to Stx phage-related disease, but there has been a report of a member of the Moraxellaceae (Acinetobacter haemolyticus) carrying an inducible Stx phage, producing Stx2, and associated with a case of bloody diarrhea in an infant (18). Understanding how Stx phages recognize commensal bacteria in the human gut should provide a platform for a therapeutic strategy aimed at limiting the potentiation of Stx production via phage conversions in the infected intestine, and the Stx phage recognition site, YaeT, provides a possible target. Expression levels of yaeT have been previously shown to fluctuate with environmental conditions (10), especially those that affect the integrity of the outer membrane. As yaeT expression is controlled by the sigma E stress regulon (10), it would not be unreasonable to expect the expression of this gene to be up-regulated in the mammalian gut environment, and this would facilitate the infection of susceptible commensal gut bacteria (14, 15) and possibly explain why infection with an Stx2-encoding phage was more successful in a porcine ileal loop model than under in vitro laboratory conditions (47).
In addition to elucidating a mechanism for Stx phage recognition of potential host cells, the antisera, phages, and YaeT variants described in this study constitute a set of tools that should be useful for studying the essential role that YaeT plays in populating the outer membrane with proteins, as well as for identifying the exact epitope(s) recognized by the phage tail-spike protein.
Acknowledgments
We thank Sarah S. Cookson for her contribution to the production of the anti-YaeT sera. We also thank the Institute Pasteur and Cheng-Hsun Chiu at the Chang Gung Children's Hospital, Chang Gung University College of Medicine, Taoyuan, Taiwan, for the kind provision of sequenced bacterial strains.
This research was supported by the Biotechnology and Biological Sciences Research Council (United Kingdom), the Department of Environment Food and Rural Affairs (United Kingdom), and the Higher Education Funding Council for England (grants to H.E.A., J.R.S., and A.J.M.). The contributions of the Natural Environment Research Council to M.J.S. and of the former Centre for Applied Microbiology Research Porton Down to studentships for M.J.S. and C.E.J. are gratefully acknowledged.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Akatsuka, H., R. Binet, E. Kawai, C. Wandersman, and K. Omori. 1997. Lipase secretion by bacterial hybrid ATP-binding cassette exporters: molecular recognition of the LipBCD, PrtDEF, and HasDEF exporters. J. Bacteriol. 179:4754-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alba, B. M., J. A. Leeds, C. Onufryk, C. Z. Lu, and C. A. Gross. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma E-dependent extracytoplasmic stress response. Genes Dev. 16:2156-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, H. E. 2007. Stx-phages: drivers and mediators of the evolution of STEC and STEC-like pathogens. Future Microbiol. 2:165-174. [DOI] [PubMed] [Google Scholar]
- 4.Allison, H. E., M. J. Sergeant, C. E. James, J. R. Saunders, D. L. Smith, R. J. Sharp, T. S. Marks, and A. J. McCarthy. 2003. Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect. Immun. 71:3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1992. Current protocols in molecular biology, vol. 1 and 2. Greene Publishing Associates and Wiley-Interscience, New York, NY.
- 6.Bagdasarian, M., R. Lurz, B. Ruckert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. [DOI] [PubMed] [Google Scholar]
- 7.Berkane, E., F. Orlik, J. F. Stegmeier, A. Charbit, M. Winterhalter, and R. Benz. 2006. Interaction of bacteriophage lambda with its cell surface receptor: an in vitro study of binding of the viral tail protein gpJ to LamB (maltoporin). Biochemistry 45:2708-2720. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 9.Climent, N., S. Ferrer, X. Rubires, S. Merino, J. M. Tomas, and M. Regue. 1997. Molecular characterization of a 17-kDa outer-membrane protein from Klebsiella pneumoniae. Res. Microbiol. 148:133-143. [DOI] [PubMed] [Google Scholar]
- 10.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 11.Doerrler, W. T., and C. R. Raetz. 2005. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J. Biol. Chem. 280:27679-27687. [DOI] [PubMed] [Google Scholar]
- 12.Friedman, D. I., E. R. Olson, C. Georgopoulos, K. Tilly, I. Herskowitz, and F. Banuett. 1984. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol. Rev. 48:299-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamage, S. D., A. K. Patton, J. F. Hanson, and A. A. Weiss. 2004. Diversity and host range of Shiga toxin-encoding phage. Infect. Immun. 72:7131-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamage, S. D., A. K. Patton, J. E. Strasser, C. L. Chalk, and A. A. Weiss. 2006. Commensal bacteria influence Escherichia coli O157:H7 persistence and Shiga toxin production in the mouse intestine. Infect. Immun. 74:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamage, S. D., J. E. Strasser, C. L. Chalk, and A. A. Weiss. 2003. Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect. Immun. 71:3107-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman, S., M. Gottesman, J. E. Shaw, and M. L. Pearson. 1981. Protein degradation in E. coli: the Ion mutation and bacteriophage lambda N and cll protein stability. Cell 24:225-233. [DOI] [PubMed] [Google Scholar]
- 18.Grotiuz, G., A. Sirok, P. Gadea, G. Varela, and F. Schelotto. 2006. Shiga toxin 2-producing Acinetobacter haemolyticus associated with a case of bloody diarrhea. J. Clin. Microbiol. 44:3838-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gyles, C. L. 1992. Escherichia coli cytotoxins and enterotoxins. Can. J. Microbiol. 38:734-746. [DOI] [PubMed] [Google Scholar]
- 20.Heymann, D. L., and R. B. Aylward. 2006. Mass vaccination: when and why. Curr. Top. Microbiol. Immunol. 304:1-16. [DOI] [PubMed] [Google Scholar]
- 21.Iguchi, A., S. Iyoda, H. Watanabe, and R. Osawa. 2007. O side chain deficiency enhances sensitivity of Escherichia coli to Shiga toxin 2-converting bacteriophages. Curr. Microbiol. 54:14-19. [DOI] [PubMed] [Google Scholar]
- 22.James, C. E., K. N. Stanley, H. E. Allison, H. J. Flint, C. S. Stewart, R. J. Sharp, J. R. Saunders, and A. J. McCarthy. 2001. Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Appl. Environ. Microbiol. 67:4335-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johannessen, G. S., C. E. James, H. E. Allison, D. L. Smith, J. R. Saunders, and A. J. McCarthy. 2005. Survival of a Shiga toxin-encoding bacteriophage in a compost model. FEMS Microbiol. Lett. 245:369-375. [DOI] [PubMed] [Google Scholar]
- 24.Johnson-Boaz, R., C. Y. Chang, and R. Young. 1994. A dominant mutation in the bacteriophage-lambda S-gene causes premature lysis and an absolute defective plating phenotype. Mol. Microbiol. 13:495-504. [DOI] [PubMed] [Google Scholar]
- 25.Karlsson, M. B., M. Pirhonen, H. T. Saarilahti, and E. T. Palva. 1991. Molecular cloning of ompRS, a regulatory locus controlling production of outer membrane proteins in Erwinia carotovora subsp. carotovora. Mol. Gen. Genet. 226:353-360. [DOI] [PubMed] [Google Scholar]
- 26.Kilno, R. K., and L. B. Rothman-Denes. 1989. Genetic analysis of N4 adsorption. J. Bacteriol. 171:3595-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl, G., G. Sironi, H. Bialy, and R. Calendar. 1970. Bacteriophage lambda; abortive infection of bacteria lysogenic for phage P2. Proc. Natl. Acad. Sci. USA 66:587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malinverni, J. C., J. Werner, S. Kim, J. G. Sklar, D. Kahne, R. Misra, and T. J. Silhavy. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61:151-164. [DOI] [PubMed] [Google Scholar]
- 31.Moldovan, R. G., E. Chapman-McQuiston, and X. L. Wu. 2007. On kinetics of phage adsorption. Biophys. J. 93:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita, M., Y. Tanji, K. Mizoguchi, T. Akitsu, N. Kijima, and H. Unno. 2002. Characterization of a virulent bacteriophage specific for Escherichia coli O157:H7 and analysis of its cellular receptor and two tail fiber genes. FEMS Microbiol. Lett. 211:77-83. [DOI] [PubMed] [Google Scholar]
- 33.Muniesa, M., J. E. Blanco, M. De Simon, R. Serra-Moreno, A. R. Blanch, and J. Jofre. 2004. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 150:2959-2971. [DOI] [PubMed] [Google Scholar]
- 34.Muniesa, M., and J. Jofre. 2000. Occurrence of phages infecting Escherichia coli O157:H7 carrying the Stx 2 gene in sewage from different countries. FEMS Microbiol. Lett. 183:197-200. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien, A. D., L. R. Marques, C. F. Kerry, J. W. Newland, and R. K. Holmes. 1989. Shiga-like toxin converting phage of enterohemorrhagic Escherichia coli strain 933. Microb. Pathog. 6:381-390. [DOI] [PubMed] [Google Scholar]
- 36.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 37.Paton, A. W., and J. C. Paton. 1996. Enterobacter cloacae producing a Shiga-like toxin II-related cytotoxin associated with a case of hemolytic-uremic sundrome. J. Clin. Microbiol. 34:463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Recktenwald, J., and H. Schmidt. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage φP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70:1896-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rietra, P. J. G. M., G. A. Willshaw, H. R. Smith, A. M. Field, S. M. Scotland, and B. Rowe. 1989. Comparison of verocytotoxin-encoding phages from Escherichia coli of human, and bovine origin. J. Gen. Microbiol. 135:2307-2318. [DOI] [PubMed] [Google Scholar]
- 40.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 41.Robinson, S. E., E. J. Wright, C. A. Hart, M. Bennett, and N. P. French. 2004. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J. Appl. Microbiol. 97:1045-1053. [DOI] [PubMed] [Google Scholar]
- 42.Saunders, J. R., H. Allison, C. E. James, A. J. McCarthy, and R. Sharp. 2001. Phage-mediated transfer of virulence genes. J. Chem. Technol. Biotechnol. 76:1-5. [Google Scholar]
- 43.Schleiff, E., and J. Soll. 2005. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 6:1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt, H., M. Montag, J. Bockemuhl, J. Heeseman, and H. Karch. 1993. Shiga-like toxin II related cytotoxins in Citrobacter freundii strains from human and beef samples. Infect. Immun. 61:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toth, I., H. Schmidt, M. Dow, A. Malik, E. Oswald, and B. Nagy. 2003. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a Shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 69:7242-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tschape, H., R. Prager, W. Streckel, A. Fruth, H. Tietre, and A. Bohme. 1995. Verotoxigenic Citrobacter freundi associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: green butter as the infection source. Epidemiol. Infect. 114:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsui, H. C. T., H. C. E. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an Hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13:35-49. [DOI] [PubMed] [Google Scholar]
- 50.van der Ley, P., P. de Graaff, and J. Tommassen. 1986. Shielding of Escherichia coli outer membrane proteins as receptors for bacteriophages and colicins by O-antigenic chains of lipopolysaccharide. J. Bacteriol. 168:449-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262-265. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe, K., and M. Sato. 1998. Plasmid-mediated gene transfer between insect-resident bacteria, Enterobacter cloacae, and plant-epiphytic bacteria, Erwinia herbicola, in guts of silkworm larvae. Curr. Microbiol. 37:352-355. [DOI] [PubMed] [Google Scholar]
- 53.Watarai, M., T. Sato, M. Kobayashi, T. Shimizu, S. Yamasaki, T. Tobe, C. Sasakawa, and Y. Takeda. 1998. Identification and characterization of a newly isolated Shiga toxin 2-converting phage from Shiga-toxin producing Escherichia coli. Infect. Immun. 66:4100-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Werner, J., and R. Misra. 2005. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 57:1450-1459. [DOI] [PubMed] [Google Scholar]
- 55.Wick, L. M., W. Qi, D. W. Lacher, and T. S. Whittam. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187:1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, T., J. Malinverni, N. Ruiz, S. Kim, T. J. Silhavy, and D. Kahne. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235-245. [DOI] [PubMed] [Google Scholar]
- 57.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]