Abstract
Riboflavin (vitamin B2) is the direct precursor of the flavin cofactors flavin mononucleotide and flavin adenine dinucleotide, essential components of cellular biochemistry. In this work we investigated the unrelated proteins YpaA from Bacillus subtilis and PnuX from Corynebacterium glutamicum for a role in riboflavin uptake. Based on the regulation of the corresponding genes by a riboswitch mechanism, both proteins have been predicted to be involved in flavin metabolism. Moreover, their primary structures suggested that these proteins integrate into the cytoplasmic membrane. We provide experimental evidence that YpaA is a plasma membrane protein with five transmembrane domains and a cytoplasmic C terminus. In B. subtilis, riboflavin uptake was increased when ypaA was overexpressed and abolished when ypaA was deleted. Riboflavin uptake activity and the abundance of the YpaA protein were also increased when riboflavin auxotrophic mutants were grown in limiting amounts of riboflavin. YpaA-mediated riboflavin uptake was sensitive to protonophors and reduced in the absence of glucose, demonstrating that the protein requires metabolic energy for substrate translocation. In addition, we demonstrate that PnuX from C. glutamicum also is a riboflavin transporter. Transport by PnuX was not energy dependent and had high apparent affinity for riboflavin (Km 11 μM). Roseoflavin, a toxic riboflavin analog, appears to be a substrate of PnuX and YpaA. We propose to designate the gene names ribU for ypaA and ribM for pnuX to reflect that the encoded proteins function in riboflavin uptake and that the genes have different phylogenetic origins.
Riboflavin consists of a ribityl side chain linked to an aromatic isoalloxazine ring structure. It is the precursor of the cofactors flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), which both are essential components of cellular metabolism. Riboflavin is phosphorylated to give FMN by flavokinase (EC 2.7.1.26), and FMN is subsequently converted to FAD by FAD synthetase (EC 2.7.7.2). Whereas free riboflavin does not have biological activity, the mostly noncovalently bound flavin cofactors FMN and FAD are the active groups of a large number of flavoproteins. These are involved in a wide range of redox reactions and catalyze the dehydrogenation of metabolites, one- and two-electron transfer reactions from and to redox centers, and hydroxylation reactions (9). Flavins are also known to act as chromophores in photoreceptors, such as the plant blue light sensors cryptochrome and phototropin (reviewed in reference 3). Moreover, flavins are the ligands of dodecin, a recently identified flavoprotein that has the highest binding affinity to lumichrome, a light-induced degradation product of riboflavin with an alloxazine ring structure lacking a ribityl side chain (13, 48).
Whereas vertebrates can generate FMN and FAD from riboflavin, they lack the enzymes to synthesize riboflavin, making this compound a vitamin (vitamin B2). In contrast, plants and most microorganisms are capable of synthesizing riboflavin from GTP and ribulose 5-phosphate (1). The Bacillus subtilis riboflavin biosynthetic genes are arranged in an operon which includes a conserved regulatory element called the RFN element (10, 21, 33, 47). This element is present within the 5′-untranslated region of the rib transcript and folds into a secondary structure that binds FMN with high affinity (33, 47). Binding of FMN prevents the expression of the downstream genes by transcriptional or translational mechanisms (46, 47). In either case, the RFN element works as a genetic switch (riboswitch) that ensures that riboflavin is not produced in higher amounts than required for flavin cofactor biosynthesis. Consistent with this function, mutations that cause riboflavin overproduction map at various positions within the RFN element (14, 21) or in ribC, the gene encoding a bifunctional flavokinase/FAD synthetase (6, 26). Moreover, the B. subtilis genome encodes a monofunctional flavokinase (ribR), but the in vivo contribution of this protein to the generation of FMN remains unclear (15, 41).
Although a requirement for riboflavin is very rare among bacteria, it is known that riboflavin is an essential growth factor for Enterococcus faecalis, Streptococcus pyogenes, Listeria monocytogenes, and some lactobacilli (22). This biosynthetic deficiency correlates with the absence of riboflavin biosynthetic genes in the genomes of these organisms (46). The sensitive growth response of Lactobacillus casei to riboflavin was used to develop one of the first microbiological assays for a vitamin (40) and is based on the presence of an efficient transport system that allows the uptake of exogenous riboflavin. Riboflavin uptake is well characterized in B. subtilis, where it was found to be a high-affinity (Km, 5 to 20 nM), carrier-mediated process requiring metabolic energy (5). Riboflavin uptake inversely correlates with the riboflavin concentration present during cell growth and increases in riboflavin-requiring mutants (5). Moreover, B. subtilis was also found to contain a membrane-associated riboflavin-binding activity with similar overall characteristics as the riboflavin transport activity, indicating that the binding component has a functional role for riboflavin uptake (5).
Identification of candidate genes for bacterial riboflavin transporters has been facilitated by comparative genomic analyses. The genome of B. subtilis contains two RFN elements, one preceding the rib operon and the other preceding ypaA, a gene encoding a hypothetical membrane protein with five putative transmembrane domains (TMDs) (10, 23, 47). Transcription profiling experiments revealed that ypaA was more strongly expressed in ribC mutants (25) and that deletion of ypaA in a riboflavin-auxotrophic mutant drastically increased the demand for riboflavin, FMN, or FAD (23). Although these data and the presence of orthologs of YpaA in riboflavin-auxotrophic species (46) suggested a role of YpaA in riboflavin uptake, this possible function has not been experimentally studied before. Here, we report the characterization of YpaA as a riboflavin transporter and also study the Corynebacterium glutamicum gene pnuX. This gene is similar to ypaA in that it is preceded by an RFN riboswitch and that it encodes a protein with multiple TMDs (46). YpaA from B. subtilis is related to the recently characterized RibU riboflavin transporter from Lactococcus lactis (4) and is a member of a protein family that includes riboflavin transporters, sodium symporters for bile acids, and exporters for arsenite from eukaryotic and bacterial organisms (29). In contrast, pnuX is not a member of the BART (bile/arsenite/riboflavin transporter) superfamily but is related to RibM from Streptomyces davawensis (12) and orthologous proteins in other sequenced gram-positive species.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The Escherichia coli strains used were BL21(DE3) [F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3)] or the riboflavin-auxotrophic ribB11 mutant BSV11 [F− glnV44(AS) λ− mcrA rfbC1 endA1 ribB11:Tn5 spoT1 thi-1 mcrB hsdR29; E. coli Genetic Stock Center no. 6991] (2). DH5α (F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG φ80dlacZΔM15 Δ(lacZYA-argF) U169 hsdR17(rK− mK+) λ−) was used for molecular cloning and for the experiment shown below in Fig. 5E. E. coli cells were grown in LB, 2TY, or M9 minimal medium supplemented with antibiotics as required (100 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, or 34 μg ml−1 chloramphenicol). For growth of BSV11, 20 mg/liter riboflavin was added.
FIG. 5.
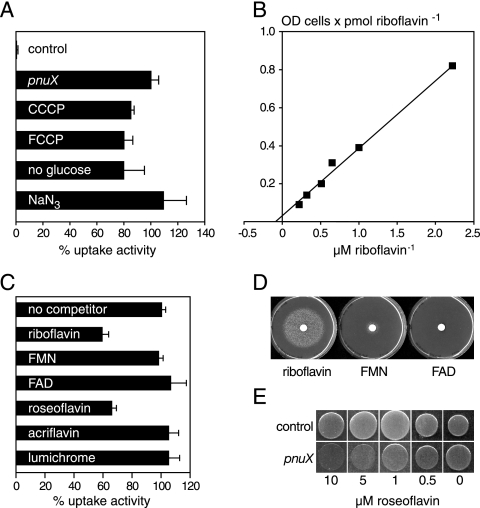
Characterization of PnuX from Corynebacterium glutamicum as a riboflavin transporter. All uptake experiments were performed in the presence of 1 mM glucose with E. coli BL21(DE3) expressing a 3HA-tagged version of pnuX from plasmid pET21a(+) or harboring an empty pET21a(+) plasmid (control). (A) E. coli BL21 cells were either measured directly (standard assay; 100% corresponds to an activity of 5.57 pmol riboflavin × the OD in cells−1 min−1), in the presence of the protonophores CCCP (130 μM) or FCCP (130 μM), without glucose, or after the addition of 1 mM sodium azide. The glucose dependency was assayed as in Fig. 3D. (B) pET21a(+)-pnuX-3HA-transformed cells were subjected to uptake experiments in the presence of various concentrations of [14C]riboflavin. The graph represents the Lineweaver-Burk plot of an experiment from which a Km value of 11 μM was calculated. Two repetitions of this experiment yielded Km values of 5 μM and 17 μM, respectively. (C) Uptake experiments were performed with 2.2 μM [14C]riboflavin in the presence of a 10-fold excess of riboflavin or riboflavin analogs. (A and C), bars represent means and error bars indicate standard deviation values of three independent determinations. The uptake activity in the absence of competitors was 5.76 pmol riboflavin × OD cells−1 min−1 (100%). (D) E. coli ribB11 mutants were plated on riboflavin-free M9 minimal medium to produce a lawn and growth was assayed as in Fig. 3B. The area of growth had a diameter 53 mm for riboflavin and 18 mm for FMN. FAD produced no growth, even after incubation for one more day. (E) E. coli DH5α cells expressing pnuX from plasmid pDG148-pnuX-6His (pnuX) or containing an empty pDG148 plasmid (control) were spotted on M9 minimal medium plates containing the indicated concentrations of roseoflavin. Growth was recorded after 1 day at 37°C.
LB medium or SP medium (8 g liter−1 nutrient broth, 1 mM MgSO4, 10 mM KCl, 0.5 mM CaCl2, 10 μM MnCl2, 4.4 mg liter−1 ferric ammonium citrate) (30) was used to propagate B. subtilis strains. All B. subtilis strains used were derived from wild-type strain 168. Strain QB5350 [trpC2 ptsH-H15A amyE::(levD-lacZ aphA3)] (44) has a point mutation in the ptsH gene and is devoid of PTS-dependent sugar transport activity. CSEG, which is C-medium (30) with added sodium succinate (6 g liter−1), potassium glutamate (8 g liter−1), and glucose (1 g liter−1), is a chemically defined medium devoid of vitamins and was used for growth and uptake assays. For energization of fructose uptake, glucose was replaced by the same amount of sorbitol. For marker selection, erythromycin/lincomycin (1 μg ml−1/5 μg ml−1), kanamycin (5 μg ml−1), or phleomycin (0.5 μg ml−1) was added to the medium. Solid 2TY medium and SP plates were prepared using 2% Agar-agar (Roth), and CSEG and M9 minimal media were solidified with 2% Bacto agar (Difco). Cells for use in plate growth assays were diluted to an optical density at 600 nm (OD600) of 0.06, and 5 μl of the cell suspension (approximately 50,000 cells) was transferred to agar plates.
DNA manipulations.
B. subtilis was transformed using a two-step protocol (24) and selected on SP plates supplemented with antibiotics as required. To create the ΔypaA:Kanr knockout strain, 1-kb flanking regions from either side of ypaA were amplified from B. subtilis genomic DNA and inserted up- or downstream of the kanamycin resistance gene in the plasmid pDG780. The plasmid was linearized with ScaI, which cuts in the Ampr gene, and transformed into B. subtilis.
To delete ribB, we amplified ribB with 1-kb up- and downstream regions and cloned it into a pUC19 derivative with a single NotI site. Making use of naturally occurring sites (EcoRV, which truncates the preceding ribD gene by 27 bp, and ClaI, which cuts within ribB), we replaced ribB with the erythromycin resistance gene. Alternatively, a ΔribB::Tetr knockout cassette was made by substituting the Ermr marker with the Tetr gene. The knockout cassettes were liberated with NotI, and B. subtilis wild-type cells or the ΔypaA::Kanr mutant was transformed with the DNA fragment. ypaA was C-terminally His tagged using a specific primer encoding eight consecutive histidine residues. The PCR product was inserted into the integrative plasmid pMUTIN-HA (19), thereby removing the hemagglutinin (HA) tag. B. subtilis wild-type cells or the ΔribB::Tetr mutant was directly transformed with the resulting plasmid followed by selection for erythromycin resistance. Correct loop-in of the plasmid into the genomic ypaA locus was verified by PCR and Western blot analysis. Overexpression of ypaA was achieved by amplification of the ypaA open reading frame with specific adaptor primers for cloning into pDG148-Stu that allows isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression (17). The promoter region of ypaA including the RFN element was amplified by PCR and fused to the lacZ reporter gene in vector pAC6 (44). The plasmid was linearized with ScaI and integrated into the amyE locus of B. subtilis wild-type cells or the ΔribB::Ermr mutant. LacZ assays were performed as described elsewhere (32).
Two plasmids were made to express the C. glutamicum transporter pnuX in E. coli. For pDG148-pnuX-6His, pnuX was amplified without the stop codon from C. glutamicum (strain ATCC 13032) genomic DNA. The PCR product was cloned into pQE60 (QIAGEN), isolated as a HindIII fragment including the His tag, and ligated into pDG148 (42). The pET21a(+)-pnuX-3HA plasmid was made by cloning a tagged version of pnuX into pET21a(+). A stop codon after the 3HA epitope tag prevented the translation into the vector-borne His tag.
Construction of GFP, phoA, and lacZ fusions to ypaA.
Topological predictions, which were the basis for the following constructs, were performed with TMpred (16). ypaA (lacking its stop codon) or the C-terminally truncated versions ypaA-4 (containing the first four TMDs, amino acids [aa] 1 to 146) or ypaA-3 (containing the first three TMDs, aa 1 to 103) were amplified and cloned into pUC18. Next, the green fluorescent protein (GFP) gene, in which the start methionine was replaced with a VDGAGA linker, or alkaline phosphatase from E. coli (phoA, starting after aa 14 to remove the signal peptide) were PCR amplified and inserted into the pUC18 plasmids containing the different forms of ypaA. For lacZ tagging, the β-galactosidase gene from E. coli lacking the first two amino acids was amplified by PCR and cloned into pUC18. Next, the various fragments of ypaA were inserted in front of lacZ. Finally, all fusion constructs were amplified by PCR using specific adaptor primers, cloned downstream of the IPTG-inducible Pspac promoter of pDG148-Stu (17), and transformed into B. subtilis 168.
Analysis of cells containing GFP-, phoA-, and lacZ-tagged ypaA.
For all assays, the cells containing the plasmids described above were grown in LB medium supplemented with 0.5 μg ml−1 phleomycin overnight, transferred to fresh medium containing 0.5 mM IPTG to an OD600 of 0.15, and aerobically cultivated at 37°C until they reached an OD600 of 1.0. For PhoA assays, 1 mM iodoacetamide was added to the culture 10 minutes before harvesting to prevent spontaneous activation of cytosolic PhoA (36). Iodoacetamide was also added to all subsequently used buffers. PhoA activity was determined with 3 ml of the cells as described elsewhere (27). The activity of β-galactosidase was measured with a standard procedure (32). For plate overlay assays, cells expressing the lacZ fusions were spotted on SP plates and grown at 37°C overnight. The overlay mixture consisted of 5 mg ml−1 low-melting-point agarose in 25 ml 50 mM K2HPO4/KH2PO4 pH 7.0, 0.05% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 7 mM β-mercaptoethanol, 0.1 mg ml−1 lysozyme, and 2% Triton X-100 (vol/vol). The plates were incubated at 30°C until the color developed. GFP was determined fluorimetrically with cells washed once with 50 mM Tris-HCl pH 8.0, 200 mM NaCl, 15 mM EDTA pH 8.0 and resuspended at an OD600 of 0.9. GFP was excited at 488 nm, and the emission at a 512-nm wavelength was recorded on a Fluoromax-2 instrument (ISA Instruments). Confocal microscopy was performed on a Zeiss LSM 510 META microscope.
Preparation of B. subtilis membranes and Western blot assays.
B. subtilis membranes were prepared from 40-ml cultures as described elsewhere (31). Briefly, after harvesting and washing with 50 mM Tris-HCl pH 8.0, the cells were resuspended in 1 ml freshly prepared and prewarmed TMS buffer (50 mM Tris-HCl pH 8.0, 16 mM MgCl2, 33% [wt/vol] sucrose). Lysozyme (final concentration, 300 μg ml−1) and 1 mM phenylmethylsulfonylfluoride were added, and the suspension was incubated for 30 min at 37°C. The protoplasts were cooled to 4°C, harvested by centrifugation (1 min, 7,500 × g), resuspended in 1 ml lysis buffer (50 mM Tris-HCl pH 8.0, 5 mM MgSO4) containing phenylmethylsulfonyl fluoride, and sonified on ice. Membranes were collected by centrifugation (15 min, 110,000 × g), washed once with 50 mM Tris-HCl pH 8.0, and resuspended in 50 μl 50 mM Tris-HCl pH 8.0. Samples of 15 μl were analyzed on 10% (see Fig. 4D and E, below) or 15% (see Fig. 2B, below) polyacrylamide gels. After transfer to nitrocellulose membranes, His-tagged YpaA was detected with the Penta-His mouse monoclonal antibody (QIAGEN), and GFP-tagged YpaA was detected with mouse anti-GFP monoclonal antibody 11E5 (Molecular Probes). Anti-mouse peroxidase-coupled secondary antibodies were obtained from Sigma (A-9044). PhoA fusion proteins were detected with rabbit anti-alkaline phosphatase antiserum (Polyscienes, Inc.), followed by peroxidase-coupled anti-rabbit antibodies (A-6154; Sigma). Chemiluminescence reagents (GE Healthcare) were used to develop the blots.
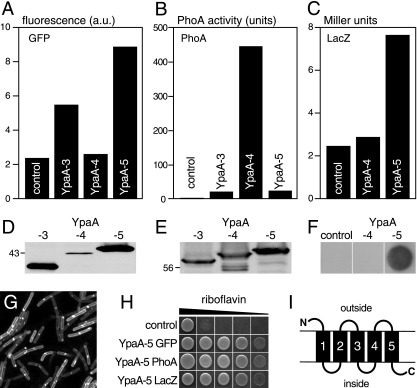
FIG. 4.
Membrane topology of YpaA. The topology of YpaA was analyzed by generating a C-terminal fusion of YpaA to GFP (A), PhoA (B), or LacZ (C) in the vector pDG148-Stu. According to the prediction of the TMpred program (16), the fusion proteins contained either three (YpaA-3; fusion after amino acid 103) or four (YpaA-4; fusion after amino acid 146) transmembrane domains of YpaA or the full-length protein (YpaA-5). Plasmids were transformed into B. subtilis wild-type cells, which were grown in LB medium and induced with IPTG prior to the analysis. The activity of GFP was determined by fluorescence spectroscopy of intact cells. The fluorescence of vector control cells was used to normalize the measurements. Arbitrary units (a.u.) are given. The activity of alkaline phosphatase was assayed in intact cells with p-nitrophenyl phosphate using a standard protocol (27). The activity of β-galactosidase is presented as Miller units and was assayed with ortho-nitrophenyl-β-d-galactoside as a substrate (32). (D) The GFP-fusion proteins used in panel A were detected in a Western blot assay with anti-GFP serum. The calculated molecular masses of the GFP fusions were as follows: 38.2 kDa for YpaA-3, 42.7 kDa for YpaA-4, and 47.6 kDa for YpaA-5. (E) A similar analysis was performed for the cells used in panel B. The calculated molecular masses of the PhoA fusions were as follows: 59.1 kDa for YpaA-3, 63.7 kDa for YpaA-4, and 68.7 kDa for YpaA-5. (F) Cells expressing fusions of LacZ to YpaA-4 and YpaA-5 were plated on an SP plate containing phleomycin and 0.5 mM IPTG. After growth, they were covered with X-Gal containing agarose and left for 3 hours at 37°C. The cells with the YpaA-5 construct turned blue, whereas the other cells did not show a color reaction. (G) The cells expressing YpaA-5 GFP were also visualized using confocal microscopy. (H) We tested the functionality of the fusion constructs used in panels A to G in B. subtilis ΔribB::Ermr ΔypaA::Kanr mutants. Cells containing the indicated plasmids were spotted on IPTG-containing CSEG plates supplemented with (from left to right) 20 mg/liter, 2 mg/liter, 200 μg/liter, 20 μg/liter, or 2 μg/liter riboflavin. Growth was recorded 1 day after plating. Fusion proteins containing YpaA-3 or YpaA-4 did not complement the growth defect of the ΔribB mutant. (I) A model of the transmembrane topology of YpaA is shown.
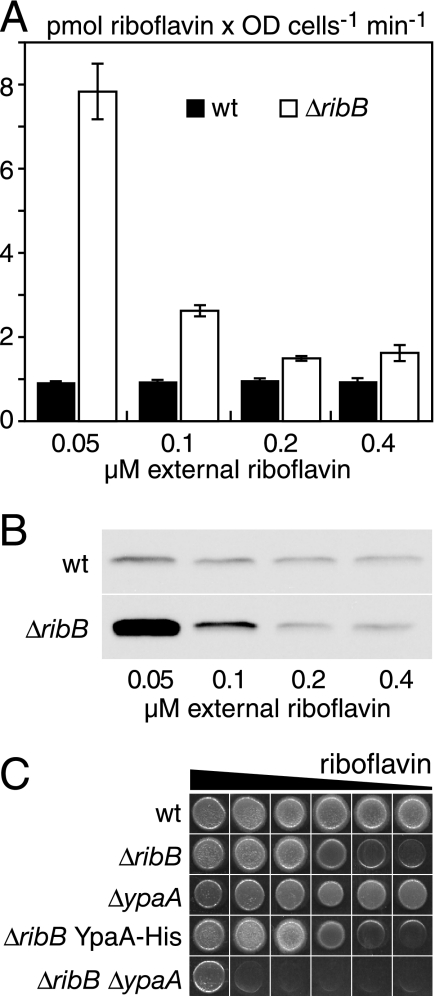
FIG. 2.
Regulation of riboflavin uptake activity by external riboflavin. (A) B. subtilis wild-type cells or a B. subtilis ΔribB:Ermr mutant cells were grown in synthetic CSEG medium to which riboflavin was added to give the desired concentrations. Standard uptake experiments with [14C]riboflavin were performed, and the uptake velocities were calculated for the first 4 minutes of the experiments. Bars represent means, and error bars indicate standard deviation values of three independent experiments. (B) YpaA was genomically tagged with a C-terminal his tag either in a wild-type strain or a strain carrying the ΔribB:Tetr disruption. After growth in CSEG medium containing the indicated concentrations of riboflavin, the cells were collected and membrane extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by transfer to a nitrocellulose membrane and detection with the Penta-His mouse monoclonal antibody. (C) Growth assays on CSEG plates containing (from left to right) 20 mg/liter, 2 mg/liter, 200 μg/liter, 20 μg/liter, 2 μg/liter, or 200 ng/liter riboflavin were performed. Each spot contained approximately 50,000 cells, and the plates were incubated for 1 day at 37°C. The exact genotype of the strains was ΔribB:Ermr, ΔypaA:Kanr, and the double mutant carried both these deletions.
Uptake experiments.
B. subtilis cells for use in riboflavin uptake experiments were grown overnight in CSEG, and E. coli cells were grown in M9 minimal medium supplemented with antibiotics, IPTG, or riboflavin as required. Medium was inoculated to an OD600 of 0.15, and the cells were grown at 37°C until they reached an OD600 of 0.5 to 0.6. B. subtilis cells were used directly, and E. coli cells were induced with 0.5 mM IPTG and grown for 3 h (OD600, 0.8). The cells were harvested, washed with ice-cold water and with transport buffer (50 mM KH2PO4/K2HPO4, 50 mM MgCl2 pH 7.0), resuspended in transport buffer (10 OD600 ml−1), and stored on ice. Uptake experiments were performed with 500 μl of cells that were vigorously stirred at 30°C. After warming for 2 min, glucose was added to a final concentration of 1 mM, and the assay was started 1 min later by adding [14C]riboflavin (specific activity, 5.54 MBq/mg; a gift of R. Krämer, Köln, Germany) for a final concentration of 1.6 or 2.2 μM. Several aliquots were removed over the next 3 or 4 minutes, filtered on 0.45-μm GN-6 membrane filters (Pall), washed with an excess of water, and analyzed by liquid scintillation counting. Roseoflavin was obtained from MPI Biochemicals, acriflavin and lumichrome were from Sigma, and FMN and FAD were from Merck. Carbonyl cyanide m-chlorophenylhydrazone (CCCP; 130 μM), carbonyl cyanide p-[tri-fluoromethoxy]phenylhydrazone (FCCP; 130 μM), or NaN3 (1 mM) was added 3 min before addition of the labeled substrate. To test fructose uptake, B. subtilis wild-type cells or ptsH mutants were grown in CSE medium containing sorbitol instead of glucose as a carbon source. The substrate, a mixture of d-[U-14C]fructose (CFB47; GE Healthcare) and unlabeled fructose, was added to a final concentration of 58 μM.
RESULTS
YpaA is involved in riboflavin uptake.
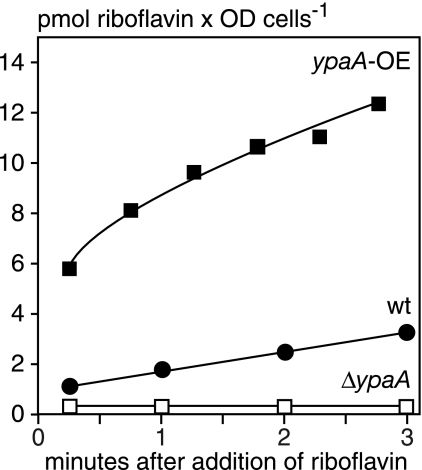
The gene ypaA from B. subtilis is regulated by an RFN element and is expressed when the cells need flavins (25). It encodes a hydrophobic protein with five putative TMDs that was predicted to catalyze riboflavin uptake across the plasma membrane (47). However, this function was not experimentally verified. To functionally characterize YpaA, we generated a B. subtilis mutant in which ypaA was deleted (ΔypaA::Kanr). In addition, we overexpressed ypaA in B. subtilis using the plasmid pDG148-Stu (17). Riboflavin uptake was monitored using [14C]riboflavin, and it was found that deletion of ypaA abolished riboflavin uptake, whereas overexpression of ypaA significantly increased riboflavin uptake relative to wild-type controls (Fig. 1). Furthermore, overexpression of ypaA resulted in a rapid increase in the cellular radioactivity after adding the labeled substrate. This may indicate that YpaA, like its L. lactis ortholog RibU, has a high binding activity for its substrate (8). These results strongly suggest that YpaA corresponds to the B. subtilis riboflavin transporter first described in 1979 (5).
FIG. 1.
Riboflavin uptake in B. subtilis depends on YpaA. Uptake assays were performed with B. subtilis strains grown in riboflavin-free CSEG medium. The strains were either wild type (wt), contained a plasmid for overexpression of ypaA (ypaA-OE), or had a deletion of the ypaA gene (ΔypaA::Kanr). The cells were resuspended in transport buffer (50 mM K2HPO4/KH2PO4, 50 mM MgCl2, pH 7.0), energized with 1 mM glucose, and the experiment was started by adding [14C]riboflavin for a final concentration of 1.6 μM. Aliquots were withdrawn, filtered, and extensively washed with water, and the radioactivity was determined by liquid scintillation counting. The experiment was performed twice with similar results.
We noted that YpaA also has weak similarity to membrane-localized subunits of phosphoenol-pyruvate:carbohydrate phosphotransferase systems (PTS). In addition, riboflavin contains a ribityl side chain and thus is structurally related to the carbohydrate substrates of PTS. Moreover, riboflavin taken up by B. subtilis is quickly converted to the phosphorylated derivatives FMN and FAD (5). These findings suggested a role for YpaA as a membrane component of a putative riboflavin-specific PTS. We performed uptake experiments in ptsH mutants that lack HPr, the universal phosphoryl donor for all membrane-bound enzymes II and thus are unable to take up PTS substrates (43). ptsH mutants displayed normal riboflavin uptake but, as expected, the uptake of the PTS sugar fructose was strongly reduced (data not shown). Thus, it is unlikely that YpaA functions as a membrane component of a PTS that mediates riboflavin uptake.
Regulation of riboflavin transport activity.
As a next step in the characterization of ypaA, we checked if the activity of riboflavin uptake was influenced by the ability of the cells to synthesize riboflavin or by the amount of riboflavin present in the growth medium. To generate a riboflavin-auxotrophic B. subtilis strain, the ribB gene encoding the α-subunit of riboflavin synthase was inactivated. The ribB deletion strain (ΔribB::Ermr) and a wild-type control were grown in a synthetic medium containing various concentrations of riboflavin, and the cells were used in uptake experiments. The experiments revealed that deletion of ribB caused an increased riboflavin uptake activity. This was most obvious in media with low riboflavin concentrations, where ΔribB mutants possessed at least eightfold more riboflavin transport activity than wild-type cells (Fig. 2A).
It was also analyzed whether the observed changes in the riboflavin uptake activity were reflected by the amount of YpaA present in the cells. To visualize YpaA in Western blot assays, a plasmid carrying a C-terminally His-tagged version of ypaA was integrated into the ypaA locus of a wild-type strain or the ΔribB::Tetr deletion strain (YpaA-His). This ensured that the modified ypaA gene was linked to the upstream RFN element and that the expression of the second untagged copy of ypaA was abolished (19). Membrane fractions were prepared from cultures growing in various concentrations of riboflavin, solubilized, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to nitrocellulose for detection with the Penta-His monoclonal antibody. This analysis revealed that the increased riboflavin uptake activity of the ΔribB strain in riboflavin-depleted medium was caused by an increased amount of YpaA. In contrast, wild-type cells had similar levels of YpaA at all riboflavin concentrations tested (Fig. 2B).
To confirm that ypaA is regulated by riboflavin, we generated a transcriptional fusion of the ypaA promoter including the RFN element to lacZ and integrated this construct into the amyE locus of wild-type cells or ΔribB::Ermr mutants. When both strains were grown in liquid medium containing 0.05 μM riboflavin, the ΔribB::Ermr mutant possessed sevenfold more β-galactosidase activity than the wild-type strain (data not shown). These findings are consistent with transcription profiling experiments that showed that ypaA is not regulated by riboflavin in a wild-type strain but that its expression is highly increased in a ribC mutant (25).
Finally, a ΔribB::Ermr ΔypaA::Kanr double mutant was generated and compared to the strains used in the experiments described above. Whereas ΔribB single mutants showed normal growth on medium containing riboflavin at concentrations of 20 μg/liter or more, ΔribB ΔypaA double mutants required at least 20 mg/liter riboflavin to grow. A ΔribB strain producing YpaA-His showed similar growth as the ΔribB strain producing untagged YpaA, demonstrating that the His tag did not interfere with the function of YpaA. As expected, ΔypaA::Kanr cells behaved like wild-type cells and were proficient for riboflavin biosynthesis (Fig. 2C). In summary, these experiments show that riboflavin deficiency leads to an increased production of YpaA and that this enhances riboflavin uptake.
Characterization of the activity of YpaA.
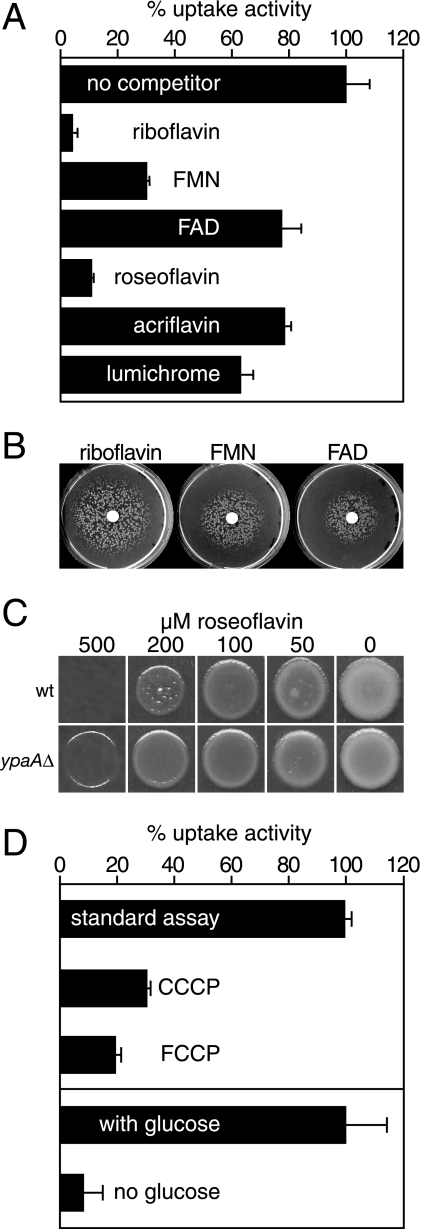
Riboflavin uptake experiments in the presence of putative competitors demonstrated that the activity was strongly reduced when an excess of riboflavin or FMN was present (Fig. 3A). Riboflavin, FMN, and FAD improved the growth of ΔribB mutants when added on a filter disk and placed on riboflavin-free plates (Fig. 3B). In these assays, riboflavin was more potent than FMN, which was more potent than FAD, reflecting the relative activities of these compounds in the uptake experiment (Fig. 3A). Roseoflavin (8-dimethyl-amino-8-demethyl-d-riboflavin), a toxic riboflavin analog produced by Streptomyces davawensis (34), strongly competed with riboflavin uptake (Fig. 3A). On plates containing high concentrations of roseoflavin, B. subtilis wild-type cells grew more poorly than ΔypaA::Kanr mutants, demonstrating that YpaA mediates roseoflavin uptake and sensitivity (Fig. 3C). Flavins lacking the ribityl side chain, such as lumichrome and acriflavin, did not significantly reduce riboflavin uptake when present in a 10-fold excess (Fig. 3A).
FIG. 3.
Characterization of the activity and energy requirements of YpaA. (A) Uptake of [14C]riboflavin (initial outside concentration, 1.6 μM) was studied in B. subtilis ΔribB:Ermr mutants grown in CSEG minimal medium containing 20 μg/liter riboflavin. Uptake was determined in the absence of competitors (mean uptake velocity, 8.29 pmol riboflavin × OD of cells−1 min−1; corresponds to 100%) or in the presence of a 10-fold excess (16 μM) of the given compounds. Bars represent means, and error bars indicate standard deviation (SD) values of three independent determinations. (B) B. subtilis ΔribB:Ermr mutants were plated on riboflavin-free CSEG minimal medium to produce a lawn. Next, a filter disc was placed in the middle of the plates and impregnated with 20 μl of a solution containing 540 μM riboflavin, FMN, or FAD. The area of growth around the disc was scored after incubation for 1 day at 37°C and had a diameter of 76 mm for riboflavin, 50 mm for FMN, and 42 mm for FAD. (C) B. subtilis wild-type cells (wt) or the ΔypaA::Kanr mutant were spotted on CSEG plates containing the indicated concentration of roseoflavin. Growth was recorded after incubation for 1 day at 42°C. (D) Uptake of [14C]riboflavin was determined using a ΔribB::Ermr mutant grown in CSEG medium containing 20 μg/liter riboflavin. The transport activity was determined without further additions (uptake velocity, 7.13 pmol riboflavin × OD cells−1 min−1; corresponds to 100%) or after adding CCCP (130 μM) or FCCP (130 μM) 3 min before starting the test by addition of the labeled substrate. To investigate if glucose stimulated riboflavin uptake, we stored the cells on ice for 2 hours in transport buffer. After this time, uptake activity was measured with [14C]riboflavin in the absence or presence of 1 mM glucose. In this experiment, 100% corresponds to an uptake activity of 5.1 pmol riboflavin × OD cells−1 min−1. Bars represent means, and error bars indicate SD values of three independent experiments.
Earlier studies in B. subtilis revealed that riboflavin uptake depended on metabolic energy and that uptake was drastically reduced in the presence of protonophors or metabolic inhibitors (5). In contrast, the recently characterized riboflavin transporter RibU could not be demonstrated to require metabolic energy for transport and likely uses a facilitated diffusion mechanism (4). These contradictory findings for two related proteins prompted us to reinvestigate riboflavin uptake in B. subtilis. To test if riboflavin uptake depends on metabolic energy, wild-type cells were stored in transport buffer lacking glucose for 2 hours, and uptake assays were performed in the presence or absence of glucose. In these experiments, the lack of glucose reduced the transport activity by more than 90% (Fig. 3D). Moreover, addition of the protonophors FCCP and CCCP caused a strong reduction of riboflavin uptake (Fig. 3D). In summary, these findings support a proton-symport mechanism for riboflavin uptake in B. subtilis.
Membrane topology of YpaA.
Computational analysis of the YpaA primary structure led to the prediction that YpaA spans the membrane with five hydrophobic α-helices. This finding is consistent with a recent phylogenetic analysis suggesting that YpaA belongs to a family of transport proteins that includes members with 5, 6, and 10 TMDs (29). Some of the six-TMD proteins contain catalytic C-terminal domains likely facing the cytoplasm, and TMDs 2 to 6 of these proteins have the highest similarity to the five TMDs of YpaA. Thus, it was concluded that the N terminus of YpaA lies on the extracellular side of the plasma membrane and that the C terminus faces the cytoplasm (29).
To investigate the topology of YpaA, we generated fusion proteins in which alkaline phosphatase (PhoA), GFP, or β-galactosidase (LacZ) was fused to the C terminus of YpaA. Alkaline phosphatase is only active when localized in the extracellular space, because it requires oxidative conditions for proper folding (27, 28). GFP is only active in the cytoplasm and does not form an active protein when exposed to the extracellular side (7). Similar to GFP, LacZ is only active in the cytoplasm (27). We found that shorter segments containing only the first or the first two TMDs of YpaA gave ambiguous results with the various tags, probably because the topological information contained in these fragments was insufficient to give the fusion proteins a distinct orientation (data not shown). However, experiments with longer fragments of YpaA, including the first three (YpaA-3) or four (YpaA-4) TMDs or the full-length protein (YpaA-5), produced conclusive results. Cells expressing YpaA-3 and YpaA-5 fused to GFP had high levels of fluorescence, whereas the YpaA-4 fusion had only background activity (Fig. 4A). The complementary pattern was obtained with the PhoA fusions, where YpaA-4 produced much more activity than YpaA-3 and YpaA-5, indicating an extracellular position of the loop following TMD4 (Fig. 4B). The pattern of GFP fluorescence was matched by β-galactosidase assays that demonstrated higher activity when LacZ was fused to YpaA-5 than to YpaA-4 (Fig. 4C). Control experiments demonstrated that the PhoA fusions described above were well expressed in the cells analyzed for PhoA activity (Fig. 4E). The GFP fusions to YpaA-3 and YpaA-5 were more abundant than the fusion to YpaA-4, likely indicating instability of the latter, probably an incorrectly folded protein (Fig. 4D). For the LacZ fusions, β-galactosidase activity was observed in a plate overlay assay only for YpaA-5 but not for YpaA-4 (Fig. 4F). As expected, confocal microscopy localized the YpaA-5 fusion to GFP to the periphery of B. subtilis cells, a position likely corresponding to the plasma membrane (Fig. 4G). To check the activity of the fusion proteins derived from YpaA-5, they were expressed in the ΔribB ΔypaA strain. Overproduction of any of the three fusion proteins allowed growth at low riboflavin concentrations, demonstrating that the fusion proteins were functional in transporting riboflavin (Fig. 4H). In summary, these results are consistent with a protein model that contains five TMDs with the N terminus localized to the extracellular space and the C terminus facing the cytoplasm (Fig. 4I).
Characterization of pnuX from Corynebacterium glutamicum.
Phylogenetic analyses of genes regulated by upstream RFN elements identified three different types of putative bacterial riboflavin transporters. These are (i) orthologs of YpaA and RibU, which are bona fide riboflavin transporters, (ii) ImpX of Fusobacterium nucleatum and Desulfitobacterium halfniense, and (iii) orthologs of PnuX which are present in the riboflavin operons of Thermonospora fusca, Streptomyces coelicor, and Streptomyces davawensis and as a solitary gene in C. glutamicum and Corynebacterium diphtheriae (12, 46). YpaA, ImpX, and PnuX have unrelated primary structures, a fact that is also reflected in the predicted protein topologies that suggest 5 TMDs for YpaA and orthologs, 6 to 7 TMDs for PnuX and orthologs, and 8 to 10 TMDs for most members of the ImpX family.
Of these proteins, PnuX of C. glutamicum (NCgl0063) was investigated with respect to riboflavin uptake. E. coli cells producing a tagged version of PnuX had increased rates of riboflavin uptake relative to control E. coli cells which, in agreement with previous observations (1), showed no endogenous riboflavin uptake activity (Fig. 5A). Expression of pnuX in E. coli allowed riboflavin uptake, which was saturable (Km, 11 ± 6 μM [mean ± standard deviation]) (Fig. 5B). The uptake experiments were repeated in the presence of putative competitors of riboflavin uptake. Of these, only roseoflavin significantly reduced [14C]riboflavin uptake, whereas FMN, FAD, acriflavin, and lumichrome had no effect. We observed that a 10-fold excess of unlabeled riboflavin reduced the uptake of [14C]riboflavin by 40%. This is explained by the fact that the concentration of labeled substrate (2.2 μM) was below the Km. In this situation the unlabeled substrate not only competes with the labeled substrate for uptake but also increases the overall transport velocity. Roseoflavin also reduced the velocity of riboflavin uptake by 40% (Fig. 5C), demonstrating that both riboflavin and roseoflavin are equally well transported by PnuX. In support of the role of PnuX in roseoflavin uptake, expression of pnuX in E. coli caused roseoflavin sensitivity (Fig. 5E). After transformation of the riboflavin auxotrophic E. coli ribB11 mutant with a pnuX-containing plasmid, we found that riboflavin caused the largest zone of growth (Fig. 5D). FMN led only to a small zone of growth in the direct vicinity of the filter disc, whereas FAD did not support growth. Similar to the experiments described above for YpaA, the energy requirements of PnuX were assayed. Riboflavin uptake was not affected by the absence of glucose or the addition of sodium azide, indicating that it is independent of metabolic energy (Fig. 5A). This finding was supported by uptake assays in the presence of the protonophors CCCP and FCCP, which both caused only a small reduction in riboflavin uptake (Fig. 5A). In summary, PnuX is a transporter for riboflavin and roseoflavin, and transport via PnuX appears to occur independently of metabolic energy. Thus, the different phylogenetic origins of the riboflavin transporters PnuX and YpaA are reflected in the biochemical properties of the two proteins.
DISCUSSION
B. subtilis is a model organism to study riboflavin uptake and biosynthesis and is used in industrial processes for riboflavin production (1, 5, 26, 35). Expression of the B. subtilis riboflavin operon is modulated by the flavin cofactor FMN, which, if available in a sufficient amount, binds to the RFN element and prevents the expression of the downstream structural genes (47). A similar RFN element is also present upstream of ypaA. This gene codes for a membrane protein that leads to an increased requirement for riboflavin when deleted in riboflavin auxotrophic mutants (23), making it a candidate riboflavin transporter.
The biochemistry of riboflavin transport in B. subtilis was thoroughly investigated in earlier studies (5). It was found that riboflavin is taken up by a high-affinity transport mechanism requiring metabolic energy. This activity can now be fully ascribed to YpaA. We characterize YpaA as a protein of the B. subtilis plasma membrane that requires metabolic energy for substrate translocation and whose expression is regulated by the availability of riboflavin. In many respects, YpaA is similar to the related riboflavin transporter RibU from L. lactis (4, 8). Although RibU is predicted to contain six TMDs, the fifth potential TMD of RibU is postulated to be part of a loop rather than a TMD (4). For YpaA, we found evidence for the presence of five TMDs and a C terminus located in the cytoplasm (Fig. 4). This topology is consistent with the finding that longer members of the superfamily of BART transporters, to which YpaA and RibU belong, have catalytic domains that would face the cytoplasm only when the position corresponding to the C terminus of YpaA is cytosolic (29). Duurkens et al. (8) demonstrated a high level of riboflavin binding to RibU, and our data (Fig. 1), as well as data from earlier experiments (5), are compatible with riboflavin binding to YpaA. A distinct difference between YpaA and RibU appears to lie in their energy requirements. Whereas RibU could not be demonstrated to require metabolic energy for transport (4), our data and the data of Ceccini et al. (5) clearly show that riboflavin uptake in B. subtilis requires metabolic energy. Although some of the cellular radioactivity in our transport assays might reflect riboflavin binding, we also found that the amount of radioactivity was strongly reduced in the presence of metabolic inhibitors such as the protonophors CCCP and FCCP or when no glucose was present during the uptake experiment. In agreement with earlier work, we thus postulate that YpaA is a proton-riboflavin symporter. YpaA is strikingly different from the C. glutamicum riboflavin transporter PnuX, which appears to be a facilitator for riboflavin. Moreover, PnuX has a lower apparent affinity for riboflavin (Km, 11 ± 6 μM) (Fig. 5B) than YpaA (Km, 5 to 20 nM) (5). As a facilitator, PnuX allows the flux of riboflavin down a concentration gradient. Since incoming riboflavin might become metabolically trapped by conversion to FMN, which does not appear to be a substrate of PnuX (Fig. 5A), PnuX is likely acting as an importer in vivo. Based on the biochemical characterization of the two proteins described above, we propose to designate the gene names ribU for ypaA and ribM for pnuX. These names reflect that both proteins function in riboflavin uptake, although they have different phylogenetic origins and no detectable sequence similarity.
It is interesting that PnuX from C. glutamicum has some homology (21% identity and 43% similarity) to transporters for nicotinamide riboside, such as PnuC from Haemophilus influenzae (39). Like riboflavin, nicotinamide riboside consists of a heterocyclic ring structure, fused to a sugar side chain, and is the building block of a redox-active cofactor, NAD+. Another bacterial transporter with a putative function in riboflavin uptake is encoded by impX. ImpX is unrelated to YpaA and PnuX and not functionally characterized (46). The identification of these bacterial proteins by comparative genomics (46) impressively demonstrates the power of computational analyses to predict protein function. Eukaryotic plasma membrane transporters for riboflavin are not related to any of these bacterial transporters. We have identified the Saccharomyces cerevisiae transporter Mch5p, which is a member of the major facilitator superfamily having 12 TMDs and working as a facilitator (38). Moreover, a mammalian riboflavin exporter has recently been identified that secretes riboflavin into milk and belongs to the ATP-binding cassette family of transport proteins (45).
With respect to the substrates transported by YpaA, we observed that riboflavin uptake is reduced in the presence of FMN or FAD (Fig. 3A). Moreover, the growth of the riboflavin-auxotrophic B. subtilis ΔribB strain was also supported by both flavin cofactors (Fig. 3B). At first sight, this indicates that FAD is transported in a YpaA-dependent fashion. However, another possibility is that FAD is degraded by extracellular enzymes and riboflavin is taken up. There are indications that this is indeed the case. First, whereas riboflavin, roseoflavin, and FMN bind with nanomolar affinities to the YpaA-related L. lactis riboflavin transporter RibU, this protein shows no binding of FAD (8). A second hint comes from the analysis of B. subtilis ribC mutants. RibC is the bifunctional B. subtilis flavokinase/FAD synthetase (26). B. subtilis ribC mutants can be complemented by FMN only when a heterologous monofunctional FAD synthetase accounts for the missing enzymatic activity (26). In contrast, FAD does not complement ribC mutants, indicating that FAD is not taken up (26). Thus, although riboflavin uptake is reduced in the presence of FAD, FAD does not appear to be a substrate of YpaA. Extracellular degradation of FAD and uptake of riboflavin is not without precedent. For example, the blood-borne bacterium Haemophilus influenzae cannot synthesize NAD+ or salvage nicotinate or nicotinamide. Instead, this organism depends on the uptake of nicotinamide riboside via the plasma membrane transporter PnuC (39). Nicotinamide riboside derives from NAD+ by the extracellular activity of NadN, an NAD+ pyrophosphatase that produces nicotinamide mononucleotide and AMP and also has nucleotide phosphatase activity to dephosphorylate nicotinamide mononucleotide and produce nicotinamide riboside (11, 20, 37). The dephosphorylation step can also be performed by e(P4), a periplasmic phosphatase with higher activity than NadN (11, 20, 37). In H. influenzae, neither NAD+ nor nicotinamide mononucleotide is transported by PnuC, which has specificity for nicotinamide riboside only (39). The NAD+ pyrophosphatase activity of H. influenzae has been studied in vitro and also has activity towards FAD, producing FMN and AMP (18). Homologs of NadN and other proteins that could act on FAD are encoded in the B. subtilis genome. These proteins have not been studied for a role in FAD degradation, but it is possible that their activity is similar to NadN. In the light of these findings, it is also questionable if FMN is transported by YpaA. Although FMN binds to RibU (8), represses the riboflavin operon in a RibU-dependent manner (4), reduces riboflavin uptake via YpaA (Fig. 3), and supports growth of B. subtilis ribB mutants (Fig. 3), these findings do not unequivocally prove that FMN is transported by YpaA. To resolve if FMN or FAD or both are substrates of bacterial riboflavin transporters, it will be necessary to study the degradation of these compounds by extracellular enzymes and to generate mutants that are defective in these pathways.
Acknowledgments
This work was supported by the DFG (GK640 and DFG STO434/2-1).
We thank Reinhard Krämer (Köln) for providing the C. glutamicum genomic DNA and [14C]riboflavin and Mary Berlin from the E. coli Genetic Stock Center for the ribB11 mutant BSV11. Plasmids were donated by Mohamed A. Mahariel (Marburg), Wolfgang Schumann (Bayreuth), and the Bacillus Genetic Stock Center. We also thank Birgit Flauger, Ina Weig-Meckl, Bert Poolman, Rainer Deutzmann, Widmar Tanner, Alfons Penzkofer, and Joachim Reidl for discussions and contributions and Peter Hegemann for initiating this project within GK640.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Bacher, A., S. Eberhardt, M. Fischer, K. Kis, and G. Richter. 2000. Biosynthesis of vitamin B2 (riboflavin). Annu. Rev. Nutr. 20:153-167. [DOI] [PubMed] [Google Scholar]
- 2.Bandrin, S. V., P. M. Rabinovich, and A. I. Stepanov. 1993. Three linkage groups involved in riboflavin biosynthesis in E. coli. Sovi. Genet. 19:1103-1109. [PubMed] [Google Scholar]
- 3.Banerjee, R., and A. Batschauer. 2005. Plant blue-light receptors. Planta 220:498-502. [DOI] [PubMed] [Google Scholar]
- 4.Burgess, C. M., D. J. Slotboom, E. R. Geertsma, R. H. Duurkens, B. Poolman, and D. van Sinderen. 2006. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J. Bacteriol. 188:2752-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cecchini, G., M. Perl, J. Lipsick, T. P. Singer, and E. B. Kearney. 1979. Transport and binding of riboflavin by Bacillus subtilis. J. Biol. Chem. 254:7295-7301. [PubMed] [Google Scholar]
- 6.Coquard, D., M. Huecas, M. Ott, J. M. van Dijl, A. P. van Loon, and H. P. Hohmann. 1997. Molecular cloning and characterisation of the ribC gene from Bacillus subtilis: a point mutation in ribC results in riboflavin overproduction. Mol. Gen. Genet. 254:81-84. [DOI] [PubMed] [Google Scholar]
- 7.Drew, D., D. Sjostrand, J. Nilsson, T. Urbig, C. N. Chin, J. W. de Gier, and G. von Heijne. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. USA 99:2690-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duurkens, R. H., M. B. Tol, E. R. Geertsma, H. P. Permentier, and D. J. Slotboom. 2007. Flavin binding to the high affinity riboflavin transporter RibU. J. Biol. Chem. 282:10380-10386. [DOI] [PubMed] [Google Scholar]
- 9.Fraaije, M. W., and A. Mattevi. 2000. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 25:126-132. [DOI] [PubMed] [Google Scholar]
- 10.Gelfand, M. S., A. A. Mironov, J. Jomantas, Y. I. Kozlov, and D. A. Perumov. 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 15:439-442. [DOI] [PubMed] [Google Scholar]
- 11.Gerlach, G., and J. Reidl. 2006. NAD+ utilization in Pasteurellaceae: simplification of a complex pathway. J. Bacteriol. 188:6719-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grill, S., H. Yamaguchi, H. Wagner, L. Zwahlen, U. Kusch, and M. Mack. Identification and characterization of two Streptomyces davawensis riboflavin biosynthesis gene clusters. Arch. Microbiol., in press. [DOI] [PubMed]
- 13.Grininger, M., K. Zeth, and D. Oesterhelt. 2006. Dodecins: a family of lumichrome binding proteins. J. Mol. Biol. 357:842-857. [DOI] [PubMed] [Google Scholar]
- 14.Gusarov, I. I., R. A. Kreneva, K. V. Rybak, D. A. Podcherniaev, V. Iomantas, L. G. Kolibaba, B. M. Polanuer, I. Kozlov, and D. A. Perumov. 1997. Primary structure and functional activity of the Bacillus subtilis ribC gene. Mol. Biol. (Moscow) 31:820-825. [PubMed] [Google Scholar]
- 15.Higashitsuji, Y., A. Angerer, S. Berghaus, B. Hobl, and M. Mack. 2007. RibR, a possible regulator of the Bacillus subtilis riboflavin biosynthetic operon, in vivo interacts with the 5′-untranslated leader of Rib mRNA. FEMS Microbiol. Lett. 274:48-54. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, K., and W. Stoffel. 1993. TMbase: a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 17.Joseph, P., J. R. Fantino, M. L. Herbaud, and F. Denizot. 2001. Rapid orientated cloning in a shuttle vector allowing modulated gene expression in Bacillus subtilis. FEMS Microbiol. Lett. 205:91-97. [DOI] [PubMed] [Google Scholar]
- 18.Kahn, D. W., and B. M. Anderson. 1986. Characterization of Haemophilus influenzae nucleotide pyrophosphatase. An enzyme of critical importance for growth of the organism. J. Biol. Chem. 261:6016-6025. [PubMed] [Google Scholar]
- 19.Kaltwasser, M., T. Wiegert, and W. Schumann. 2002. Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl. Environ. Microbiol. 68:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kemmer, G., T. J. Reilly, J. Schmidt-Brauns, G. W. Zlotnik, B. A. Green, M. J. Fiske, M. Herbert, A. Kraiss, S. Schlor, A. Smith, and J. Reidl. 2001. NadN and e(P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae. J. Bacteriol. 183:3974-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kil, Y. V., V. N. Mironov, I. Gorishin, R. A. Kreneva, and D. A. Perumov. 1992. Riboflavin operon of Bacillus subtilis: unusual symmetric arrangement of the regulatory region. Mol. Gen. Genet. 233:483-486. [DOI] [PubMed] [Google Scholar]
- 22.Koser, S. A. 1968. Vitamin requirements of bacteria and yeasts. Charles C. Thomas, Springfield, IL.
- 23.Kreneva, R. A., M. S. Gelfand, A. A. Mironov, A. Iomantas, I. Kozlov, A. S. Mironov, and D. A. Perumov. 2000. Study of the phenotypic occurrence of ura gene inactivation in Bacillus subtilis. Genetika 36:1166-1168. [PubMed] [Google Scholar]
- 24.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, J. M., S. Zhang, S. Saha, S. Santa Anna, C. Jiang, and J. Perkins. 2001. RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol. 183:7371-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mack, M., A. P. van Loon, and H. P. Hohmann. 1998. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J. Bacteriol. 180:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 34:61-75. [DOI] [PubMed] [Google Scholar]
- 28.Manoil, C., J. J. Mekalanos, and J. Beckwith. 1990. Alkaline phosphatase fusions: sensors of subcellular location. J. Bacteriol. 172:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansour, N. M., M. Sawhney, D. G. Tamang, C. Vogl, and M. H. Saier, Jr. 2007. The bile/arsenite/riboflavin transporter (BART) superfamily. FEBS J. 274:612-629. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Verstraete, I., M. Debarbouille, A. Klier, and G. Rapoport. 1990. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J. Mol. Biol. 214:657-671. [DOI] [PubMed] [Google Scholar]
- 31.Merchante, R., H. M. Pooley, and D. Karamata. 1995. A periplasm in Bacillus subtilis. J. Bacteriol. 177:6176-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Mironov, A. S., I. Gusarov, R. Rafikov, L. E. Lopez, K. Shatalin, R. A. Kreneva, D. A. Perumov, and E. Nudler. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111:747-756. [DOI] [PubMed] [Google Scholar]
- 34.Otani, S., M. Takatsu, M. Nakano, S. Kasai, and R. Miura. 1974. Roseoflavin, a new antimicrobial pigment from Streptomyces. J. Antibiot. (Tokyo) 27:86-87. [PubMed] [Google Scholar]
- 35.Perkins, J., and J. Pero. 2002. Biosynthesis of riboflavin, biotin, folic acid, and cobalamin, p. 271-276. In A. Sonenshine, J. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 36.Rapp, M., D. Drew, D. O. Daley, J. Nilsson, T. Carvalho, K. Melen, J. W. De Gier, and G. Von Heijne. 2004. Experimentally based topology models for E. coli inner membrane proteins. Protein Sci. 13:937-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reidl, J., S. Schlor, A. Kraiss, J. Schmidt-Brauns, G. Kemmer, and E. Soleva. 2000. NADP and NAD utilization in Haemophilus influenzae. Mol. Microbiol. 35:1573-1581. [DOI] [PubMed] [Google Scholar]
- 38.Reihl, P., and J. Stolz. 2005. The monocarboxylate transporter homolog Mch5p catalyzes riboflavin (vitamin B2) uptake in Saccharomyces cerevisiae. J. Biol. Chem. 280:39809-39817. [DOI] [PubMed] [Google Scholar]
- 39.Sauer, E., M. Merdanovic, A. P. Mortimer, G. Bringmann, and J. Reidl. 2004. PnuC and the utilization of the nicotinamide riboside analog 3-aminopyridine in Haemophilus influenzae. Antimicrob. Agents Chemother. 48:4532-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snell, E. E., and F. M. Strong. 1939. A microbiological assay for riboflavin. Ind. Eng. Chem., Anal. ed. 11:346-350. [Google Scholar]
- 41.Solovieva, I. M., R. A. Kreneva, D. J. Leak, and D. A. Perumov. 1999. The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon. Microbiology 145:67-73. [DOI] [PubMed] [Google Scholar]
- 42.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 43.Stülke, J., I. Martin-Verstraete, V. Charrier, A. Klier, J. Deutscher, and G. Rapoport. 1995. The HPr protein of the phosphotransferase system links induction and catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177:6928-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 45.van Herwaarden, A. E., E. Wagenaar, G. Merino, J. W. Jonker, H. Rosing, J. H. Beijnen, and A. H. Schinkel. 2007. Multidrug transporter ABCG2/breast cancer resistance protein secretes riboflavin (vitamin B2) into milk. Mol. Cell. Biol. 27:1247-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winkler, W. C., S. Cohen-Chalamish, and R. R. Breaker. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 99:15908-15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zirak, P., A. Penzkofer, T. Schiereis, P. Hegemann, A. Jung, and I. Schlichting. 2006. Photodynamics of the small BLUF protein BlrB from Rhodobacter sphaeroides. J. Photochem. Photobiol. B 83:180-194. [DOI] [PubMed] [Google Scholar]