Abstract
Mycoplasma pneumoniae is a major cause of bronchitis and atypical pneumonia in humans. This cell wall-less bacterium has a complex terminal organelle that functions in cytadherence and gliding motility. The gliding mechanism is unknown but is coordinated with terminal-organelle development during cell division. Disruption of M. pneumoniae open reading frame MPN311 results in loss of protein P41 and downstream gene product P24. P41 localizes to the base of the terminal organelle and is required to anchor the terminal organelle to the cell body, but during cell division, MPN311 insertion mutants also fail to properly regulate nascent terminal-organelle development spatially or gliding activity temporally. We measured gliding velocity and frequency and used fluorescent protein fusions and time-lapse imaging to assess the roles of P41 and P24 individually in terminal-organelle development and gliding function. P41 was necessary for normal gliding velocity and proper spatial positioning of new terminal organelles, while P24 was required for gliding frequency and new terminal-organelle formation at wild-type rates. However, P41 was essential for P24 function, and in the absence of P41, P24 exhibited a dynamic localization pattern. Finally, protein P28 requires P41 for stability, but analysis of a P28− mutant established that the MPN311 mutant phenotype was not a function of loss of P28.
With a minimal genome as small as ∼ 580 kbp and a cell volume about 5% of that of model bacteria such as Escherichia coli or Bacillus subtilis, the mycoplasmas are among the smallest and simplest organisms capable of self-replication. Unusual cell wall-less bacteria, the mycoplasmas are widespread in nature as commensal or parasitic symbionts of specific eukaryotic hosts. Several species, including Mycoplasma pneumoniae, are human pathogens, with M. pneumoniae infections of the respiratory tract commonly resulting in tracheobronchitis and primary atypical or “walking,” pneumonia. M. pneumoniae is the leading cause of pneumonia in older children and young adults and accounts for up to 30% of all cases of pneumonia requiring hospitalization; extrapulmonary manifestations can also occur, in some cases reflecting a capacity to spread (23).
In addition to its considerable medical significance, M. pneumoniae is striking from a biological perspective, with a remarkable cellular organization for a minimalist microorganism, highlighted by a complex, polar, membrane-bound terminal organelle that functions in cell division, cytadherence, and gliding motility (2, 10, 15, 21). Although the molecular mechanisms that regulate and power gliding are unknown, coordination of motor activity with adherence and terminal-organelle development is required for normal cell division (8). During mycoplasma replication, motile cells cease gliding coincident with the formation of a nascent terminal organelle adjacent to an existing structure. Significantly, assembled terminal organelles generally do not initiate gliding for several hours, during which time an existing terminal organelle typically resumes gliding, thereby displacing the nascent structure to the trailing cell pole prior to cytokinesis (8).
We previously isolated M. pneumoniae gliding-deficient transposon insertion mutants in order to identify mycoplasma genes specifically associated with gliding motility and elucidate the relationship between gliding, terminal-organelle development, and cell division (11). Characterization of two such mutants revealed distinct transposon insertions in open reading frame MPN311, resulting in loss of cytoskeletal protein P41 and the downstream MPN312 product, protein P24 (10) (Fig. 1A). Both P41 and P24 localize to the base of the terminal organelle in wild-type M. pneumoniae (14), and while the timing of P24 incorporation into developing structures has not been previously assessed, newly synthesized P41 localizes to developing terminal organelles prior to gliding cessation (8) and may function early in the hierarchy of events in terminal-organelle assembly (16).
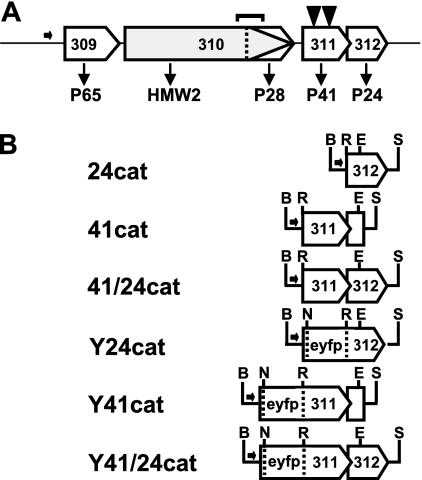
FIG. 1.
Organization of the P65 operon and recombinant P24 and P41 alleles thereof. (A) The gene products are indicated below each open reading frame of the P65 operon (3, 17). The horizontal arrow indicates the promoter for the operon, while transposon insertions corresponding to residues 22 and 161 of P41 are shown as inverted solid triangles. P28 is a product of internal translation initiation of the MPN310 transcript in the same reading frame as HMW2; antibodies directed against the C-terminal domain of HMW2 recognize P28 (4). The bracket above MPN310 indicates a region deleted in P28− mutant C1R1 (1, 4). (B) Recombinant MPN311 and MPN312 alleles with or without translational fusion to the EYFP gene were engineered in transposon vector Tn4001cat (7). B, BamHI; E, EcoRI; N, NcoI; R, BsrGI; S, StuI.
Originally identified on the basis of highly filamentous satellite growth (11), MPN311 mutants commonly appear as chains of coccoid cells (10). Individual cells are motile but have gliding velocities and frequencies typically only 10 to 30% of wild-type levels (11). MPN311 mutants also fail to regulate properly the positioning and gliding activity of nascent terminal organelles, which often form at sites well separated from existing structures and initiate gliding very soon after incorporation of adhesin protein P30 in the assembly process (8, 10). Significantly, terminal organelles frequently detach from the cell body of MPN311 (or terminal-organelle detachment [TOD]) mutants and often remain motile in a cell-independent state for up to 1 h, demonstrating unequivocally that this structure constitutes the gliding motor (10). Recombinant P41 alone restores wild-type cell morphology and integrity, suggesting a structural role in anchoring the terminal organelle to the mycoplasma cell body (10), but the role of P41 in terminal-organelle development and gliding motility is not known, and the function of P24 was not apparent from morphological examination alone. Here we characterized M. pneumoniae TOD mutants carrying recombinant wild-type MPN311 or MPN312 alleles, effectively constituting P24− and P41− mutants, respectively. By using fluorescent protein fusions and time-lapse microcinematography, we demonstrated that P24 requires P41 for localization to the terminal organelle, contributes to initiation of nascent terminal-organelle assembly during cell division, and functions in regulating activation of the gliding motor. Furthermore, we established that P41 is required for wild-type gliding velocity and for directing the formation of new terminal organelles adjacent to existing structures.
MATERIALS AND METHODS
Mycoplasma strains, genetic engineering, and protein profiling.
Wild-type M. pneumoniae M129, the P28− mutant C1R1, and TOD mutants 311-22 and 311-161 were described previously (4, 10, 11, 18), as was the construction and activity of a P30-yellow fluorescent protein (YFP) translational fusion in transposon vector Tn4001cat in M. pneumoniae (7-10). Construction of recombinant MPN311 and MPN312 alleles individually or in tandem under control of their normal promoter (17) in Tn4001cat (designated here 24cat, 41cat, and 41/24cat, respectively [Fig. 1B]), with or without P30-YFP, and their delivery into TOD mutants were described previously (10).
Vectors pEYFP-MPN311, pEYFP-MPN312, and pEYFP-MPN311/312 (8, 10) have translational fusions of YFP at the N terminus of P41 or P24 and contain an engineered StuI site downstream. Here each was digested with NcoI and BamHI, opening the vector at the 5′ end of the enhanced YFP (EYFP) gene. A BamHI-NcoI fragment containing the P65 promoter region (P65-prom) from a pMT85 derivative (8) was cloned into the corresponding sites of pEYFP-MPN311, pEYFP-MPN312, and pEYFP-MPN311/312 and then digested with BamHI and StuI to liberate P65-prom-EYFP-MPN311, P65-prom-EYFP-MPN312, or P65-prom-EYFP-MPN311/312, which were cloned individually into the BamHI and SmaI sites of transposon vector Tn4001cat to yield Y24cat, Y41cat, and Y41/24cat, respectively (Fig. 1B). To analyze the locations of P41 or P24 relative to terminal-organelle protein P30 in P24− or P41− mutant backgrounds, P30-cyan fluorescent protein (CFP) was excised with XbaI from pECFP-P30 (8) and cloned into the corresponding site in pEYFP containing the P65-prom-EYFP-MPN312 described above. The proper orientation of the P30-CFP insert, as determined by sequence analysis, generated BamHI sites at both ends of the P30-CFP insert, which was excised with BamHI and cloned into the corresponding site of Y24cat, Y41cat, and Y41/24cat, placing P30-CFP upstream of the P65 promoter region for each. The fidelity of all constructs was established by sequence analysis before delivery by electroporation (12) into M. pneumoniae. Multiple transformants for each were selected, filter cloned, expanded, and stocked (11). Mutants 311-22 and 311-161 were maintained under gentamicin selection (18 μg/ml), while chloramphenicol was included (30 μg/ml) for transformants with Tn4001cat derivatives. Protein profiles were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting as described previously (9), using rabbit antisera to P1 (24), P65 (20), P24 (4), P41 (4), and HMW2 (25) and monoclonal green fluorescent protein-specific antibody (BD Biosciences, San Jose, CA).
Gliding motility.
Satellite growth of mutants and transformants was assessed relative to that of wild-type M. pneumoniae as described previously (9), with digital images from phase-contrast microscopy (40× objective) recorded at 12-h intervals for up to 6 days. Cell gliding was quantitated as detailed previously (9), with gliding velocities and frequencies of >250 individual cells averaged from two filter-cloned populations for each transformant and normalized to the wild-type values.
Microscopy.
Time-lapse phase-contrast/fluorescence microscopy to assess the roles of proteins P41 and P24 in M. pneumoniae terminal-organelle development was conducted as detailed previously (8, 10). Briefly, log-phase cultures of mycoplasma transformants producing P30-YFP or producing P30-CFP plus Y41cat, Y24cat, or Y41/24cat were diluted 1:40 in fresh medium and incubated overnight in chamber slides. For P30-YFP and YFP-P24 localization, digital phase-contrast and fluorescence images were captured with an exposure time of 1.0 s and 2- to 30-min frame intervals on a Leica DM IRB epifluorescence inverted microscope (Wetzlar, Germany) enclosed within an incubation chamber set at 37°C (Solent Scientific, Ltd., Portsmouth, United Kingdom) and coupled to an Orca ER charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan), merged, quantitated, and rendered in three dimensions by Openlab imaging software v4.5-5.02 (Improvision, Lexington, MA). Colocalization of CFP and YFP fusions was likewise determined using 1-s exposures with fluorescence excitation of both signals recorded in rapid succession by alternating between dichroic EYFP and ECFP filter cubes (Chroma Technology Corp., McHenry, IL).
RESULTS
Recombinant P24− and P41− mutants.
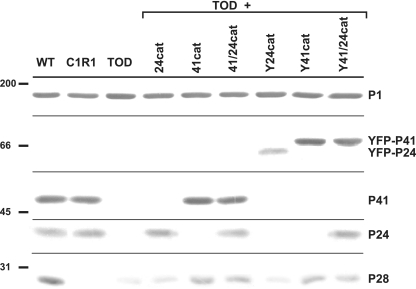
The delivery of wild-type P41 and P24 alleles into TOD mutants was described previously, with recombinant P41 restoring wild-type satellite growth and cell morphology but P24 function not readily apparent (10). In the current study we characterized further the phenotype of TOD mutants producing recombinant P24, P41, or N-terminal YFP translational fusions thereof (Fig. 1B) in order to elucidate their functions. Multiple transformants for each construct were examined for production of the recombinant proteins. Transformants with Y24cat or Y41cat produced YFP-P24 or YFP-P41, respectively, at approximately wild-type levels (Fig. 2). Protein P28, a product of MPN310 immediately upstream (Fig. 1A) that requires P41 for stability (10), rebounded substantially with Y41/24cat or Y41cat (Fig. 2). Furthermore, Y41/24cat or Y41cat, but not Y24cat, restored wild-type cell morphology and satellite growth (data not shown), indicating that the YFP fusion did not affect P41 function in cell architecture. Finally, the P28− mutant was indistinguishable from the wild type in satellite growth and cell morphology (data not shown), and therefore the loss of P28 did not appear to contribute to the MPN311 mutant phenotype.
FIG. 2.
Western immunoblot analysis of recombinant P41 and P24. M. pneumoniae strains and recombinant alleles they carry are indicated across the top. Protein size standards are shown to the left, with specific protein bands labeled to the right. P1 was included as an internal control. Protein profiles are representative of multiple filter clones of transformants of MPN311-22 and MPN311-161 (TOD). Horizontal lines indicate where the membrane was cut prior to probing with primary antisera. WT, wild type.
P24 and gliding motility.
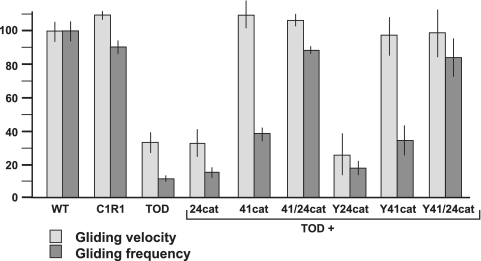
TOD mutants are motile but at gliding velocities and frequencies about 30% and 10% of wild-type levels, respectively (10, 11). To assess the contributions of P41 and P24 individually in M. pneumoniae cell gliding, TOD mutants producing recombinant P24 or P41 alone were examined by time-lapse digital imaging (Fig. 3). Recombinant P41 and P24 together restored cell gliding parameters to wild-type levels, as expected (10). Recombinant P41 alone or with fusion to YFP conferred a wild-type gliding velocity but only partially restored gliding frequency (Fig. 3), but 24cat or Y24cat alone had little impact on gliding velocity or frequency. The reductions in gliding frequency and velocity in TOD mutants were not attributable to loss of P28, as mutant C1R1 exhibited near-wild-type gliding parameters (Fig. 3). Taken together, these data indicate that P41 is sufficient to restore gliding velocity, while P41 and P24 are both required for normal gliding frequency.
FIG. 3.
Role of P24 and P41 in gliding motility parameters. Mean gliding frequency and velocity were determined for at least 250 cells of the indicated strains. Gliding velocities were calculated as the total distance traveled by a cell divided by total time of the field interval minus the amount of time spent in resting periods (i.e., corrected gliding velocity, as described in detail previously [9]). Gliding frequencies were calculated as the percentage of cells exhibiting gliding in each observation interval (11). Means and standard errors were determined from clonal transformants of MPN311-22 and MPN311-161 mutants (TOD). Error bars, standard errors of the means. WT, wild type.
P24 and terminal-organelle development.
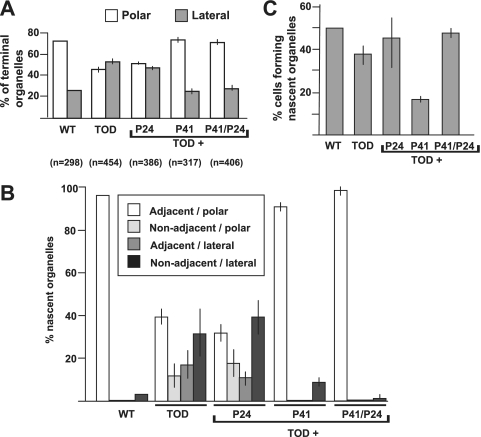
We monitored growing cultures of TOD mutants producing recombinant P24 or P41 and terminal-organelle protein P30-YFP (8, 9) by time-lapse digital imaging to determine if nascent terminal organelles formed adjacent to existing structures or at distal sites, like wild-type M. pneumoniae or the TOD mutant alone, respectively. The P30-YFP focal pattern at time zero and cell development pattern throughout for TOD mutants plus 24cat were similar to those for the TOD mutants alone (10), with P30 foci distributed approximately equally among polar and lateral sites (Fig. 4A). Furthermore, newly formed terminal organelles initiated gliding rapidly and often subsequently separated from the cell body (data not shown). In contrast, new terminal-organelle formation in TOD mutants plus P41cat alone was comparable in many ways to that of wild-type M. pneumoniae or TOD mutants plus 41/24cat (Fig. 4 and data not shown). Thus, with P41 present, about 75% of P30-YFP foci were polar (Fig. 4A) and >90% of nascent terminal organelles formed adjacent to an existing, polar structure (Fig. 4B). In addition, like the wild type (8), P41-producing cells ceased gliding coincident with new terminal-organelle formation for the few motile cells observed. Surprisingly, however, the percentage of P41+/P24− cells initiating terminal-organelle development during the 2-h observations was less than that in wild-type M. pneumoniae, the parental TOD mutants, TOD mutants complemented with P24 alone or with both P41 and P24 (Fig. 4C), or mutant C1R1 (data not shown).
FIG. 4.
Terminal-organelle location in P41− and P24− mutants. (A) Localization of terminal organelles in the indicated M. pneumoniae strains by using P30-YFP as a marker. P41 was required for terminal-organelle localization predominantly at a cell pole (polar) as in the wild type (WT) (8), rather than at sites along the cell body (lateral) as described previously for the parental P41−/P24− TOD mutants (10). (B) Images from time-lapse phase-contrast/fluorescence microscopy over 2-h intervals were used to establish the relative locations of newly synthesized terminal organelles. In the presence of P41, new terminal organelles formed primarily adjacent to an existing structure at one cell pole, as described previously (8). In contrast, in the absence of P41, new terminal organelles frequently appeared at lateral sites as defined above and not adjacent to an existing structure. (C) The frequency of terminal-organelle development was determined based on the percentage of cells forming a new terminal organelle over 2-h intervals for the indicated strains, from images obtained by time-lapse phase-contrast/fluorescence microscopy. Error bars, standard errors of the means.
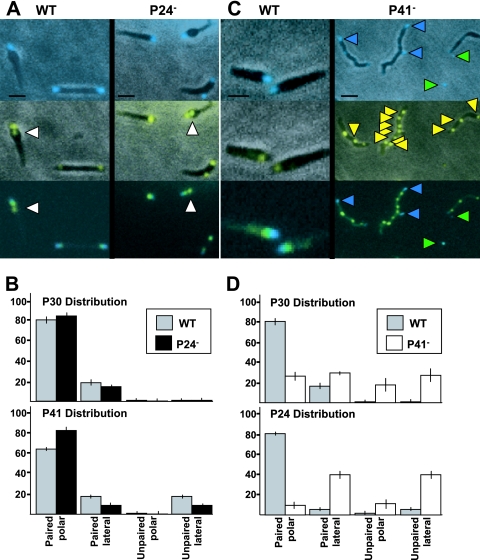
Analysis of gliding frequency and terminal-organelle development in P24− M. pneumoniae suggested a role for P24 in coordinating these processes in a P41-dependent manner. Both P41 and P24 localize to the base of the terminal organelle (14), but it was not known whether either required the other to do so. Therefore, we examined YFP fusions of each in TOD mutants producing P30-CFP as a terminal-organelle marker. P30-CFP and YFP-P41 localized to the distal end and base of the terminal organelle, respectively, in wild-type M. pneumoniae, as expected (8, 14), and in the C1R1 and TOD mutants (Fig. 5A and data not shown). Thus, neither P28 nor P24 is required for P41 to localize properly. Each lateral P30 focus was paired with P41, likely representing nascent terminal organelles in the process of displacement to the opposite cell pole. As expected (8), some cells having a polar P30-P41 pair had a second, unpaired P41 focus directly proximal (Fig. 5A), indicating that P41 foci form prior to incorporation of P30. Loss of P28 did not affect frequency of nascent terminal-organelle development, as unpaired P41 foci represented 18 to 20% of all P41 foci in wild-type M. pneumoniae and mutant C1R1 (Fig. 5B), as well as TOD mutants plus Y41/24cat (data not shown), consistent with published values (8). In contrast, we observed a >50% decrease in unpaired P41 foci in the absence of P24 (Fig. 5B), suggesting less frequent initiation of terminal-organelle formation and reinforcing a role for P24 in initiation of new terminal-organelle development.
FIG. 5.
Localization of P41 and P24 relative to P30 in wild-type (WT) and mutant M. pneumoniae. The positions of YFP-P24 and YFP-P41 were analyzed relative to P30-CFP as a terminal-organelle marker by using phase-contrast/fluorescence microscopy in the absence of P24 (A and B) or P41 (C and D). (A) P30-CFP (top panels), YFP-P41 (middle panels), and merged fluorescence images (bottom panels). In the absence of P24 there was no qualitative difference in the relative positions of P41 and P30 compared to the wild type, with some cells having an unpaired P41 focus proximal to a paired P41/P30 focus, as expected (8) (white arrowheads). (B) Quantitation of the relative positions of P41 and P30 in the absence of P24 (n > 50). Top panel, P30-CFP distribution in the absence of P24; bottom panel, P41 distribution in the absence of P24. (C) P30-CFP (top panels), YFP-P24 (middle panels), and merged fluorescence images (bottom panels). In the absence of P41, detached terminal organelles containing P30-CFP were common, as expected (10) (top and bottom panels, green arrowheads), although many had little detectable P24. YFP-P24 foci were distributed along the length of the mycoplasma cell, often unpaired with an obvious P30 (yellow arrowheads). Blue arrowheads, P30 foci paired with P24. (D) Quantitation of the relative positions of P24 and P30 in the absence of P41 (n > 50). Top panel, P30 distribution in the absence of P41 (10); bottom panel, P24 distribution in the absence of P41. Scale bars, 1 μm (A) and 2 μm (C). Error bars, standard errors of the means.
P41 is required for proper localization of P24.
P24 localized to the base and P30 to the distal end of the terminal organelle as expected in wild-type M. pneumoniae (14) (Fig. 5C) and in mutant C1R1 (data not shown). P30 foci were always observed paired with P24 foci, whether polar or lateral; unpaired P24 foci were occasionally evident directly proximal to a P30/P24 pair at the terminal organelle and accounted for about 7% of all P24 foci, compared to about 18% for P41 (Fig. 5B and D and data not shown). In the absence of P41, P24 also formed discrete foci typically spaced at irregular intervals along short and long chains of cells, and unpaired P24 foci accounted for approximately 50% of total P24 foci (Fig. 5C and D). Furthermore, only approximately 40% of the detached terminal organelles exhibited a YFP-P24 signal, and unlike for P30-CFP foci, fluorescence intensities of YFP-P24 foci were typically weaker in detached terminal organelles than when cell associated (Fig. 5C), raising the possibility that terminal-organelle detachment contributes in part to the high number of unpaired, cell-associated P24 foci. We were unable to colocalize P24 and P30 over multiple time-lapse images to investigate this possibility further due to the rapid photobleaching and apparent toxicity of P30-CFP upon excitation, as described previously (8).
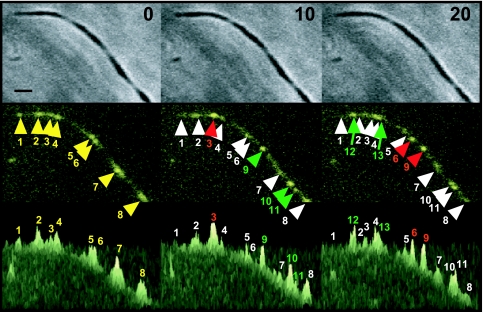
Weak or transient interactions between P24 and the terminal organelle might explain the high incidence of unpaired P24 foci in the absence of P41. When observed at 10-min intervals, P24 foci appeared to migrate or, alternatively, to form and dissipate in the mycoplasma cell body in both motile and nonmotile cells (Fig. 6). For example, in the nonmotile cells shown, focus 3 was more intense and flanking foci 2 and 4 were less intense after 10 min, but by 20 min, focus 3 had decreased in intensity and new foci 12 and 13 had formed. We could not distinguish between true lateral displacement and dissipation/reformation of P24 foci, even when examined at 2-min intervals (data not shown), and attempts to monitor in real time were unsuccessful due to photobleaching. Significantly, this dynamic was not evident for YFP-P24 foci in wild-type or C1R1 cells or for P30-YFP foci in the absence of P41, suggesting a requirement for P41 both to anchor the terminal organelle to the M. pneumoniae body and to sequester P24 at this structure.
FIG. 6.
Time-lapse analysis of P24 localization in the absence of P41. Top panels, phase-contrast images; middle panels, YFP-P24 fluorescence images; bottom panels, three-dimensional representation of fluorescence intensities from the middle panels. Time points for time-lapse analysis are given at the top. At 0 min, eight major YFP-P24 foci were observed (numbered and yellow arrowheads). At 10 min, one focus had appeared to increase in intensity (red numbers and arrowheads) and seven foci to decrease in intensity (white numbers and arrowheads), while three new foci had emerged (green numbers and arrowheads). By 20 min, two foci had increased in intensity (red arrowheads) and nine decreased in intensity (white arrowheads), while two new foci had emerged (green arrowheads). Bar, 1 μm.
DISCUSSION
In the current study we explored the contributions of P41 and P24 individually in gliding and terminal-organelle development. In addition to anchoring the M. pneumoniae terminal organelle to the cell body (10), P41 is specifically required to achieve a wild-type gliding velocity. As detached terminal organelles glide at near-wild-type velocities, it is likely that decreased gliding velocity in the absence of P41 does not reflect a defect in the gliding motor itself. We envision that P41 links the bowl structure (13) at the base of the terminal organelle with the filamentous cytoskeletal network that radiates through the cell body (6, 19). In the absence of this linkage, inefficient force transmission, increased drag from the cell body, or both might account for reduced gliding velocity.
MPN311 mutants complemented only with recombinant P41 exhibited a wild-type cell morphology, terminal-organelle linkage, and gliding velocity, but the percentage of cells gliding was typically only 30 to 40% of that of wild-type cells (Fig. 3), indicating a requirement for P24 for gliding to occur at wild-type frequencies. However, P24 alone was not sufficient to restore gliding frequency to wild-type levels, suggesting that P24 function was P41 dependent. We were unable to establish to what extent, if any, terminal-organelle detachment contributed to the low percentage of motile cells in the absence of P41. However, the observation that mutants lacking only P24 glided at reduced frequencies yet did not exhibit the TOD phenotype argues against this possibility.
We explored whether a reciprocal requirement exists for the localization of P41 and P24 to the base of the terminal organelle by using YFP fusions of each in combination with P30-CFP. Most P24 foci were paired with P30 in wild-type M. pneumoniae, but the presence of unpaired P24 foci suggested that, like P41 (8), P24 incorporates into developing terminal organelles before P30. The percentage of unpaired P24 foci was about half that of unpaired P41 foci, indicating that P41 might precede P24 in the assembly sequence, but time-lapse analysis with combinations of these proteins is required to confirm their order of assembly. P41 localized like the wild type in the absence of P24, but without P41, multiple P24 foci were observed spaced at irregular intervals; many were unpaired with P30 and may represent sites of previous terminal-organelle detachment. However, in time-lapse images P24 exhibited a dynamic behavior in the absence of P41 that we believe reflects slow dissociation and reformation along the cell axis, perhaps including weak or transient association with the terminal organelle. It is not clear whether P24 might interact directly with P41 or perhaps with a P41-dependent protein in wild-type M. pneumoniae.
P41 was sufficient to direct terminal-organelle assembly to the cell pole, but in the absence of P24 new terminal organelles formed at a frequency of <40% of wild type (Fig. 4C). We anticipate that formation of terminal organelles at a lower frequency is reflected in a lower growth rate but have not explored this quantitatively. Somewhat surprisingly, however, new terminal organelles developed at a wild-type frequency with the loss of both P41 and P24, suggesting that P41 or a P41-dependent protein may restrict the initiation of terminal-organelle formation in the absence of P24. The reductions in gliding frequency and initiation of terminal-organelle formation were similar in the absence of P24, which may be coincidental or perhaps reflects a common mechanism, and therefore it is tempting to speculate that P24 functions as a gatekeeper during terminal-organelle assembly. Thus, P41 appears early in terminal-organelle development and is required for proper positioning of the assembling organelle adjacent to an existing structure. As P24 is not required to coordinate gliding cessation with these early events, we believe that P41 also directs the subsequent localization of P24 to the developing terminal organelle. In the absence of P24, cells having ceased gliding coincident with nascent terminal-organelle formation are inhibited from completing that process. Moreover, the twofold reduction in the number of P41 foci unpaired with P30 in the absence of P24 (Fig. 5A) raises the possibility that conversely, prelocalized P24 (for example, associated with the existing terminal organelle) may promote the initial condensation of P41 foci in the formation of a new structure.
In the absence of P41, new terminal organelles formed at sites separate from rather than adjacent to existing structures, as with wild-type M. pneumoniae. This pattern is consistent with new terminal-organelle development de novo (8) and reflects a role for P41 in the spatial positioning of the assembly process, which might be achieved by at least two mechanisms. First, P41 might normally recognize and bind a protein component of the existing terminal organelle. Alternatively, there is growing evidence in model prokaryotes that the bacterial chromosome exhibits a higher-ordered structure such that the origin and terminus are consistently positioned at specific sites within the cell (5), and therefore the mycoplasma chromosome might provide the framework for P41 binding and proper terminal-organelle localization, thereby perhaps coordinating terminal-organelle function with cell division. A correlation between nucleoid staining intensity and the number and location of terminal organelles (22) underscores the coordination of chromosome replication and terminal-organelle development. A potential linkage between the chromosome and the terminal organelle is consistent with the possibility that gliding initiation following chromosome and terminal-organelle duplication (8) drives chromosome segregation, but additional studies are required to evaluate possible chromosome-terminal-organelle interactions in M. pneumoniae.
Acknowledgments
We thank M. B. Hamrick for technical assistance.
This work was supported by Public Health Service research grants AI22362 and AI49194 from the National Institute of Allergy and Infectious Diseases to D.C.K.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Balish, M. F., S. M. Ross, M. Fisseha, and D. C. Krause. 2003. Deletion analysis identifies key functional domains of the cytadherence-associated protein HMW2 of Mycoplasma pneumoniae. Mol. Microbiol. 50:1507-1516. [DOI] [PubMed] [Google Scholar]
- 2.Bredt, W. 1968. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol. Microbiol. (Basel) 32:321-326. [DOI] [PubMed] [Google Scholar]
- 3.Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sanchez-Pulido, B. Snel, M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisseha, M., H. W. Gohlmann, R. Herrmann, and D. C. Krause. 1999. Identification and complementation of frameshift mutations associated with loss of cytadherence in Mycoplasma pneumoniae. J. Bacteriol. 181:4404-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gitai, Z., M. Thanbichler, and L. Shapiro. 2005. The choreographed dynamics of bacterial chromosomes. Trends Microbiol. 13:221-228. [DOI] [PubMed] [Google Scholar]
- 6.Göbel, U., V. Speth, and W. Bredt. 1981. Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J. Cell Biol. 91:537-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn, T. W., E. A. Mothershed, R. H. Waldo III, and D. C. Krause. 1999. Construction and analysis of a modified Tn4001 conferring chloramphenicol resistance in Mycoplasma pneumoniae. Plasmid 41:120-124. [DOI] [PubMed] [Google Scholar]
- 8.Hasselbring, B. M., J. L. Jordan, R. W. Krause, and D. C. Krause. 2006. Terminal organelle development in the cell wall-less bacterium Mycoplasma pneumoniae. Proc. Natl. Acad. Sci. USA 103:16478-16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasselbring, B. M., J. L. Jordan, and D. C. Krause. 2005. Mutant analysis reveals a specific requirement for protein P30 in Mycoplasma pneumoniae gliding motility. J. Bacteriol. 187:6281-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasselbring, B. M., and D. C. Krause. 2007. Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae. Mol. Microbiol. 63:44-53. [DOI] [PubMed] [Google Scholar]
- 11.Hasselbring, B. M., C. A. Page, E. S. Sheppard, and D. C. Krause. 2006. Transposon mutagenesis identifies genes associated with Mycoplasma pneumoniae gliding motility. J. Bacteriol. 188:6335-6345. (Erratum, 189:1179, 2007.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedreyda, C. T., K. K. Lee, and D. C. Krause. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30:170-175. [DOI] [PubMed] [Google Scholar]
- 13.Henderson, G. P., and G. J. Jensen. 2006. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 60:376-385. [DOI] [PubMed] [Google Scholar]
- 14.Kenri, T., S. Seto, A. Horino, Y. Sasaki, T. Sasaki, and M. Miyata. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186:6944-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krause, D. C., and M. F. Balish. 2001. Structure, function and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett. 198:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 17.Krause, D. C., T. Proft, C. T. Hedreyda, H. Hilbert, H. Plagens, and R. Herrmann. 1997. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J. Bacteriol. 179:2668-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipman, R. P., W. A. Clyde, Jr., and F. W. Denny. 1969. Characteristics of virulent, attenuated, and avirulent Mycoplasma pneumoniae strains. J. Bacteriol. 100:1037-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng, K. E., and R. M. Pfister. 1980. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J. Bacteriol. 144:390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proft, T., and R. Herrmann. 1994. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol. Microbiol. 13:337-438. [DOI] [PubMed] [Google Scholar]
- 21.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldo, R. H., III, and D. C. Krause. 2006. Synthesis, stability, and function of cytadhesin P1 and accessory protein B/C complex of Mycoplasma pneumoniae. J. Bacteriol. 188:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willby, M. J., M. F. Balish, S. M. Ross, K. K. Lee, J. L. Jordan, and D. C. Krause. 2004. HMW1 is required for stability and localization of HMW2 to the attachment organelle of Mycoplasma pneumoniae. J. Bacteriol. 186:8221-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]