Abstract
Integrative and conjugative elements (ICEs), also known as conjugative transposons, are mobile genetic elements that can transfer from one bacterial cell to another by conjugation. ICEBs1 is integrated into the trnS-leu2 gene of Bacillus subtilis and is regulated by the SOS response and the RapI-PhrI cell-cell peptide signaling system. When B. subtilis senses DNA damage or high concentrations of potential mating partners that lack the element, ICEBs1 excises from the chromosome and can transfer to recipients. Bacterial conjugation usually requires a DNA relaxase that nicks an origin of transfer (oriT) on the conjugative element and initiates the 5′-to-3′ transfer of one strand of the element into recipient cells. The ICEBs1 ydcR (nicK) gene product is homologous to the pT181 family of plasmid DNA relaxases. We found that transfer of ICEBs1 requires nicK and identified a cis-acting oriT that is also required for transfer. Expression of nicK leads to nicking of ICEBs1 between a GC-rich inverted repeat in oriT, and NicK was the only ICEBs1 gene product needed for nicking. NicK likely mediates conjugation of ICEBs1 by nicking at oriT and facilitating the translocation of a single strand of ICEBs1 DNA through a transmembrane conjugation pore.
Mobile genetic elements are ubiquitous in bacteria and can contain genes for antibiotic resistance, symbiosis, and virulence; their dissemination contributes to bacterial evolution by conferring new genes and phenotypes to their recipients (reviewed in references 8 and 21). The most common mobile genetic elements are phages, plasmids, and integrative and conjugative elements (ICEs), also known as conjugative transposons. Conjugative plasmids and ICEs are transferred directly from cell to cell and generally encode their own conjugation systems (6, 25).
ICEBs1 is an ICE that is found integrated into the trnS-leu2 genes of some Bacillus subtilis strains (Fig. 1A) (3, 7). Detailed analyses of ICEBs1 have been aided by its efficient transfer, its site-specific integration, and the ease of genetic manipulations in B. subtilis (2, 3; C. A. Lee, J. M. Auchtung, R. E. Monson, and A. D. Grossman, submitted for publication). When induced, ICEBs1 excises from the chromosome and can transfer to recipient cells. ICEBs1 gene expression and excision are induced by the SOS response or when cells are at high density surrounded by neighbors that do not contain a copy of ICEBs1 (3). Regulation by population density and recognition of self are mediated by the regulator RapI and the pentapeptide PhrI (3).
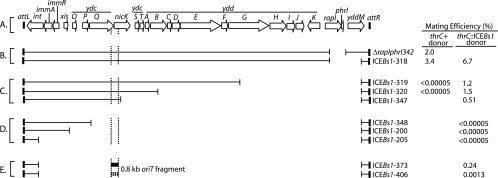
FIG. 1.
Effect of deletions in ICEBs1 on transfer and mobilization. (A) The genetic map of ICEBs1, indicating genes as open arrows and the flanking 60-bp repeats at attL and attR as thin black rectangles. The vertical dotted lines indicate the region of ICEBs1 oriT. (B to E) Thin lines below the map of ICEBs1 indicate the regions of ICEBs1 between attL and attR that are present in the various ΔICEBs1 mutations. Open spaces represent regions that are missing. Mating efficiencies are indicated to the right. Donor cells were induced with MMC and mixed with recipient strain CAL264, an ICEBs10 recipient strain that expresses int from the Pspank promoter. Donor strains either contained the indicated ΔICEBs1 allele alone (thrC+) or also carried an immobilized ICEBs1 at thrC {thrC325::[ICEBs1(ΔattR::tet)]} that supplied all of the ICEBs1 excision and conjugation functions in trans but is unable to excise due to the deletion of attR. Mating efficiency was calculated as the percentage of transconjugant CFU per donor cell. The mean from at least two independent assays is reported. Mating efficiencies for the Δ(rapI-phrI)342::kan donor strain ranged from 0.81% to 3.7% in six independent assays and gave a mean of 2.0% with a standard deviation of 1.2%. Except for donor strains that gave no detectable transconjugants, mating efficiencies for other donor strains had similar amounts of variability. (E) Thick lines indicate that two derivatives of ΔICEBs1-205::kan carry an ∼0.8-kb oriT fragment from the ydcQ-nicK region (wild-type fragment, solid; mutant fragment, dashed).
Both DNA damage and RapI-PhrI regulation affect the activity of the ICEBs1 immunity repressor ImmR (2), and inactivation of ImmR causes increased ICEBs1 gene expression, production of the excisionase Xis, and excision of ICEBs1 (2, 3; Lee et al., submitted). Integration into and excision from the chromosome by site-specific recombination is mediated by a lambda-like integrase, Int, encoded in ICEBs1 (Lee et al., submitted). Excision requires both Xis and Int, whereas integration requires only Int (Lee et al., submitted).
Once excised from the chromosome, some ICEs transfer to other cells by using mechanisms similar to those of conjugative plasmids (reviewed in references 6, 11, 20, 25, 34, 36, and 50). Transfer of conjugative plasmids typically initiates from a specific site in the plasmid, the origin of transfer, oriT. oriT functions in cis and is required for efficient transfer. A relaxase, usually encoded by the plasmid, recognizes oriT, makes a single-strand DNA break (a nick) in oriT, and covalently attaches to the 5′ end of the nicked DNA strand via a phosphotyrosyl linkage (9, 34). Some conjugal relaxases have a helicase domain, which unwinds the single strand of DNA for transfer from the donor into the recipient (39, 45). In the absence of a cognate helicase activity, conjugative plasmids can use leading-strand DNA synthesis (rolling-circle replication) from the nicked 3′ end to promote strand displacement and single-strand DNA transfer (9, 34). In either case, the covalently attached relaxase interacts with a coupling protein in the bacterial membrane that targets the single strand of plasmid DNA to a transmembrane conjugation pore (26, 37, 57, 58). The attached relaxase may transfer into the recipient cell, while another relaxase monomer may remain bound to the plasmid DNA in the donor cell (17, 22, 37). The DNA relaxase terminates transfer by precisely rejoining the ends of the plasmid and releasing a single-stranded DNA circle into the recipient (37, 48). Synthesis of the complementary strand of the transferred circle initiates primarily at an origin of plasmid replication (53).
In contrast to those of conjugative plasmids, origins of transfer and the cognate relaxases from only a few ICEs have been identified and characterized (1, 12, 56, 62). Where characterized, oriT on an ICE is required in cis for transfer but usually not for excision, although there are possible exceptions (60). Like plasmids, ICEs typically encode a relaxase that binds to the cognate oriT, nicks the DNA, and becomes covalently attached. In some cases, there appears to be an additional protein providing specificity to the relaxase. For example, Tn916, an ICE from Enterococcus faecalis, contains a cis-acting origin of transfer, oriT (31), and encodes a DNA nuclease, the orf20 gene product (56). In vitro, Orf20 protein from Tn916 requires the transposon integrase for strand and site specificity. In the absence of the integrase, the Orf20 protein functions as an endonuclease cleaving both strands of Tn916 oriT DNA “at several distinct sites favoring GT dinucleotides” (56).
We have identified and characterized the origin of transfer, oriT, of ICEBs1. We found that induction of ICEBs1 gene expression leads to nicking in a GC-rich inverted repeat in oriT. We also found that ydcR (renamed nicK) is required for nicking and transfer of ICEBs1 and that NicK is the only ICEBs1 gene product needed for specific nicking at oriT. The oriT nicking site is actually located within the nicK open reading frame (ORF). Nicking of oriT by NicK likely facilitates the transfer of one strand of ICEBs1 into recipient cells.
MATERIALS AND METHODS
Media and growth conditions.
B. subtilis was grown in LB or defined minimal glucose medium at 37°C (27). The following antibiotics and other chemicals were used: isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM), mitomycin C (MMC) (1 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) (80 μg/ml), chloramphenicol (5 μg/ml), kanamycin (5 μg/ml), spectinomycin (100 μg/ml), streptomycin (100 μg/ml), and erythromycin (0.5 μg/ml) and lincomycin (12.5 μg/ml) together to select for macrolide-lincosamide-streptogramin B resistance (mls or erm).
B. subtilis strains and alleles.
B. subtilis strains are listed in Table 1. B. subtilis strains were constructed by natural transformation or conjugation (2, 3, 27; Lee et al., submitted). comK::cat is an insertion of mini-Tn10 (cat) and prevents competence development (41). The spontaneous streptomycin resistance allele (str-84, most likely in rpsL) was from strain CAL84 and is often used as a counterselective marker in mating experiments (2, 3). ICEBs10 indicates that the strain is cured of ICEBs1. rapI was overexpressed from Pspank(hy)-rapI integrated into amyE, amyE::{[Pspank(hy)-rapI] spc}, to induce ICEBs1 gene expression and excision. int was expressed from amyE::[(Pspank-int) spc] to provide integrase when needed (2; Lee et al., submitted). Δ(rapI-phrI)342::kan is a deletion-insertion (3).
TABLE 1.
Bacillus subtilis strains used
| Strain | Genotypea |
|---|---|
| CAL223 | ΔICEBs1-200::kan |
| CAL224 | ΔICEBs1-205::kan |
| CAL264 | ICEBs10str-84 amyE::[(Pspank-int) spc] comK::cat |
| CAL306 | ΔnicK306 Δ(rapI-phrI)342::kan amyE::{[Pspank(hy)-rapI] spc} |
| CAL321 | ΔICEBs1-318::kan |
| CAL322 | ΔICEBs1-319::kan |
| CAL323 | ΔICEBs1-320::kan |
| CAL326 | ΔICEBs1-318::kan thrC325::[(ICEBs1-311 ΔattR::tet) mls] |
| CAL327 | ΔICEBs1-319::kan thrC325::[(ICEBs1-311 ΔattR::tet) mls] |
| CAL328 | ΔICEBs1-320::kan thrC325::[(ICEBs1-311 ΔattR::tet) mls] |
| CAL332 | ΔICEBs1-200::kan thrC325::[(ICEBs1-311 ΔattR::tet) mls] |
| CAL346 | ΔnicK306 Δ(rapI-phrI)342::kan amyE::{[Pspank(hy)-rapI] spc} thrC329::[(Pxis-nicK-lacZ) mls] |
| CAL347 | ΔICEBs1-347::kan |
| CAL348 | ΔICEBs1-348::kan |
| CAL349 | ΔICEBs1-205::kan thrC325::[(ICEBs1-311 ΔattR::tet) mls] |
| CAL350 | ΔICEBs1-347::kan thrC325::[(ICEBs1-311 ΔattR::tet) mls] |
| CAL351 | ΔICEBs1-348::kan thrC325::[(ICEBs1-311 ΔattR::tet) mls] |
| CAL381 | ΔICEBs1-373::kan |
| CAL386 | ΔICEBs1-373::kan thrC325::[(ICEBs1-311 ΔattR::tet) mls] |
| CAL413 | ΔICEBs1-406::kan |
| CAL417 | ΔICEBs1-406::kan thrC325::[(ICEBs1-311 ΔattR::tet) mls] |
| CAL419 | ICEBs10str-84 comK::cat |
| CAL501 | ICEBs10amyE501::{[Pspank(hy)-nicK477] spc} |
| CAL502 | ICEBs10amyE502::{[Pspank(hy)-nicK488] spc} |
| IRN342 | Δ(rapI-phrI)342::kan |
| JMA168 | Δ(rapI-phrI)342::kan amyE::{[Pspank(hy)-rapI] spc} |
All strains are derived from JH642 (55) and contain the pheA1 and trpC2 mutations.
Construction of two large deletions at the endogenous ICEBs1 locus, ΔICEBs1-205::kan and ΔICEBs1-200::kan (Fig. 1D), was described previously (Lee et al., submitted). The deletion in ΔICEBs1-205::kan disrupts every ORF in ICEBs1, replacing all but 651 bp of int and 157 bp of yddM with the kanamycin resistance gene from pGK67 (35). The deletion in ΔICEBs1-200::kan starts 86 bp downstream of the xis ORF. Five additional deletions in ICEBs1 (Fig. 1B to D), each extending to the same 3′ endpoint in the yddM ORF, were constructed as described previously (Lee et al., submitted). The deletion in ΔICEBs1-348::kan starts in ydcQ, leaving 222 bp of the 1,440-bp ydcQ ORF. ΔICEBs1-347::kan starts in nicK, leaving 367 bp of the 1,050-bp nicK ORF. ΔICEBs1-320::kan starts in yddB, leaving 491 bp of the 1,062-bp yddB ORF. ΔICEBs1-319::kan starts in yddG, leaving 1,784 bp of the 2,445-bp yddG ORF. ΔICEBs1-318::kan starts in rapI, leaving 586 bp of the 1,173-bp rapI ORF. The deletion in ΔICEBs1-318::kan starts at almost the same position as that in the Δ(rapI-phrI)342::kan allele, which leaves 587 bp of rapI (3).
Hybrid derivatives of the ΔICEBs1-205::kan element that contain a 802-bp oriT fragment at the PstI site between the int and kan gene sequences were constructed. The oriT fragment is in its native orientation, relative to attL and attR. The 802-bp fragment includes 378 bp upstream and 400 bp downstream of the 24-bp sequence ACCCCCCCACGCTAACAGGGGGGT, which is located 17 bp downstream of the start of the nicK ORF. ΔICEBs1-373::kan contains the wild-type fragment, while ΔICEBs1-406::kan contains a mutant fragment, which was generated by the splice-overlap-extension PCR method (29).
An ICEBs1 element was immobilized at thrC, which allowed us to stably express all of the ICEBs1 gene products in trans to ICEBs1 derivatives located at the endogenous chromosomal locus. thrC325::[(ICEBs1-311 ΔattR100::tet) mls] contains the entire ICEBs1 element except for 161 bp at the right-hand end, which were removed by the ΔattR100::tet mutation. This attR mutation prevents excision (Lee et al., submitted). The thrC325::ICEBs1-311 allele also includes sequences that usually flank the 60-bp direct repeats that mark the left and right ends of ICEBs1 in its normal attachment site in the chromosome. Thus, 206 bp of chromosomal DNA upstream of the left direct repeat and 768 bp of chromosomal DNA downstream of the ΔattR100::tet mutation are included.
Construction of thrC325::[(ICEBs1-311 ΔattR100::tet) mls] involved many steps. First, we inserted the ICEBs1 attB site at thrC. This was accomplished by replacing all of the ICEBs1 genes, except for immR, immA, and int, with the cat gene. The ΔICEBs1-117::cat element, including 206 bp upstream and 823 bp downstream of the flanking 60-bp direct repeats, was cloned and inserted into the thrC locus (pDG795 vector, a gift of P. Stragier). Excision of the ΔICEBs1-117::cat element at thrC was induced by expressing xis from amyE168::[(Pspank-xis) spc] (Lee et al., submitted). By screening for those cells that had lost chloramphenicol resistance, we obtained thrC213::(attB-117 mls), in which the ICEBs1 attB region is inserted at thrC. A functional kanamycin-resistant ICEBs1 Δ(rapI-phrI)342::kan was integrated into attB at thrC by mating JMA168 donors with thrC213::(attB-117 mls) recipients that lacked the native attB region (ΔattB::cat) (Lee et al., submitted). Finally, the conjugation-proficient ICEBs1 in thrC229::{[ICEBs1 Δ(rapI-phrI)342::kan] mls} was converted to an excision-defective rapI-phrI+ derivative by recombination with a DNA fragment containing rapI-phrI+ and the ΔattR100::tet allele (Lee et al., submitted), yielding the desired tetracycline-resistant, kanamycin-sensitive thrC325::[(ICEBs1-311 ΔattR100::tet) mls] allele.
ΔnicK306 is an unmarked, in-frame 519-bp deletion, which fuses the first 125 codons of nicK to the last 54 codons. ΔnicK306 deletes most of the NicK-coding region that corresponds to the conserved pfam02486 Rep_trans domain, but it appears to leave the cis-acting oriT region of ICEBs1 intact. A 2.2-kb DNA fragment containing the ΔnicK306 allele was obtained by the splice-overlap-extension PCR method (29) and cloned into the EcoRI and BamHI sites of the chloramphenicol-resistant vector pEX44 (a gift from E. Küster-Schöck) (15) with the promoterless spoVG-lacZ ORF in pEX44 placed downstream of the ICEBs1 ORFs on the 2.2-kb insert. The resulting plasmid, pCAL285, was used to replace the nicK gene with the ΔnicK306 allele in the chromosome of JMA168, as described previously (Lee et al., submitted).
The amyE501::{[Pspank(hy)-nicK477] spc} and amyE502::{[Pspank(hy)-nicK488] spc} alleles were designed to express nicK from the IPTG-inducible Pspank(hy) promoter (pDR111 vector, a gift of D. Rudner) (5). The amyE501::{[Pspank(hy)-nicK477] spc} and amyE502::{[Pspank(hy)-nicK488] spc} alleles contain 87 bp and 393 bp from the region upstream of the nicK ORF and include 92 bp and 398 bp of the 1,440-bp ydcQ ORF, respectively.
The thrC329::[Pxis-(nicK-lacZ) mls] allele was designed to express nicK from the xis promoter (Pxis) of ICEBs1. nicK was cloned into the BamHI site between Pxis and lacZ in pKG1, which had previously been used to construct thrC::[(Pxis-lacZΩ343) mls] (2). The resultant plasmid, pCAL178, was linearized and used to introduce thrC329::[Pxis-(nicK-lacZ) mls] into the B. subtilis chromosome.
ICEBs1 excision assays.
Excision of ICEBs1 in RapI-induced or MMC-induced cells was assayed by detecting the excised circular intermediate and repaired chromosomal junctions, as described previously (3; Lee et al., submitted).
ICEBs1 mating assays.
Matings were done essentially as described previously (3; Lee et al., submitted). Equal numbers of donor (Kanr) and recipient (Strr) cells were mixed and filtered onto cellulose nitrate filters. The filters were placed on plates comprised of Spizizen's minimal salts (27) and 1.5% agar for 3 h at 37°C. The mean of mating efficiencies from at least two independent experiments is reported. Mating efficiencies for RapI-induced donors were calculated as the percentage of Kanr Strr transconjugant CFU per Kanr donor CFU recovered postmating. Since MMC treatment reduced the recovery of donor cells postmating, mating efficiencies for MMC-induced donors were calculated as the percentage of Kanr Strr transconjugant CFU recovered postmating per donor cell present in the initial mating mixture. In this case, the number of donor cells was determined using a value of 1.65 × 108 cells per ml for cultures grown to an optical density at 600 nm of 1.
Identification of the site of nicking within ICEBs1.
B. subtilis genomic DNA was purified on QIAGEN DNeasy minicolumns from cell lysates treated with RNase A and proteinase K in the optional lysis buffer for gram-positive bacteria (QIAGEN). The DNA was digested with HindIII, bound to QIAGEN PCR purification minicolumns, and washed three times with PB buffer and once with PE buffer, before elution with EB buffer (all buffers from QIAGEN). Five hundred nanograms of digested DNA was used as a template with Taq polymerase (Roche) and 2 pmol 32P-labeled primer in 50-μl primer extension reaction mixtures incubated for 20 cycles of 94°C for 30 s, 54°C for 2 min, and 72°C for 3 min. Primers (50 pmol) were end labeled with T4 polynucleotide kinase (New England Biolabs), as per the manufacturer's instructions, with 150 μCi [γ-32P]ATP (6,000 Ci/mmol; Perkin-Elmer) and then purified on QIAGEN nucleotide removal columns. 32P-labeled primers were also used in dideoxy-DNA sequencing reactions (Promega fmol sequencing system), which were run with primer extension products on 8% polyacrylamide-Tris-borate-EDTA-urea gels. Primers CLO75 and CLO76 were designed to detect breaks in the oriT region by hybridizing on opposite strands in the ydcQ-nicK region, 61 bp upstream and 72 bp downstream of the 24-bp GC-rich inverted repeat sequence, respectively. Controls showed that each primer could detect cleaved templates generated by restriction enzyme digestion.
RESULTS
Rationale and experimental design.
We set out to identify the origin of transfer, oriT, of ICEBs1 and the gene encoding the ICEBs1 relaxase. Our expectation was that ICEBs1 contains a single oriT that is required, in cis, for transfer and is nicked in induced donor cells. We started by making a series of deletions of ICEBs1 starting from near the right end and extending to different left endpoints (Fig. 1B to D). We had previously shown that the only ICEBs1 genes needed for excision were int and xis. int and xis are necessary and sufficient for excision, and int is necessary for integration (Lee et al., submitted). Also, we found that DNA near the ends of the integrated ICEBs1 was sufficient for excision (Lee et al., submitted). Since int is at the far left end and xis is the fourth gene from the left, we expected that nested deletions starting from the right end (leaving attR intact) might affect conjugation but not excision. We tested these nested deletions for mating and the ability to be mobilized by complementation in trans. Since oriT should be needed in cis, inactivation of oriT should render ICEBs1 unable to transfer to recipients even though the element excises and all other ICEBs1 functions are provided in trans by complementation.
In complementation experiments, trans-acting functions of ICEBs1 were provided by ICEBs1 located at thrC (see Materials and Methods). The element at thrC was unable to excise (was “locked in”) due to loss of the right end of ICEBs1 (ΔattR), but it was able to mobilize an otherwise defective ICEBs1 located at the normal attachment site in the chromosome. We used recipients that expressed int, encoding integrase, because some of the donor ICEBs1 mutants did not contain their own int. Expression of int in the recipient is sufficient to complement loss of int on the donor element (2; Lee et al., submitted).
ICEBs1 gene expression and excision were induced by adding MMC to induce the SOS response. Induced ICEBs1-containing strains were mixed with recipients cured of ICEBs1 (ICEBs10) but that expressed ICEBs1 int from a heterologous promoter. After mixing potential donors and recipients, cells were filtered, incubated for 3 h to allow mating, and then plated selectively to detect transconjugants.
Induction of ICEBs1 with MMC is less efficient and a bit more variable than induction by overproduction of RapI (3; Lee et al., submitted). However, we used MMC and not overproduction of RapI because strains containing ICEBs1 at the normal attachment site, the “locked-in” ICEBs1 at thrC, and the Pspank(hy)-rapI construct were unstable, even without IPTG, likely because low-level expression of genes from the “locked-in” ICEBs1 and nicking of oriT at thrC cause defects in cell viability and ICEBs1 maintenance at the normal attachment site (2; Lee et al., submitted).
Genes at the right end of ICEBs1 that are not required for mating.
Previously, we found that rapI and phrI are not needed for mating (3). The ICEBs1 deletion ΔICEBs1-318 removes yddM in addition to rapI and phrI (Fig. 1B). The mating frequency of ΔICEBs1-318 was normal (Fig. 1B, thrC+ donor), indicating that yddM is not required for mating. The function of yddM is unknown, and YddM does not yet appear to be homologous to any other protein.
Deletions from the right end of ICEBs1 that are defective in mating.
ICEBs1 deletions ΔICEBs1-319 and ΔICEBs1-320, which remove additional ydd genes (Fig. 1C), were defective for mating (Fig. 1C, thrC+ donors). These two ICEBs1 deletion mutants and even larger deletions are capable of normal excision (data not shown) (Lee et al., submitted). Despite this, we were unable to detect any transconjugants when these mutants were used as donors without complementation.
The results with ΔICEBs1-319 and ΔICEBs1-320 indicate that at least one gene in the yddGHIJK region is required for transfer of ICEBs1 and may encode a component of the conjugation apparatus. YddG and YddH are similar to proteins encoded by other ICEs and are predicted to be membrane proteins with eight transmembrane spanning domains and one transmembrane spanning domain, respectively (3, 7) (TopPred http://bioweb.pasteur.fr/seqanal/interfaces/toppr.html [14, 63]). YddH contains a domain (cd00254 LT_GEWL [42]) that is found in murein hydrolases (61) and may facilitate ICEBs1 transfer by degrading the peptidoglycan barrier.
When ICEBs1 functions were provided in trans, the defects in mating of ΔICEBs1-319 and ΔICEBs1-320 were largely complemented (Fig. 1C, thrC::ICEBs1 donor), indicating that the ICEBs1 at thrC, although not capable of excising due to the loss of attR, was capable of mobilizing the defective ICEBs1 at the chromosomal attachment site. In addition, a larger deletion that extends into nicK (ydcR), ΔICEBs1-347, and leaves intact only seven ORFs at the left end of ICEBs1 was mobilized when ICEBs1 functions were provided in trans (Fig. 1C, ICEBs1-347). These results indicate that oriT lies somewhere to the left of the endpoint in this deletion.
The ability of the “locked-in” ICEBs1 at thrC to complement these ICEBs1 mutants indicates that excision and circularization are not required for ICEBs1 gene expression and production of a functional conjugation apparatus. However, the mating efficiencies of the three ICEBs1 derivatives (ΔICEBs1-319, -320, and -347) were consistently lower than the mating efficiencies of those that did not require complementation for mating (Fig. 1B and 1C). We suspect that this is due to a combination of effects; perhaps expression of the ICEBs1 genes from the excision-defective construct at thrC is not completely normal. Also, it is possible that having two copies of some of the ICEBs1 genes in the merodiploid alters the stoichiometry and assembly of a functional conjugation apparatus. In addition, perhaps some of the ICEBs1 proteins function better in cis than in trans (e.g., the relaxase).
Identification of a cis-acting region of ICEBs1 required for its mobilization.
We tested three additional deletions in ICEBs1 for their ability to be mobilized by functions provided in trans. These deletions, ΔICEBs1-348, ΔICEBs1-200, and ΔICEBs1-205, all extend past nicK and into or past ydcQ (Fig. 1D). Despite the presence of ICEBs1 ΔattR at thrC, these deletions could not be mobilized (Fig. 1D, thrC::ICEBs1 donor). Combined with the finding that the deletion mutant ΔICEBs1-347 can be mobilized, these results indicate that there is a cis-acting element needed for transfer in the 1.5-kb region that is present in ΔICEBs1-347 and absent in ΔICEBs1-348 (Fig. 1C and D).
A 0.8-kb fragment of ICEBs1 contains oriT and an essential GC-rich inverted repeat.
Since the oriTs of conjugative and mobilizable plasmids often contain inverted repeats (19, 34), we searched for an inverted repeat in the 1.5-kb ydcQ-nicK region and found a 24-bp sequence, ACCCCCCCACGCTAACAGGGGGGT, comprised of a perfect 7-bp GC-rich inverted repeat (underlined) and an intervening 10 bp. This 24-bp sequence seemed particularly noteworthy since oriT sequences are often in close proximity to the genes encoding their cognate DNA relaxases (19, 20, 36, 49), and this 24-bp sequence is located in the 5′ end of nicK, which is predicted to encode a DNA relaxase. In addition, a nearly identical sequence, ACCCCCCgtatCTAACAGGGGGGT (four mismatches are in lowercase), is located in the oriT region of Tn916, 330 bp upstream of orf20, which encodes an enzyme with endonuclease activity in vitro (56).
We found that a 0.8-kb fragment of ICEBs1 containing this 24-bp sequence confers mobility to the nonmobilizable mutant ΔICEBs1-205. We cloned this 0.8-kb fragment into ΔICEBs1-205, generating ΔICEBs1-373 (Fig. 1E). In contrast to ΔICEBs1-205, this element (ΔICEBs1-373) could be mobilized when ICEBs1 functions were provided in trans (Fig. 1D and E, thrC::ICEBs1 donor).
We also constructed ΔICEBs1-406, which is identical to the mobilizable ΔICEBs1-373 but contains four point mutations in the 24-bp sequence (ACCaCCaCACGCTAACAGaGGaGT) (mutations are in lowercase). We found that these mutations reduced mating activity conferred by the 0.8-kb fragment by greater than 100-fold (Fig. 1E, thrC::ICEBs1 donor). These results narrow down the location of the oriT of ICEBs1 to a 0.8-kb fragment, which contains a GC-rich inverted repeat that is important for oriT function.
oriT is nicked within the GC-rich inverted repeat after activation of ICEBs1.
Transfer of conjugative plasmids requires nicking of one strand of their oriT, often several base pairs downstream of an inverted repeat (34, 38). We used primer extension assays designed to detect nicks near the 24-bp sequence. Primers CLO75 and CLO76 are complementary to opposite strands in the ydcQ-nicK region, 61 bp upstream and 72 bp downstream of the 24-bp inverted repeat, respectively. Controls showed that each primer could detect cleaved templates generated by restriction enzyme digestion in vitro (data not shown).
We identified a nick in the top strand of ICEBs1 in primer extension reactions using CLO76 as a primer and B. subtilis DNA as the template. ICEBs1 was induced by overexpression of RapI, and DNA was purified and subjected to primer extension analysis. Two primer extension products were detected in reactions using end-labeled primer CLO76 (Fig. 2A, lane 1). The lower band likely corresponds to the primer extension product terminated at the nick in ICEBs1, whereas the upper band likely corresponds to the same extension product with an extra base added by the Taq polymerase terminal transferase activity (13). By running the primer extension reactions in the same lanes as the DNA sequencing ladder (data not shown), the nic site was found to be located between the repeated elements in the inverted repeat in nicK, a sequence that is also conserved in Tn916 (Fig. 2C).
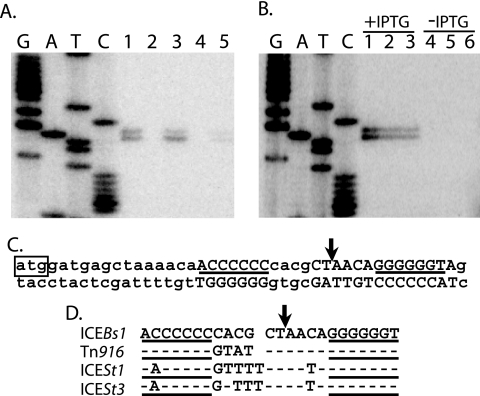
FIG. 2.
NicK-dependent nicking of ICEBs1 between the GC-rich inverted repeat in oriT. (A and B) Primer extension products generated using end-labeled CLO76 and B. subtilis genomic DNA are shown along with DNA sequencing reactions (GATC). (A) Lanes 1 to 3, nicK+ (JMA168); lane 4, ΔnicK306 (CAL306); lane 5, ΔnicK306 Pxis-nicK (CAL346). Strains contained the IPTG-inducible Pspank(hy)-rapI and were grown without IPTG (lane 2) or with IPTG for 1 h (lanes 1 and 3 to 5). (B) Lanes 1 and 4, control ICEBs1+ Pspank(hy)-rapI (JMA168); lanes 2 and 5, ICEBs10 Pspank(hy)-nicK488 (CAL502); lanes 3 and 6, ICEBs10 Pspank(hy)-nicK477 (CAL501). Strains were grown for 1 h with (lanes 1 to 3) or without (lanes 4 to 6) IPTG. (C) Diagram of the double-stranded DNA sequence showing the nicK start codon (ATG in box), inverted repeats (horizontal lines), location of the nic site (vertical arrow), and base pairs conserved in the Tn916 oriT region (uppercase). (D) Alignment of the top strands of the conserved sequence in ICEBs1, Tn916, ICESt1, and ICESt3. A gap in the top two sequences indicates that ICEBs1 and Tn916 have one less base pair than ICESt1 and ICESt3 in the intervening region. Dashes indicate identity with ICEBs1 sequence. The lines and arrow are as in panel C.
Induction of ICEBs1 was necessary for efficient nicking. We did not detect any nicking in the absence of induction of RapI expression with IPTG (Fig. 2A, lane 2). Nicking did not require excision of ICEBs1; we detected RapI-dependent nicking of oriT in a nonexcisable ΔattR100::tet derivative of ICEBs1 (data not shown). We did not find any nicks on the bottom strand of ICEBs1 using primer CLO75 (data not shown). These results indicate that activation of ICEBs1 gene expression leads to nicking within an inverted repeat that is important for oriT activity. Analogous to conjugative and mobilizable plasmids that are nicked on one strand of their oriTs, only one strand of the excised ICEBs1 may be transferred to recipient cells, (52). Furthermore, our results indicate that nicking of ICEBs1 does not require excision and circularization of the element.
nicK is necessary for nicking and transfer of ICEBs1.
NicK is homologous to Orf20 of Tn916, which nicks the oriT of Tn916 in vitro and may facilitate the transfer of a single strand of Tn916 to recipient cells (56). NicK and Orf20 are also homologous to DNA relaxases involved in rolling-circle replication of the pT181 family of plasmids (32, 46).
We found that nicK is necessary for cleavage within the ICEBs1 oriT, located within the nicK ORF. We constructed a deletion of nicK (ΔnicK306) that starts 332 bp downstream of the 24-bp inverted repeat and leaves almost the entire 0.8-kb sequence that contains oriT intact. This mutation abolished detectable nicking at oriT (Fig. 2A, lane 4).
Expression of nicK in trans restored nicking (Fig. 2A, lane 5). However, since the nicking assay was based on primer extension with a primer that detects both the oriT associated with the nicK306 allele and the oriT associated with the ectopic nicK+ allele, we could not distinguish whether nicking was restored at the ΔnicK306 locus or was just occurring within oriT in nicK+.
To test whether nicking was restored in the ICEBs1 ΔnicK306 mutant, we measured mating efficiencies. After induction of wild-type ICEBs1 by overproduction of RapI (donor strain JMA168), the mating efficiency (into recipient CAL419) was ∼7%. In contrast, when the ICEBs1 ΔnicK306 mutant was used as the donor, mating was undetectable (<0.0002%). This defect in mating was not due to a defect in excision (data not shown), consistent with previous results showing that the only ICEBs1 genes necessary and sufficient for excision are int and xis (Lee et al., submitted).
The mating defect caused by the ΔnicK306 mutation was partially complemented by providing relaxase in trans. We fused nicK to the promoter that drives xis (Pxis-nicK). This promoter is normally repressed by the ICEBs1 immunity repressor ImmR and induced when ICEBs1 is activated by RapI or DNA damage (2). When the ICEBs1 ΔnicK306 mutant was used as a donor and relaxase was provided from Pxis-nicK (donor CAL346), the mating efficiency was significantly restored, to ∼1% from undetectable levels (<0.0002% in the absence of the Pxis-nicK fusion).
Whereas expression of nicK in trans significantly restored mating, the efficiency was not up to levels seen with nicK+ ICEBs1. This significant but partial complementation might be due to poor expression of nicK from the ectopic Pxis-nicK fusion. Alternatively, it might indicate that, while not destroying oriT, the ΔnicK306 might delete part of oriT. A third possibility is that relaxase might function preferentially in cis. Nonetheless, taken together, our results indicate that nicK is required for nicking and that oriT in the ICEBs1 ΔnicK306 mutant is mostly or completely functional.
nicK is the only ICEBs1 gene product needed for nicking at oriT.
We found that expression of nicK is sufficient to cause nicking within oriT. We made two fusions of nicK to the IPTG-inducible promoter Pspank(hy), Pspank(hy)-nicK477 and Pspank(hy)-nicK488, which extend 87 and 393 bp upstream of the nicK ORF, respectively. The Pspank(hy)-nicK488 construct contains the entire 0.8-kb sequence that confers mobility to ΔICEBs1-205, and the Pspank(hy)-nicK477 construct is missing 274 bp at the 5′ end of the 0.8-kb sequence. In strains cured of ICEBs1 (ICEBs10), nicking occurred in both constructs (Fig. 2B, lanes 2 and 3) and was not observed in the absence of induction with IPTG (Fig. 2B, lanes 5 and 6). These results indicate that NicK is the only ICEBs1 gene product needed for specific nicking within the GC-rich inverted repeat in nicK and that the same site is nicked in the intact ICEBs1, the “locked-in” element, and the isolated nicK.
DISCUSSION
We found that oriT of ICEBs1 is contained on a 0.8-kb DNA fragment overlapping ydcQ and nicK. This fragment was sufficient to allow mobilization of a mutant derivative of ICEBs1 that contains DNA only from the ends of the element. Furthermore, this fragment contains a GC-rich inverted repeat internal to nicK that is necessary for full oriT function. When ICEBs1 was activated, the top strand of the element was nicked between the arms of the repeat. Nicking required NicK and no other ICEBs1 gene products and did not require excision of ICEBs1 from the chromosome. We propose that increased expression of NicK, induced when ICEBs1 is activated by RapI or DNA damage, leads to nicking of ICEBs1 at oriT and covalent attachment of NicK to one strand of ICEBs1, analogous to homologous relaxases. This form of ICEBs1 is likely the substrate for transfer of a single strand of ICEBs1 DNA to mating partners.
Conserved relaxases and sequences in oriT in ICEs from B. subtilis, E. faecalis, and Streptococcus thermophilus.
The relaxase and oriT from ICEBs1 are similar to those from Tn916, ICESt1, and ICESt3 (3, 7, 54, 56). oriT of Tn916 from E. faecalis is located near orf21 and orf20 (31), homologs of ICEBs1 ydcQ and nicK, respectively. ICESt1 and the closely related ICESt3 from S. thermophilus are predicted to contain oriT in the intergenic region between orfK and orfJ (7, 54), which are homologs of ydcQ and nicK. All four oriT regions contain a highly conserved sequence comprised of a GC-rich inverted repeat and an intervening 10 or 11 bp (Fig. 2D). In ICEBs1, the conserved sequence is located within nicK. In Tn916, ICESt1, and ICESt3, the conserved sequence is located in the intergenic region upstream of their nicK homologs.
The orf20 gene product from Tn916 has been purified and characterized in vitro (56). The purified Orf20 has endonuclease activity that is relatively nonspecific. However, addition of the Tn916 integrase protein results in specific cleavage in the spacer sequences in an AT-rich inverted repeat 55 bp downstream from the GC-rich inverted repeat. DNase I footprinting results indicate that integrase protects part of the conserved GC-rich inverted repeat, and it was proposed that binding of integrase to the Tn916 oriT might coordinate excision and conjugation (28).
The in vitro specificity of the ICEBs1 nicK gene product is not known. However, in vivo, ICEBs1 oriT was nicked at the same site in the presence or absence of ICEBs1 integrase. Furthermore, nicking occurred at the same site in ICEBs1 derivatives that could excise and in constructs containing only nicK and oriT (in the absence of other parts of ICEBs1). It is not known if the in vivo activity of ICEBs1 NicK is indicative of that of Tn916 Orf20. Orf20 homologs were divided into two groups based on the amount of sequence identity to Orf20 (56). NicK (YdcR) of ICEBs1 belongs to the group with less overall identity and similarity. It is possible that relaxases more similar to Orf20 require a specificity factor and that those more similar to ICEBs1 NicK do not. It is also possible that the in vivo activity of Orf20 is not identical to that in vitro.
DNA translocases and conjugation.
The gene upstream of nicK, ydcQ, encodes a homolog of FtsK and SpoIIIE (pfam01580 FtsK_SpoIIE [4]). In addition, the genes upstream of orf20 and orfK of Tn916 and ICESt1 (and ICESt3), respectively, also encode FtsK/SpoIIIE homologs. FtsK and SpoIIIE are DNA translocases that are distantly related to the coupling proteins of plasmid conjugation systems (10, 18, 30). Coupling proteins interact with their cognate conjugal DNA relaxase and bind to both single- and double-stranded DNA (10, 47, 58). Coupling proteins likely form transmembrane pores and may facilitate the translocation of the conjugal DNA relaxase and the attached single strand of DNA through the membrane (17, 23, 24, 36, 40, 58).
It seems likely that, analogous to the case for conjugal relaxases and coupling proteins, NicK directly interacts with the putative coupling protein YdcQ to promote the 5′-to-3′ transfer of a single strand of ICEBs1 through a transmembrane conjugation pore and into the recipient cell. Our model predicts that a large 3′ portion of the nicK ORF will be transferred first and that ydcQ and the 5′ region of nicK will be transferred last. The strand- and site-specific nicking of oriT of Tn916 by the Orf20 endonuclease in the presence of Tn916 Int similarly indicates that the 5′-to-3′ transfer of a single strand of Tn916 initiates with orf20 and terminates with orf21 (56).
DNA relaxases for plasmid conjugation and rolling-circle replication.
DNA relaxases involved in plasmid conjugation and rolling-circle replication have common features. These relaxases attach to the 5′ end of the nicked DNA strand via a phosphotyrosyl covalent linkage (9, 46). Their genes and cognate nic sites are often in close proximity to each other (19, 33). Rolling-circle replication relaxases often nick between an inverted repeat, while conjugal relaxases often nick between an inverted repeat or several base pairs downstream of an inverted repeat (19, 33, 34, 38, 46). Rolling-circle replication relaxases recruit replication factors to the double-strand origin of plasmid replication so that leading-strand synthesis can proceed from the nicked 3′ end (16, 33). In plasmid conjugation systems, replication factors may also be recruited to the nicked 3′ ends of oriTs so that leading-strand synthesis can replace the transferred strand in the donor and unwind the single strand for transfer (34).
Some of the DNA relaxases involved in conjugation have diverged from those involved in rolling-circle replication by acquiring additional functions specific for DNA transfer (9). For example, the mobilizable plasmid R1162 produces a DNA relaxase that has primase activity, which may facilitate complementary-strand synthesis of the transferred single strand in certain recipients (51). F TraI, required for F plasmid conjugation, has both DNA relaxase and DNA helicase activities, as well as domains that allow it to interact with other conjugation proteins (44, 45, 57).
Complementary-strand synthesis.
For both rolling-circle replication and conjugation, the nicking, unwinding, and recircularization of a single strand of DNA is followed by complementary-strand synthesis (32, 53). For rolling-circle replication, complementary-strand synthesis is initiated from a single-strand origin of replication, which, like lagging-strand synthesis, requires RNA priming (16, 32). For conjugative and mobilizable plasmids, complementary-strand synthesis of the transferred circularized single strand occurs in the recipient (53). Some conjugative plasmids encode primases that are transferred into the recipient and are important for complementary-strand synthesis, while others appear to rely on host-encoded primase activities (34, 51). In either case, complementary-strand synthesis is initiated primarily at the normal origin of replication on the transferred plasmid (53).
Unlike conjugative and mobilizable plasmids, ICEBs1 resides in the host chromosome. It excises to form a circular intermediate. If ICEBs1 is transferred as a single strand, then complementary-strand synthesis in the recipient is likely to be required to generate the double-stranded circular form of ICEBs1 for integration. Complementary-strand synthesis is probably also required before Int can even be produced in the recipient, since most promoters are active only in double-stranded DNA (43), and we predict that the transferred single strand corresponds to the nontemplate strand of int. It is not clear if or how much replication of the excised ICEBs1 circle occurs in the donor, but most characterized ICEs are not known to replicate autonomously.
Acknowledgments
We thank Jennifer Auchtung, Melanie Berkmen, and Bijou Bose for helpful comments on the manuscript.
This work was supported by NIH PHS grant GM50895.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Adams, V., D. Lyras, K. A. Farrow, and J. I. Rood. 2002. The clostridial mobilisable transposons. Cell Mol. Life Sci. 59:2033-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auchtung, J. M., C. A. Lee, K. L. Garrison, and A. D. Grossman. 2007. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 64:1515-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auchtung, J. M., C. A. Lee, R. E. Monson, A. P. Lehman, and A. D. Grossman. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. USA 102:12554-12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 7.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77-97. [DOI] [PubMed] [Google Scholar]
- 8.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376-386. [DOI] [PubMed] [Google Scholar]
- 9.Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 25:1011-1022. [DOI] [PubMed] [Google Scholar]
- 10.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, I., P. J. Christie, and D. Dubnau. 2005. The ins and outs of DNA transfer in bacteria. Science 310:1456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Churchward, G. 2002. Conjugative transposons and related mobile elements, p. 177-191. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 13.Clark, J. M. 1988. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 16:9677-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 15.Comella, N., and A. D. Grossman. 2005. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 57:1159-1174. [DOI] [PubMed] [Google Scholar]
- 16.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Draper, O., C. E. Cesar, C. Machon, F. de la Cruz, and M. Llosa. 2005. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl. Acad. Sci. USA 102:16385-16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Errington, J., J. Bath, and L. J. Wu. 2001. DNA transport in bacteria. Nat. Rev. Mol. Cell. Biol. 2:538-545. [DOI] [PubMed] [Google Scholar]
- 19.Francia, M. V., A. Varsaki, M. P. Garcillan-Barcia, A. Latorre, C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 20.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 22.Garcillan-Barcia, M. P., P. Jurado, B. Gonzalez-Perez, G. Moncalian, L. A. Fernandez, and F. de la Cruz. 2007. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol. Microbiol. 63:404-416. [DOI] [PubMed] [Google Scholar]
- 23.Gomis-Ruth, F. X., F. de la Cruz, and M. Coll. 2002. Structure and role of coupling proteins in conjugal DNA transfer. Res. Microbiol. 153:199-204. [DOI] [PubMed] [Google Scholar]
- 24.Gomis-Ruth, F. X., G. Moncalian, R. Perez-Luque, A. Gonzalez, E. Cabezon, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 25.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton, C. M., H. Lee, P. L. Li, D. M. Cook, K. R. Piper, S. B. von Bodman, E. Lanka, W. Ream, and S. K. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. Wiley, Chichester, United Kingdom
- 28.Hinerfeld, D., and G. Churchward. 2001. Specific binding of integrase to the origin of transfer (oriT) of the conjugative transposon Tn916. J. Bacteriol. 183:2947-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 30.Iyer, L. M., K. S. Makarova, E. V. Koonin, and L. Aravind. 2004. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 32:5260-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaworski, D. D., and D. B. Clewell. 1995. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J. Bacteriol. 177:6644-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan, S. A. 2005. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53:126-136. [DOI] [PubMed] [Google Scholar]
- 33.Khan, S. A. 1997. Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol Rev. 61:442-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 35.Lemon, K. P., I. Kurtser, and A. D. Grossman. 2001. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llosa, M., and F. de la Cruz. 2005. Bacterial conjugation: a potential tool for genomic engineering. Res. Microbiol. 156:1-6. [DOI] [PubMed] [Google Scholar]
- 37.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz Fd. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 38.Llosa, M., G. Grandoso, and F. de la Cruz. 1995. Nicking activity of TrwC directed against the origin of transfer of the IncW plasmid R388. J. Mol. Biol. 246:54-62. [DOI] [PubMed] [Google Scholar]
- 39.Llosa, M., G. Grandoso, M. A. Hernando, and F. de la Cruz. 1996. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J. Mol. Biol. 264:56-67. [DOI] [PubMed] [Google Scholar]
- 40.Luo, Y., Q. Gao, and R. C. Deonier. 1994. Mutational and physical analysis of F plasmid traY protein binding to oriT. Mol. Microbiol. 11:459-469. [DOI] [PubMed] [Google Scholar]
- 41.Luttinger, A., J. Hahn, and D. Dubnau. 1996. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19:343-356. [DOI] [PubMed] [Google Scholar]
- 42.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masai, H., and K. Arai. 1997. Frpo: a novel single-stranded DNA promoter for transcription and for primer RNA synthesis of DNA replication. Cell 89:897-907. [DOI] [PubMed] [Google Scholar]
- 44.Matson, S. W., and H. Ragonese. 2005. The F-plasmid TraI protein contains three functional domains required for conjugative DNA strand transfer. J. Bacteriol. 187:697-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matson, S. W., J. K. Sampson, and D. R. Byrd. 2001. F plasmid conjugative DNA transfer: the TraI helicase activity is essential for DNA strand transfer. J. Biol. Chem. 276:2372-2379. [DOI] [PubMed] [Google Scholar]
- 46.Novick, R. P. 1998. Contrasting lifestyles of rolling-circle phages and plasmids. Trends Biochem. Sci. 23:434-438. [DOI] [PubMed] [Google Scholar]
- 47.Panicker, M. M., and E. G. Minkley, Jr. 1992. Purification and properties of the F sex factor TraD protein, an inner membrane conjugal transfer protein. J. Biol. Chem. 267:12761-12766. [PubMed] [Google Scholar]
- 48.Pansegrau, W., and E. Lanka. 1996. Mechanisms of initiation and termination reactions in conjugative DNA processing. Independence of tight substrate binding and catalytic activity of relaxase (TraI) of IncPalpha plasmid RP4. J. Biol. Chem. 271:13068-13076. [DOI] [PubMed] [Google Scholar]
- 49.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 50.Parker, C., E. Becker, X. Zhang, S. Jandle, and R. Meyer. 2005. Elements in the co-evolution of relaxases and their origins of transfer. Plasmid 53:113-118. [DOI] [PubMed] [Google Scholar]
- 51.Parker, C., and R. Meyer. 2005. Mechanisms of strand replacement synthesis for plasmid DNA transferred by conjugation. J. Bacteriol. 187:3400-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parker, C., and R. J. Meyer. 2002. Selection of plasmid molecules for conjugative transfer and replacement strand synthesis in the donor. Mol. Microbiol. 46:761-768. [DOI] [PubMed] [Google Scholar]
- 53.Parker, C., X. L. Zhang, D. Henderson, E. Becker, and R. Meyer. 2002. Conjugative DNA synthesis: R1162 and the question of rolling-circle replication. Plasmid 48:186-192. [DOI] [PubMed] [Google Scholar]
- 54.Pavlovic, G., V. Burrus, B. Gintz, B. Decaris, and G. Guedon. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150:759-774. [DOI] [PubMed] [Google Scholar]
- 55.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 56.Rocco, J. M., and G. Churchward. 2006. The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20. J. Bacteriol. 188:2207-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sastre, J. I., E. Cabezon, and F. de la Cruz. 1998. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 180:6039-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott, J. R., F. Bringel, D. Marra, G. Van Alstine, and C. K. Rudy. 1994. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11:1099-1108. [DOI] [PubMed] [Google Scholar]
- 60.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 2000. Multiple gene products and sequences required for excision of the mobilizable integrated Bacteroides element NBU1. J. Bacteriol. 182:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 62.Vedantam, G., S. Knopf, and D. W. Hecht. 2006. Bacteroides fragilis mobilizable transposon Tn5520 requires a 71 base pair origin of transfer sequence and a single mobilization protein for relaxosome formation during conjugation. Mol. Microbiol. 59:288-300. [DOI] [PubMed] [Google Scholar]
- 63.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]