Abstract
Clostridium cellulovorans, an anaerobic bacterium, produces a small nonenzymatic protein called HbpA, which has a surface layer homology domain and a type I cohesin domain similar to those found in the cellulosomal scaffolding protein CbpA. In this study, we demonstrated that HbpA could bind to cell wall fragments from C. cellulovorans and insoluble polysaccharides and form a complex with cellulosomal cellulases endoglucanase B (EngB) and endoglucanase L (EngL). Synergistic degradative action of the cellulosomal cellulase and HbpA complexes was demonstrated on acid-swollen cellulose, Avicel, and corn fiber. We propose that HbpA functions to bind dockerin-containing cellulosomal enzymes to the cell surface and complements the activity of cellulosomes.
Many clostridia produce a cellulolytic complex called the cellulosome (11), which consists basically of a nonenzymatic scaffolding protein and cellulases and hemicellulases (2, 4). Cellulosomes can degrade plant cell materials very efficiently. There has been increasing interest in cellulosomes both for their inherent cellulosic activity and their possible use for converting plant biomass into sugars and biofuels.
In Clostridium cellulovorans, the hbpA gene is a component of a large cellulosomal gene cluster which contains the genes for the scaffolding protein CbpA, for exoglucanase ExgS, for mannanase ManA, a number of endoglucanases, and HbpA (19). HbpA is a small nonenzymatic protein with a molecular weight of 24,940 and contains a surface layer homology (SLH) domain and a cohesin domain that is similar to the cohesins found in the scaffolding protein CbpA (16). However the role of HbpA has remained unclear. A similar protein found in Clostridium cellulolyticum, OrfXp, was found to contain a cohesin and was localized to the membrane of C. cellulolyticum and not detected in the cellulosome (14). OrfXp was able to interact with all the cellulosomal enzymes, although its binding of the enzymes was weaker than the binding of type I cohesins present in CipA. It was proposed that OrfXp played some role either in the assembly of the cellulosome or in the degradation of the substrate.
In order to obtain a better understanding of the role of HbpA, we have analyzed its interaction with two cellulosomal enzymes and its ability to bind to polysaccharides and the cell wall fraction from C. cellulovorans. We have demonstrated that HbpA can bind to cellulosomal cellulases and enhance cellulase activity towards microcrystalline cellulose. We propose that HbpA attached to the cell surface binds cellulosomal enzymes and complements the activity of the cellulosomes.
MATERIALS AND METHODS
Bacterial strains.
C. cellulovorans strain ATCC 35296 was used for the isolation of the genomic DNA. Escherichia coli strain C600 (22) and Bacillus subtilis WB800 (20, 21) were used in this study.
Plasmid construction for HbpA-His, miniCbpA1-His, EngB-His, and EngL-His.
The plasmid vectors used in this study were pWB1000 and pDG148 (1). To generate pWB1000, a fragment which contains an amp gene and an ori replicable in E. coli was amplified by PCR from pBR322 using a primer pair (GAACAATTGCAGGAAAGAACATGTGAGC and TGAGAATTCTTGAAGACGAA). The fragment was digested with MunI and EcoRI (New England Biolabs) and then ligated with pWB980, which was digested with EcoRI. The resulting vector, pWB1000, was designed as a shuttle vector. The plasmids used to produce HbpA-His, miniCbpA1-His, EngB-His, and EngL-His were constructed as follows. DNA fragments encoding each gene were amplified by PCR from the C. cellulovorans genomic DNA with LA Taq DNA polymerase (Takara), with the use of appropriate combinations of primers: hbpA-His6, GTCGTCGACGTCAAACAGTAAAGATTCTAGTAGT and GCAGCATGCTCAGTGGTGGTGGTGGTGGTGCTTTCCATTTGTAGTTTTTACTTC; minicbpA1-His6, GTCGTCGACATGAGGGGGAGCAATTATGCA and GCAGCATGCTCAGTGGTGGTGGTGGTGGTGGATAGTTACTGTTCCT; engB-his6, GTCGTCGACGACAAATAATATATTTTGGAGGA and GCAGCATGCTCAGTGGTGGTGGTGGTGGTGGCTTAAAAGCATTTTTTTAAGAAC; engL-his6, GTCGTCGACGACTTAGAAATATATAAATAGGGGGAT and GCAGCATGCTCAGTGGTGGTGGTGGTGGTGACCAAGAAGTAACTTTTTAAGAAG.
The fragments contained SalI and SphI sites (underlined) for inserting the PCR fragments into pWB1000 (hbpA-his) and pDG148 (minicbpA1-his, engB-his, and engL-his). For cloning these genes, E. coli C600 clones with introduced plasmids were selected on Luria-Bertani agar plates containing 50 μg/ml ampicillin. After cloning, the DNA sequences were checked with sequences in the NCBI database.
Transformation of B. subtilis.
Transformation of B. subtilis WB800 was carried out basically by the method of Spizizen (17). To obtain transformants, 30 μg/ml of kanamycin (for pWB1000) or 20 μg/ml of neomycin (for pDG148) was used in the selection plates.
Purification of HbpA-His, miniCbpA1-His, EngB-His, and EngL-His.
To produce and secrete miniCbpA1-His and cellulosomal endoglucanases, B. subtilis recombinants were grown to early log phase (optical density at 600 nm = 0.2) in 500 ml of Super rich medium at 30°C and then 0.5 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) was added. After an additional incubation of 6 h at 30°C, growth medium supernatants were obtained by centrifugation. In the case of the production and secretion of HbpA-His, B. subtilis harboring pWB-hbpA-his was grown in 500 ml of Super rich medium for 8 h. Then growth medium supernatant was obtained by centrifugation. The supernatants were loaded onto Ni-nitrilotriacetic acid-agarose columns. After elution, the proteins were concentrated by a polyethersulfone centrifugation concentrator (Viva Science, Goettingen, Germany).
Determination of protein concentration.
Protein concentration was measured by using the method of Bradford (3) with a protein assay kit from Bio-Rad, with bovine serum albumin as the standard.
Assembly of HbpA or miniCbpA1 and cellulosomal endoglucanase.
The purified HbpA-His or miniCbpA1-His and the recombinant cellulosomal subunits (equal molar) were mixed in 15 μl of binding buffer (25 mM sodium acetate buffer [pH 6.0], 10 mM CaCl2) and kept for 1 h at 4°C. The assembly of HbpA-His6 and the cellulosomal subunit was confirmed by native polyacrylamide gel electrophoresis (PAGE) as described below.
SDS-PAGE.
Sodium dodecyl sulfate (SDS)-PAGE was performed with 10% polyacrylamide by the method of Laemmli (10). Proteins were visualized by Bio-Safe Coomassie staining (Bio-Rad).
Native PAGE.
Native PAGE was performed with 4 to 15% polyacrylamide (Bio-Rad).
Enzyme assays.
Enzyme activity was assayed in the presence of a 0.5% (wt/vol) concentration of each substrate at 37°C in 50 mM sodium phosphate buffer (pH 7.0). Enzyme concentrations were 10 nM (HbpA-His), 5 nM (EngB-His), and 4.25 nM (EngL-His). To investigate synergistic effects, HbpA-His was added to 0, 5, 10, 20, and 50 nM for EngB-His or 0, 4.25, 8.5, 17, and 42.5 nM for EngL-His. Reaction samples (500 μl) were collected at appropriate times and immediately mixed with 500 μl of chilled 0.38 M sodium carbonate containing 1.8 mM cupric sulfate and 0.2 M glycine. The reducing sugars were determined by reductometry by the Dygert method (5). Incubations with acid-swollen cellulose (ASC), Avicel, and corn fiber were for 6 h, 12 h, and 24 h, respectively. All assays were repeated three times.
Interaction of HbpA-His with cell wall fragments of C. cellulovorans.
Preparation of cell wall fragments of C. cellulovorans followed the method of Kosugi et al. (8). Binding experiments were carried out by incubating and cosedimenting each recombinant protein with cell wall fragments. One microgram of HbpA-His was mixed with 50 μl of cell wall fraction, and reaction volume was increased to 200 μl with 50 mM sodium phosphate buffer (pH 7.0). The reaction mixtures were incubated for 1 h at 37°C with shaking. The bound and free proteins were separated by centrifugation at 16,000 × g for 20 min at room temperature. The supernatant consisted of the soluble fraction. A wash fraction was obtained after washing the cell wall with 50 mM sodium phosphate buffer (pH 7.0). The pellet, consisting of the insoluble cell wall fragments and attached proteins, was washed with the same buffer and then resuspended with 200 μl of the same buffer. Each fraction was analyzed by SDS-PAGE and then Western blotting, as described below.
Affinity of HbpA for insoluble polysaccharides.
One microgram of HbpA-His was incubated for 1 h at 37°C with shaking in 300 μl of 50 mM sodium phosphate buffer (pH 7.0) containing Avicel, chitin (Sigma), and oat-spelled xylan (Sigma) at a concentration of 25 mg/ml as insoluble polysaccharides. Free HbpA-His was separated by centrifugation at 16,000 × g for 5 min at room temperature. The supernatants were labeled as supernatant fractions. The pellets were suspended in 300 μl of 50 mM sodium phosphate buffer (pH 7.0) and labeled as precipitate fractions. Each fraction was analyzed by SDS-PAGE and then Western blotting as described below.
Western blotting.
Western blotting was performed by using anti-His antibody conjugated with horseradish peroxidase (HRP) (Invitrogen). After electrophoresis, the gel was soaked in transfer buffer (25 mM Tris, 192 mM glycine, 10% methanol). Proteins in the gel were transferred onto a polyvinylidene difluoride membrane (Millipore) using the semidry blotting method and then neutralized with phosphate saline buffer (PBS). Skim milk (1%) in PBS was used for blocking at room temperature for 1 h, and then anti-His antibody conjugated with HRP was added (1:5,000), and the mixture was incubated at room temperature for 2 h. The membrane was washed with PBS for 5 min three times. To detect the signal, ECL-plus (Amersham) was used. The signals were exposed onto X-ray film (Fuji film) for appropriate times.
RESULTS AND DISCUSSION
Comparison of domains of HbpA with CbpA and EngE.
Since the HbpA sequence revealed domains that were similar to those of surface layer proteins, a study comparing of the SLH domains in HbpA, CbpA, and EngE and comparing the cohesin domain of HbpA with those of CbpA was done. The SLH domain in HbpA had 16.7% to 19.7% similarity to the SLH domains in CbpA and EngE. SLH domains within CbpA (about 48%) and EngE (from 50 to 70%) have higher similarity to each other than to the SLH domain in HbpA. The SLH domains in CbpA (7) and EngE (18) are able to bind to the cell surface and have been implicated in the binding of the cellulosome to the cell surface.
Since cohesins are involved in binding cellulosomal enzymes through their dockerin domains to form cohesin-dockerin interactions, we compared the cohesin sequence of HbpA to those of the nine cohesins present in CbpA. The cohesin domain in HbpA had various degrees of similarity to the cohesin domains in CbpA, from 26.9% to 32.1%. As with the SLH domain, cohesins in CbpA are very similar (about 80%) except for cohesin 9, but the cohesin of HbpA is very different from the cohesins in CbpA. From these comparisons, the origin of the SLH and cohesin domains of HbpA appears to be different from the origin of those of CbpA and EngE. These results also suggest that the properties of the HbpA domains might be different from those of the CbpA and EngE domains.
Construction of plasmids for expression of C. cellulovorans genes in B. subtilis WB800.
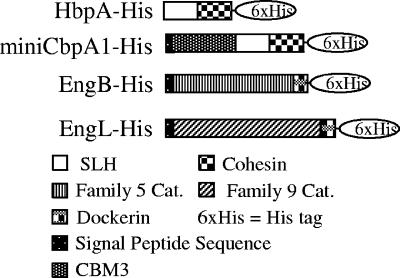
In order to test whether the cohesin domain of HbpA was able to bind cellulosomal enzymes, we constructed expression plasmids for HbpA, miniCbpA1, and the cellulosomal enzymes EngB and EngL (see Materials and Methods) (Fig. 1). The vector pWB1000 is an E. coli-B. subtilis shuttle vector containing a P43 promoter, a sacB sequence encoding a signal peptide, a multiple cloning site, an ampicillin resistance marker from E. coli, and a kanamycin resistance marker from B. subtilis. HbpA-His was expressed with pWB1000. The His tag was added for ease of purification of the expressed protein.
FIG. 1.
Molecular architecture of HbpA, miniCbpA1, EngB, and EngL. SHL, SLH domain; Cat., catalytic domain; CBM, carbohydrate binding module.
The vector pDG148 is also a shuttle vector containing a SalI-SphI cloning site and an IPTG-inducible promoter. minicbpA1, engB, and engL were cloned into pDG148; their products had their original Shine-Dalgarno and signal peptide sequences and His6 tags at their C termini. Expression of the C. cellulovorans genes had no effect on the growth rate of B. subtilis (data not shown). The C. cellulovorans genes were expressed, and their products were secreted successfully by B. subtilis (Fig. 2). These results indicated that the Shine-Dalgarno and signal peptide sequences from C. cellulovorans also worked in B. subtilis and that the secreted proteins were folded into active forms.
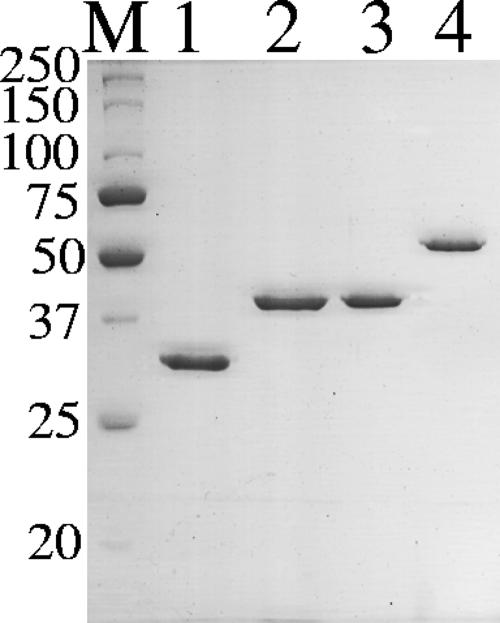
FIG. 2.
Coomassie blue staining of SDS-PAGE gel of purified HbpA-His, miniCbpA1-His, EngB-His, and EngL-His. Each lane contained 1 μg of protein. Lane M, molecular mass markers; lane 1, HbpA-His; lane 2, miniCbpA1-His; lane 3, EngB-His; lane 4, EngL-His in a 10% polyacrylamide gel. The positions of molecular mass markers (in kilodaltons) are indicated on the left.
Interaction between HbpA-His and cellulosomal cellulases and their synergistic effect.
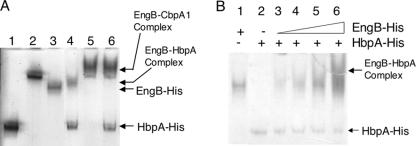
To demonstrate interaction between HbpA-His and EngB-His, we mixed the proteins in binding buffer and then performed native PAGE to look for the formation of an HbpA-EngB complex. miniCbpA1-His was also used as a binding competitor of HbpA-His. In native gel, each protein formed a single band, except that the EngL-His band was very diffuse (data not shown). As shown in Fig. 3A, lane 4, Eng B formed a complex with HbpA in vitro. In Fig. 3A, lane 5, EngB also formed a complex with miniCbpA1 as expected. However, the complex which consisted of HbpA-His and EngB disappeared when miniCbpA-His was added to the reaction (Fig. 3A, lane 6). In Fig. 3B, mixtures of EngB-His and HbpA-His were analyzed for formation of the EngB-His-HbpA-His complex. These results indicate that the cohesin domain of HbpA has the ability to bind to the dockerin domain of EngB but that the binding is weaker than that with the cohesin in miniCbpA1-His. These properties are similar to those of OrfXp (14).
FIG. 3.
Complex formation between HbpA-His and cellulosomal cellulases analyzed by native PAGE. (A) Lane 1, HbpA-His; lane 2, miniCbpA1-His; lane 3, EngB-His; lane 4, EngB-His and HbpA-His; lane 5, miniCbpA1-His and EngB-His; lane 6, miniCbpA1-His, HbpA-His, and EngB-His. Each protein was present at 25 pmol. (B) Lane 1, EngB-His (25 pmol); lane 2, HbpA-His (25 pmol); lane 3, 1:1 EngB-His-HbpA-His (25 pmol each); lane 4, 2:1 EngB-His-HbpA-His (50 pmol of EngB-His-25 pmol HbpA-His); lane 5, 4:1 EngB-His-HbpA-His (100 pmol EngB-His-25 pmol HbpA-His); lane 6, 10:1 EngB-His-HbpA-His (250 pmol EngB-His-25 pmol HbpA-His).
Synergistic effects between CbpA and cellulosomal cellulases and hemicellulases were observed (9, 12, 13). For the purpose of determining enzymatic activity and any synergism between HbpA and the cellulosomal enzymes, we measured the glucanase activity of the complexes against ASC, Avicel, and corn fiber. There was no apparent synergistic effect when endoglucanases and HbpA-His were present at a 1:1 molar ratio, but, at a higher ratio of HbpA-His to enzymes, the glucanase activity increased (Table 1). The synergistic activities of HbpA and EngB or EngL reached 1.58- and 1.25-fold for ASC, 2.19- and 1.74-fold for Avicel, and 1.83- and 2.40-fold for corn fiber when 10× HbpA-His was added in the reaction solution. HbpA and EngL had higher activity towards cellulose and corn fiber than HbpA and EngB. These results indicated not only that HbpA was capable of binding cellulosomal cellulases but also that formation of the complex resulted in higher activity of the bound enzymes. No synergy was observed when carboxymethyl cellulose was used as the substrate (data not shown).
TABLE 1.
Activity of cellulases as a function of HbpA concentrationa
| Substrate | Enzyme | Free enzyme concn (μM) | 1× HbpA-His
|
2× HbpA-His
|
4× HbpA-His
|
10× HbpA-His
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Amtb (μM) | SFc | Amt (μM) | SF | Amt (μM) | SF | Amt (μM) | SF | |||
| ASC | HbpA-His | NDd | ||||||||
| EngB-His | 25.7 ± 2.8 | 26.1 ± 5.7 | 1.01 | 28.8 ± 2.2 | 1.12 | 31.4 ± 1.5 | 1.22 | 40.7 ± 2.5 | 1.58 | |
| EngL-His | 56.4 ± 4.5 | 57.3 ± 4.7 | 1.02 | 59.1 ± 3.4 | 1.05 | 61.9 ± 3.1 | 1.10 | 70.3 ± 2.6 | 1.25 | |
| Avicel | HbpA-His | ND | ||||||||
| EngB-His | 5.4 ± 1.1 | 5.1 ± 0.5 | 0.95 | 6.2 ± 0.5 | 1.15 | 8.2 ± 1.6 | 1.53 | 11.8 ± 0.5 | 2.19 | |
| EngL-His | 7.9 ± 1.7 | 8.4 ± 1.4 | 1.06 | 8.7 ± 0.7 | 1.10 | 9.9 ± 0.9 | 1.25 | 13.7 ± 1.3 | 1.74 | |
| Corn fiber | HbpA-His | ND | ||||||||
| EngB-His | 20.1 ± 4.1 | 20.3 ± 1.1 | 1.01 | 19.0 ± 4.7 | 0.95 | 28.8 ± 3.7 | 1.43 | 36.9 ± 7.4 | 1.83 | |
| EngL-His | 16.7 ± 7.3 | 24.4 ± 7.8 | 1.46 | 23.7 ± 6.5 | 1.42 | 34.4 ± 1.7 | 2.05 | 40.2 ± 0.2 | 2.40 | |
See Materials and Methods for assay conditions.
Amount of reducing sugar produced from ASC in 6 h, from Avicel in 12 h, and from corn fiber in 24 h.
SF, synergy factor.
ND, not detected.
Analysis of binding ability of HbpA-His for C. cellulovorans cell wall fragments.
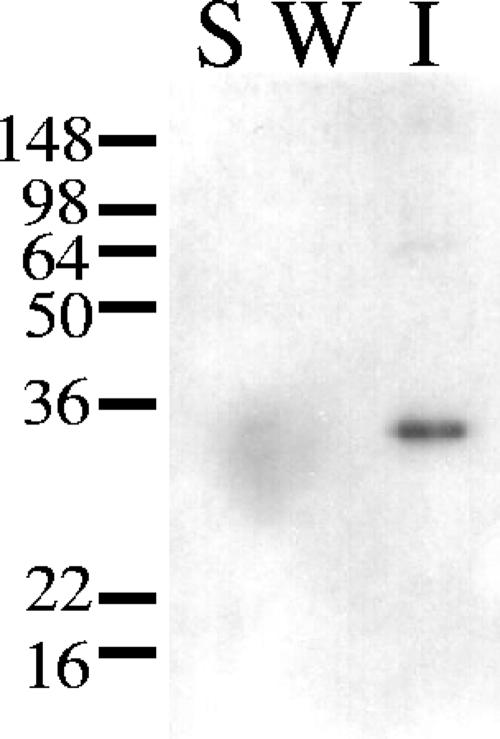
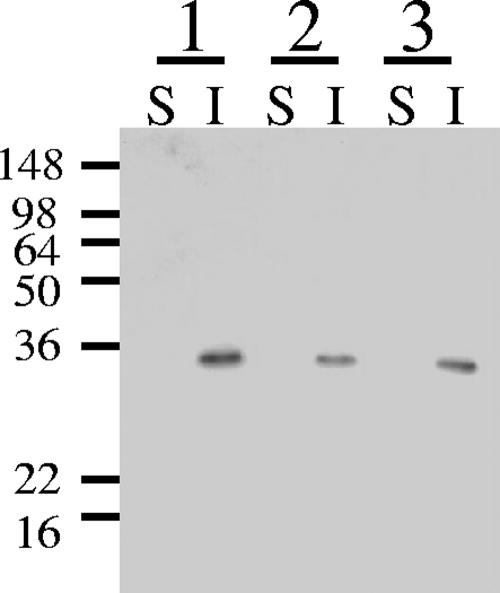
Kosugi et al. showed that SLH domains of CbpA and EngE had the ability to attach these proteins to the cell wall of C. cellulovorans (7, 8). HbpA has also an SLH domain, but its amino acid sequence has low similarity to those of EngE. Therefore we investigated whether HbpA has the ability to bind to the cell wall of C. cellulovorans. As shown in Fig. 4, HbpA did bind to the cell wall fragments of C. cellulovorans. In spite of the low similarity of the amino acid sequences of the SLH domains of HbpA and EngE, the results indicate that the SLH domain of HbpA plays a role similar to that of the SLH domains of EngE, i.e., to bind the protein to C. cellulovorans cell walls.
FIG. 4.
Interaction between HbpA-His and the C. cellulovorans cell wall fraction. The materials were loaded on 10% polyacrylamide-SDS gels and blotted onto a membrane. Shown is the binding of HbpA-His to the cell wall fraction as determined by immunoblotting with anti-His antibody. Lane S, soluble fraction; lane W, wash fraction; lane I, insoluble fraction. The positions of molecular mass markers (in kilodaltons) are indicated on the left.
Binding of HbpA-His to several polysaccharides.
Table 1 showed that HbpA might have the ability to bind polysaccharides. To prove that hypothesis, we examined whether HbpA-His could bind to cellulose, chitin, and xylan. As shown in Fig. 5, HbpA-His did bind to those polysaccharides. From these results, HbpA could bind not only to the cell wall of C. cellulovorans but also to polysaccharides such as cellulose, chitin, and xylan. Thus, HbpA can bind not only to cellulosomal enzymes but also to the cell wall and to the substrate. These properties are very similar to those attributed to CbpA (7).
FIG. 5.
Binding of HbpA-His to cellulose, chitin, and xylan. Purified HbpA-His was incubated with cellulose, chitin, and xylan. After centrifugation, proteins in supernatant (lane S) and in the precipitate (lane I) were loaded on 10% polyacrylamide-SDS gels and blotted onto a membrane. The binding of HbpA-His to the polysaccharides was determined by immunoblotting with anti-His antibody. Lanes 1, binding to cellulose; lanes 2, binding to chitin; lanes 3, binding to xylan. The positions of molecular mass markers (in kilodaltons) are indicated on the left.
In C. cellulolyticum, the production of ORFXp is induced by the presence of cellulose (14). The authors suggested that ORFXp had an important role in cellulose degradation or in facilitating cellulosome assembly in the vicinity of the membrane. In C. cellulovorans, hbpA is located in a large cellulosomal gene cluster and seems to be in an operon with engL (19). Han et al. showed that many cellulases were induced by cellulose (6). As in C. cellulolyticum, HbpA might be induced by the presence of cellulose. A paralogue also exists in a large gene cluster in Clostridium acetobutylicum (15). C. acetobutylicum cannot grow in a medium which contains cellulose as a sole carbon source. However, by analogy to C. cellulovorans and C. cellulolyticum, the OrfXp of C. acetobutylicum might work cooperatively with other cellulosomal subunits.
Here we demonstrated that HbpA bound to the cell wall of C. cellulovorans and to polysaccharides such as Avicel, chitin, and xylan. HbpA also formed a complex with EngB and EngL via a cohesin-dockerin interaction, and the HbpA-enzyme complex had higher activity. This synergistic activity is particularly interesting, since it implies that either the formation of the complex results in a conformational change of the enzyme resulting in higher activity or the HbpA-bound enzyme complex may be bound more efficiently to the substrate, resulting in higher activity.
In any case, we suggest that the role of HbpA is to anchor cellulosomal cellulases to the cell surface and to bind to polysaccharides to facilitate cellulosomal enzyme activity. Furthermore the HbpA-enzyme complexes may complement the activity of the cellulosome.
Acknowledgments
We thank Kei Asai at Saitama University (Japan) for kindly providing plasmid pDG148 and Helen Chan for skillful technical assistance.
The research was supported in part by U.S. Department of Energy grant DE-FG02-04ER15553 and by the Research Institute of Innovative Technology for the Earth.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Asai, K., H. Yamaguchi, C. M. Kang, K. Yoshida, Y. Fujita, and Y. Sadaie. 2003. DNA microarray analysis of Bacillus subtilis sigma factors of extracytoplasmic function family. FEMS Microbiol. Lett. 220:155-160. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., J. P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Doi, R. H., and A. Kosugi. 2004. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2:541-551. [DOI] [PubMed] [Google Scholar]
- 5.Dygert, S., L. H. Li, D. Florida, and J. A. Thoma. 1965. Determination of reducing sugar with improved precision. Anal. Biochem. 13:367-374. [DOI] [PubMed] [Google Scholar]
- 6.Han, S.-O., H.-Y. Cho, H. Yukawa, M. Inui, and R. H. Doi. 2004. Regulation of expression of cellulosomes and noncellulosomal (hemi)cellulolytic enzymes in Clostridium cellulovorans during growth on different carbon sources. J. Bacteriol. 186:4218-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosugi, A., Y. Amano, K. Murashima, and R. H. Doi. 2004. Hydrophilic domains of scaffolding protein CbpA promote glycosyl hydrolase activity and localization of cellulosomes to the cell surface of Clostridium cellulovorans. J. Bacteriol. 186:6351-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosugi, A., K. Murashima, Tamaru, Y., and R. H. Doi. 2002. Cell-surface-anchoring role of N-terminal surface layer homology domains of Clostridium cellulovorans EngE. J. Bacteriol. 184:884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koukiekolo, R., H.-Y. Cho, A. Kosugi, M. Inui, H. Yukawa, and R. H. Doi. 2005. Degradation of corn fiber by Clostridium cellulovorans cellulases and hemicellulases and contribution of scaffolding protein CbpA. Appl. Environ. Microbiol. 71:3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Synergistic effects on crystalline cellulose degradation between cellulosomal cellulases from Clostridium cellulovorans. J. Bacteriol. 184:5088-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murashima, K., A. Kosugi, and R. H. Doi. 2003. Synergistic effects of cellulosomal xylanase and cellulases from Clostridium cellulovorans on plant cell wall degradation. J. Bacteriol. 185:1518-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pages, S., A. Belaich, H.-P. Fierobe, C. Tardif, C. Gaudin, and J.-P. Belaich. 1999. Sequence analysis of ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabathe, F., A. Belaich, and P. Soucaille. 2002. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 217:15-22. [DOI] [PubMed] [Google Scholar]
- 16.Shoseyov, O., M. Takagi, M. A. Goldstein, and R. H. Doi. 1992. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A (CbpA). Proc. Natl. Acad. Sci. USA 89:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamaru, Y., and R. H. Doi. 1999. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit, EngE. J. Bacteriol. 181:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu, S.-C., J. C. Yeung, Y. Duan, R. Ye, S. J. Szarka, H. R. Habibi, and S.-L. Wong. 2002. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl. Environ. Microbiol. 68:3261-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, X.-C., W. Lee, L. Tran, and S.-L. Wong. 1991. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J. Bacteriol. 173:4952-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young, R. A., and R. W. Davis. 1983. Efficient isolation of genes by using antibody probes. Proc. Natl. Acad. Sci. USA 80:1194-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]