Abstract
We fully annotated two large plasmids, pMOL28 (164 open reading frames [ORFs]; 171,459 bp) and pMOL30 (247 ORFs; 233,720 bp), in the genome of Cupriavidus metallidurans CH34. pMOL28 contains a backbone of maintenance and transfer genes resembling those found in plasmid pSym of C. taiwanensis and plasmid pHG1 of C. eutrophus, suggesting that they belong to a new class of plasmids. Genes involved in resistance to the heavy metals Co(II), Cr(VI), Hg(II), and Ni(II) are concentrated in a 34-kb region on pMOL28, and genes involved in resistance to Ag(I), Cd(II), Co(II), Cu(II), Hg(II), Pb(II), and Zn(II) occur in a 132-kb region on pMOL30. We identified three putative genomic islands containing metal resistance operons flanked by mobile genetic elements, one on pMOL28 and two on pMOL30. Transcriptomic analysis using quantitative PCR and microarrays revealed metal-mediated up-regulation of 83 genes on pMOL28 and 143 genes on pMOL30 that coded for all known heavy metal resistance proteins, some new heavy metal resistance proteins (czcJ, mmrQ, and pbrU), membrane proteins, truncated transposases, conjugative transfer proteins, and many unknown proteins. Five genes on each plasmid were down-regulated; for one of them, chrI localized on pMOL28, the down-regulation occurred in the presence of five cations. We observed multiple cross-responses (induction of specific metal resistance by other metals), suggesting that the cellular defense of C. metallidurans against heavy metal stress involves various regulons and probably has multiple stages, including a more general response and a more metal-specific response.
Cupriavidus (formerly Ralstonia) metallidurans CH34 is a facultatively hydrogenotrophic, metal-resistant bacterium isolated from the sludge of a zinc decantation tank in Belgium that was contaminated with high concentrations of several heavy metals (2, 15, 23, 25). The Joint Genome Institute (http://genome.jgi-psf.org/finished_microbes/ralme/ralme.home.html) sequenced the entire genome of this strain, which revealed four replicons: the chromosome (3.9 Mb), a megaplasmid (2.6 Mb), and two large plasmids, pMOL28 (171 kb) and pMOL30 (234 kb). C. metallidurans CH34 displays a variety of responses or resistance to several heavy metals; plasmid curing and transfer experiments demonstrated that the two plasmids are involved in metal resistance.
Plasmid pMOL28 was associated with resistance to Ni(II), Co(II), CrO42−, and Hg(II), and pMOL30 was associated with resistance to Ag(I), Cd(II), Co(II), Cu(II), Hg(II), Pb(II), and Zn(II). Cloning and sequencing of several fragments from both plasmids revealed many determinants of resistance (cnr, chr, and mer for pMOL28; czc, pbr, mer, sil, and cop for pMOL30) (1, 9, 18, 22, 27, 30). The mechanisms of resistance included chemoosmotic efflux of cations with proton antiporters (HME-RND family) encoded by czcCBA, cnrCBA, and silCBA, cation diffusion facilitators such as CzcD and CnrT, and P-type ATPases for cytoplasmic detoxification (pbrA, copF, and czcP). The large copVTMKNS1R1A1B1C1D1IJGFLQHE cluster on pMOL30 [resistance to Cu(II)] is involved in cytoplasmic (via copF) copper detoxification; similarly, in the periplasm (27) the copS1R1A1B1C1D1 genes, which have numerous equivalents in other bacterial plasmids and genomes, are involved in copper detoxification (3, 21, 24, 27).
Based on a sequence analysis, we described the complete genetic contents of both plasmids and their organization. A transcriptomic approach, using both reverse transcription-PCR and microarrays, targeting differential expression of genes in cultures challenged by heavy metals allowed us to identify many new genes involved in the response to heavy metals.
MATERIALS AND METHODS
Media, strains, plasmids, and culture conditions.
We grew C. metallidurans CH34 and its derivatives in mineral salts liquid 284 medium (25, 36) supplemented with 0.2% (wt/vol) gluconate. Cultures were grown at 30°C on a rotary shaker at 150 rpm. The four reference strains that we used were the wild-type strain C. metallidurans CH34(pMOL28, pMOL30) and strains AE128, AE126, and AE104 lacking pMOL28, pMOL30, and both plasmids, respectively (25).
Sequence determination and genome data mining.
The C. metallidurans CH34 genome was sequenced at the Joint Genome Institute, Department of Energy (United States), and the data are available at the website http://genome.jgi-psf.org/finished_microbes/ralme/ralme.home.html.
Annotation of the two plasmids.
Sequence analysis and annotation were performed using the integrated annotation tool (iANT) described for Ralstonia solanacearum (35), except that the probabilistic hidden Markov model for coding regions (as predicted by the FrameD software) was generated using C. metallidurans gene sequences obtained from public databases. Each predicted open reading frame (ORF) was reviewed manually by gene annotators to assign start codons. The outputs of a Prosite search and BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/) analysis for the corresponding products also were individually reviewed by experts to generate the proposed annotations.
The insertion sequence (IS) element families and groups were investigated using the BLAST tool from the IS Finder database at http://www-is.biotoul.fr/is.html.
Plasmid maps.
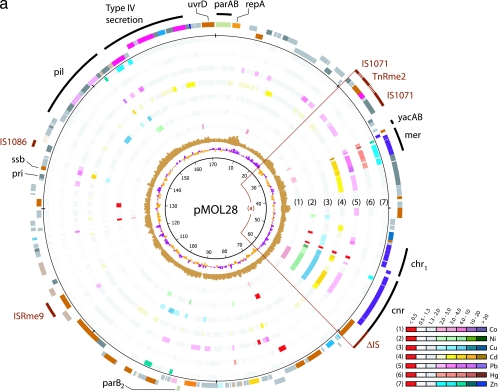
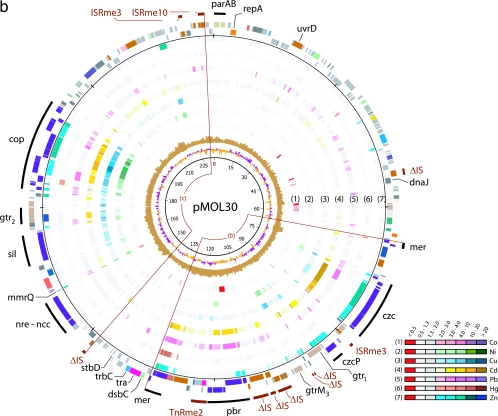
Plasmid maps displaying the results for gene expression, GC skew, G+C content, and COG function (Fig. 1) were drawn using GenomeViz v1.1 (14); all the data for these maps were prepared separately for each type of information, using the TAG and MAP file formats. In particular, the expression results were linked to color codes to display ranges of expression ratios (Fig. 1). Further, we added an inner circle indicating the coordinates of each gene (within a 10-kb window) to facilitate retrieving it in the annotation/expression data tables (see Table S2 in the supplemental material).
FIG. 1.
Expression analysis of pMOL28 (a) and pMOL30 (b). Plasmids pMOL28 and pMOL30 display gene expression under various metal conditions. The sequences of both plasmids start at the ATG codon of the parA gene that is oriented clockwise. The innermost circle is a GC deviation plot and represents the mean centered G+C content (purple, above mean; orange, below mean) using a window size of 500 nucleotides and a 250-nucleotide window overlap. The next circle (light brown) is a GC skew plot in which the (G-C)/(G+C) ratios are shown in sliding windows of 500 nucleotides with a window overlap of 250 nucleotides. The next seven circles (circles 1 through 7) correspond to the results of gene expression for the seven different metals listed in the color-coded diagram. The common color red indicates a negative expression ratio (i.e., down-regulation), while the common colors gray and white indicate expression ratios (range, 0.5 to 2.0) that are considered to be within the background threshold. Up-regulated genes (expression ratio, >2.0) appear in different shades from light to dark corresponding to the degree of increased expression. The two circles on the outside display the genes in functional categories according to the COG color scheme (from NCBI, Bethesda, MD; http://www.ncbi.nlm.nih.gov/COG/grace/fiew.cgi), with genes in the outer circle oriented clockwise and genes in the inner circle oriented counterclockwise. Special features are shown as bars that are either black (for metal resistance genes) or dark brown (for IS elements). The circular positions are indicated at intervals of 10,000 bp. The arcs of circle displayed are (a) the cnr-chr-mer island, (b) the mer-pbr-czc island, and (c) the copper island.
Microarray analyses of heavy metal resistance gene expression. (i) Culture induction and RNA extraction.
Liquid cultures were grown at 30°C in cylindroconical flasks with 10 ml of 284 medium and shaken at 120 rpm on an orbital shaker until the optical density at 660 nm reached 0.3 (early exponential phase). Ten-milliliter samples of cultures were transferred into Falcon tubes, supplemented with the desired heavy metals [final concentrations, 0.4 mM Pb(II), 0.6 mM Ni(II), 0.6 mM Cd(II), 2 mM Co(II), 0.8 mM Zn(II), 5 μM Hg(II), and 0.1 mM Cu(II)], and then incubated further at 30°C for 30 min before centrifugation. Induction and RNA extraction were performed in duplicate using two independent cultures.
(ii) Spotting.
We used the draft genome sequence (November 2003 version) of C. metallidurans to design 60-mer aminosilane-modified oligonucleotide probes corresponding to 6,205 ORFs. The oligonucleotides were synthesized by Eurogentec S. A. (Liège, Belgium). They were suspended at the appropriate concentration in the spotting buffer (3× SSC [1× SSC is 0.15 M NaCl plus 0.15 M sodium citrate]) and spotted in duplicate onto a glass slide (UltraGAPS; Corning, United States) using a MicroGrid system (BioRobotics, United Kingdom). The spotted slides were cross-linked and placed in the presoaking solutions from a Pronto kit (Corning-Promega, United States).
(iii) Labeling and hybridization.
Total RNA was extracted using a QIAGEN RNeasy midi kit. RNAprotect (QIAGEN, United Kingdom) was added to the cultures to stop the synthesis of RNA and prevent its degradation. Extracted RNAs were stored at −80°C. The quality of total RNA was monitored using a BioAnalyzer 2100 (Agilent, United States). Ten micrograms of high-quality RNA was reverse transcribed by random priming according to the Pronto kit's instructions (Corning-Promega, United States) and labeled by incorporation of Cy3-dCTP or Cy5-dCTP nucleotides (Amersham Bioscience, United Kingdom). Labeled cDNA derived from the different bacterial cultures was resuspended in the universal hybridization buffer (Pronto kit; Corning-Promega, United States), mixed, and added to the spotted slide for overnight hybridization at 42°C. The slide was washed according to the Pronto kit's protocol.
(iv) Scanning and analysis.
The array was scanned with a laser scanner at 532 and 635 nm (Genepix 4100A; Axon, Instruments, The Netherlands). The image was analyzed with Genepix Pro software. Spot intensities were measured, and artifacts due to the circularity, shape, and background were removed. Statistical analyses were performed using “S-PLUS.” PrintTipLoess normalization was applied to the different slides. From the four values obtained for each condition (two technical assays and two biological assays) the mean average of ratios (red fluorescence/green fluorescence) was determined, and the P value was calculated using the SAM (significance analysis of microarrays) test. Genes with a P value of <0.05 and a degree of freedom of 3 were considered significant. Known metal resistance genes (based on transcriptomic and proteomic data and/or mutant analyses) were considered down- or up-regulated when the expression ratios were at least −2 or 2. For the numerous hypothetical or unknown genes, we focused mainly on those with expression ratios less than −3 and greater than 3.
The expression ratios for pMOL28 and pMOL30 are shown in Table S2 in the supplemental material.
Microarray accession number.
Array data have been deposited at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE7272.
Nucleotide sequence accession numbers.
The sequences obtained for plasmids pMOL28 and pMOL30 of C. metallidurans CH34 have been deposited in the GenBank database under accession numbers NC_007972 and NC_007971, respectively.
RESULTS
General features of plasmids pMOL28 and pMOL30.
Plasmids pMOL28 (171,459 bp) and pMOL30 (233,720 bp) have average G+C contents of 60.4 and 60.1%, respectively, while the chromosome and megaplasmid have an average G+C content of 63.0%. A total of 164 ORFs were identified on pMOL28, and 247 ORFs were identified on pMOL30; the percentages of coding sequences were 87% for pMOL28 and 80% for pMOL30. The annotation revealed genes involved in plasmid replication, stability, pilus biosynthesis, and conjugative transfer, as well as the presence of several mobile genetic elements and many hypothetical genes (conserved or unique). Detailed annotations are shown in Table S2 in the supplemental material and include, for every annotated ORF, the microarray expression data from cultures grown in the presence and in the absence of seven different metals.
Plasmid pMOL28. (i) pMOL28 shares a common core for basic plasmid functions with pHG1 (chemolithoautotrophy; Ralstonia eutropha H16) and pSym (legume symbiosis; Ralstonia taiwanensis).
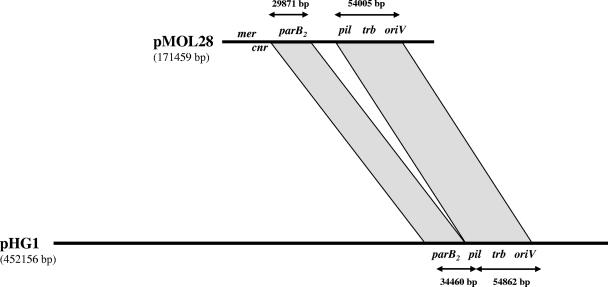
pMOL28 contains genes for DNA replication (39), maintenance, and plasmid transfer (trb and pil) (26, 33) (Fig. 1a) that are mostly syntenic and have a high level of identity with the corresponding genes of pHG1 (13, 38) (Fig. 2) and the pSym plasmid of R. taiwanensis LMG19424 (4; C. Masson, personal communication). Seventy-nine pMOL28 genes correspond to a continuous syntenic piece of pHG1, beginning with repA parB parA uvrD1, trb genes, pil genes, ssb, pri, and parB2 and ending with sbcD sbcC. Yet the level of similarity between the repA parB and parA orthologs is quite low (around 27%), while for most of the other genes of the synteny the level of similarity is around 80% (ssb, pri, and most trb genes). As shown in Fig. 1a, a 13-gene insertion in pMOL28 separates the sbcC-parB2 cluster from the ssb cluster. This insertion contains a xerD gene (encoding a site-specific recombinase) and ends with three rhs-like genes rich in YD motifs whose function is unknown (RHS elements are proteins having nonessential functions believed to play an important role in the cell's natural ecology [12]). The protein sequences include a highly conserved 141-kDa domain containing multiple tandem 22-residue repeats, followed by divergent C-terminal domains (12, 16). The 22-residue repeats contain a YD dipeptide, the most strongly conserved motif of the repeat.
FIG. 2.
Synteny between plasmid pMOL28 from C. metallidurans CH34 and pHG1 from R. eutropha H16. Plasmid pMOL28 from C. metallidurans CH34 and plasmid pHG1 from R. eutropha H16 display large regions of synteny. The syntenic region contains the pil genes (involved in pilus biosynthesis) (see Fig. S1 in the supplemental material), the trb genes (involved in conjugative plasmid transfer) (see Fig. S1 in the supplemental material), the origin of replication, and two IS elements. On pMOL28, mer cnr indicates the heavy metal resistance region. The syntenic analysis was performed using tools from MAGE (http://www.genoscope.cns.fr/agc/mage).
The organization of the cluster of pilus biosynthesis genes largely resembles that of the Salmonella enterica serovar Typhi plasmid R64 pil operon (34, 38). The region for conjugative gene transfer resembles the organization of the plasmid RP4 Tra2 region (38), although the level of protein sequence similarity between the putative orthologs is low. For pilus biosynthesis, pSym and pHG1 are completely syntenic, while pMOL28 lacks pilO and pilS. The organization of the three plasmids (pMOL28, pSym, and pHG1) indicates that there is a common family of elements having very different functions but having a common backbone with basic plasmid functions (Fig. 2).
Numerous data reflecting the effects of heavy metals on the regulation of the plasmid were obtained. pilQ (encoding a hexameric ATPase required for pilus assembly [38]) was up-regulated 4.35-fold in the presence of Cu(II); pilT (encoding a lytic transglycosylase involved in pilus biosynthesis [38]) was induced 6.77-, 10.4-, and 11.33-fold in the presence of Cd(II), Pb(II), and Zn(II), respectively; trbN (also encoding a lytic transglycosylase) was up-regulated 6.6-fold in the presence of Cu(II); trbI was up-regulated 3.51-fold in the presence of Ni(II); trbG (involved in mating pair formation) was up-regulated 6.36-fold in the presence of Pb(II); and trbJ (encoding an “entry exclusion protein” in other known plasmid transfer systems) was up-regulated 3.42-fold when Cd(II) was present.
(ii) Island with heavy metal resistance genes.
All the genetic determinants involved in heavy metal resistance in pMOL28 were grouped in a 34-kb region. The entire 42-kb region containing all pMOL28 heavy metal resistance genes is flanked by a partial IS element from the Tn3 family on the cnrT side and by IS1071 from the same Tn3 family on the other side (near merR). The latter element, IS1071 (10, 29), has a complex structure with a 21-bp deletion of its right inverted repeat sequence and a duplication of the internal end of its left extremity. Furthermore, the transposase gene of this element is disrupted by a conserved cluster of four genes: two genes encoding hypothetical proteins, a gspA gene encoding a protein in the type II secretion system, and a recombinase gene. Eleven other copies of this cluster occur in the genome, and one is inserted into the pbrU gene of pMOL30. This four-gene element may correspond to a new mobile element carrying a gene involved in transport or secretion and provisionally is designated TnRme2.
Besides the rearranged IS1071, plasmid pMOL28 contains one copy of IS1086 (11) and one copy of ISRme9 (see Table S1 in the supplemental material). The yacA and yacB genes, which are very similar to putative orthologs found in the IncPβ Β8 plasmid (37) coding for a toxin/antitoxin system, also were identified between the IS1071-based structure and merR. The function of these genes could be related to maintaining the stability of the genomic island rather than that of the plasmid.
(iii) Expression of pMOL28 metal resistance genes.
The entire cnr cluster was induced in the presence of Ni(II) and, surprisingly, also by Cu(II) and Cd(II). We noted induction of cnrX, cnrA, and cnrB after addition of Pb(II) and induction of cnrH with Zn(II). Yet we observed no involvement of the cnr cluster in resistance to Cu(II) or Pb(II), although cultures fully induced by Ni(II) or Co(II) displayed some resistance to Zn(II) and Cd(II) (5). However, it was found that insertion of IS elements into the regulatory gene cnrY or cnrX resulted in constitutive expression of cnrCBAT (5, 42) that led to very high resistance to Co(II) and Ni(II) and also to medium resistance to Zn(II).
Low induction of chrC1A1B1 was observed in the presence of Cu(II), and low induction of chrFEC1 was observed in the presence of Ni(II). The last gene of this cluster, chrI, which putatively is involved in regulating chr (18), was one of the very few genes that was down-regulated in the presence of Cd(II), Cu(II), Ni(II), or Pb(II). The protein encoded by chrI is predicted to be anchored to the inner cell membrane and has two lipoprotein domains, suggesting that it is a membrane-bound regulatory protein (like the proteins encoded by cnrY and cnrX) (42).
The merRTPADE genes involved in mercury resistance show 100% sequence identity between transposons Tn4378 and Tn4380. The tnpA and tnpR genes involved in transposition and the orf-2 gene have levels of sequence similarity of 69, 88, and 93%, respectively, to their Tn4380 counterparts. The divergence of tnpA, tnpR, and orf-2 in the two transposons has enabled the design of transposon-specific probes, although the induction levels for the mer genes cannot be discriminated. The mer genes were the only genes that were induced by mercury. However, they also were strongly up-regulated by Zn(II), Cd(II), and Pb(II). The genes involved in transposition (tnpA, tnpR, and orf-2) also were induced by Cd(II) and Pb(II).
(iv) Responses to heavy metals of hypothetical and unknown genes in pMOL28.
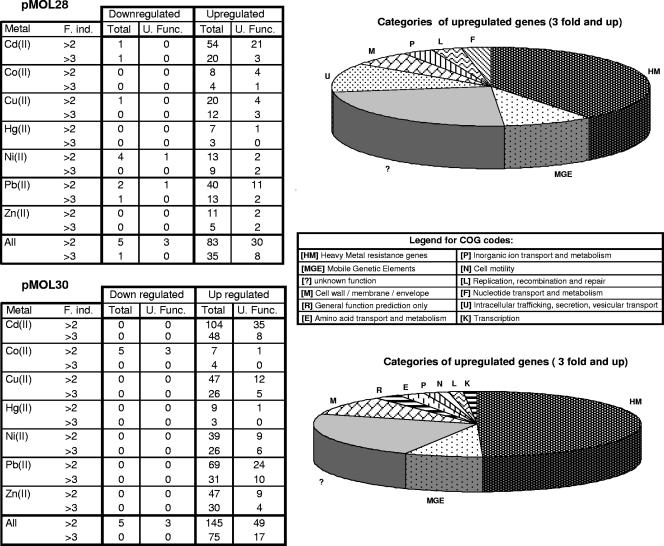
Besides the genes described above, 50 other genes were up-regulated more than twofold after exposure to a metal; 12 of these genes were up-regulated more than threefold (Fig. 3).
FIG. 3.
Numbers of down- and up-regulated genes on the two plasmids according to metal exposure. The tables summarize the numbers of down- and up-regulated genes with two thresholds: more than twofold induction and more than threefold induction. Results are given for both plasmids and for all metals tested [Cd(II), Co(II), Cu(II), Hg(II), Ni(II), Pb(II), and Zn(II)]. “All” represents the number of down- or up-regulated genes in the presence of at least one of the metals. The pie graphs display the proportion of genes up-regulated more than threefold as a function of the COG class. F.ind., fold induction; U.Func., unknown function.
Plasmid pMOL30. (i) Plasmid maintenance and replication genes.
The parA, parB, and repA genes (Fig. 1b) differ substantially (11 to 19% sequence similarity) from their pMOL28 orthologs. Plasmids pMOL28 and pMOL30 clearly belong to very different plasmid incompatibility (Inc) groups, as both of these plasmids are stably maintained in each other's presence. pMOL30 parAB are very similar to their equivalents in Burkholderia vietnamiensis G4, as well as to a helicase gene (uvrD). Other genes important for plasmid maintenance and mobility were found on a 15-kb fragment located ∼140 kb from the parAB genes, as follows: (i) stdB encodes an antitoxin protein, but so far the associated toxin gene has not been found; (ii) three genes, traW, traU, and trbC (orthologs of IncF plasmids), may be considered related to plasmid transfer (plasmid pMOL30 itself was observed to transfer at a very low frequency); and (iii) the dsbC gene encodes a disulfur bond isomerase located on pMOL30, which could be involved in the resistance to copper (17; M. Berkmen, personal communication), yet no metals have induced dsbC under any conditions.
(ii) Mobile genetic elements and genomic islands.
pMOL30 contains two copies of ISRme3 (the most represented IS element in the C. metallidurans genome, with 10 copies), one copy of ISRme10, and one copy of TnRme2 that we describe above for pMOL28. The TnRme2 copy is inserted in the pbrU gene. Between the czc and pbr operons, there is also a recombinase-rich region containing ISRme3, three genes encoding a Ser/Tyr recombinase, and nine ORFs from five truncated IS elements (Fig. 1b). pMOL30 contains 12 ORFs from truncated IS elements. Surprisingly, some of them were induced by heavy metals. orf-102 was induced by Zn(II) (7.3-fold), Cd(II) (20.1-fold), Cu(II) (2.4-fold), Ni(II) (5.9-fold), and Pb(II) (3.5-fold). orf-103 was induced by Zn(II) (2.4-fold), Cd(II) (3.6-fold), Ni(II) (2.5-fold), and Pb(II) (2-fold). And orf-157 and orf-158 were also induced by Cd(II).
IS3 elements contain two ORFs: one encoding a protein with a defined DNA-binding domain and one that encodes the catalytic site with characteristic aspartyl and glutamyl residues. orf-103 and orf-157 display a fairly intact DNA-binding motif, while the catalytic site is largely deleted in orf-102 and orf-158.
As in pMOL28, genes involved in heavy metal resistance seem to be concentrated in areas delimited by genes belonging to mobile genetic elements. The first region, containing the czc and pbr clusters, is flanked by the fully active mercury transposon Tn4380 on one side and on the other side by three mer genes that might be remnants of former rearrangements. Hypothetically, the mer transposon might have had a role in the final assembly of this island. The second region, containing the cop, sil, and nre-ncc clusters, is flanked by a complete ISRme10 element and a strongly ISRme10-related remnant (orf-157 and orf-158) with an extremity very similar to the ISRme10 inverted repeats; partial deletion of one of the IS elements might have stabilized this acquisition.
(iii) Updating pMOL30 heavy metal resistance genes and main features in differential gene expression in the presence of heavy metals. (a) czc region.
The entire czc cluster of genes was induced mainly by Cd(II) and Zn(II). The czcA, czcB, and czcC genes were the genes that were induced the most, with 13- to 26-fold induction. czcN and czcA also were induced by Cu(II), czcN and czcI also were induced by Ni(II), and czcN, czcI, czcC, and czcB also were induced by Pb(II). Furthermore, Pb(II)-mediated induction of czcR had been observed previously in a czcR::Tn4431 construction with luxEDCBA as reporting genes (7). Upstream of czcN, czcM, which was formerly called mgtC, exhibits protein sequence similarities with the ATPases involved in Mg(II) transport. Another paralog of czcM (with 28.9% similarity) was found on the C. metallidurans megaplasmid close to a czc-like cluster (31).
Downstream of czcDRSE, czcJ (D. Nies, personal communication) was strongly induced by Cd(II) (26.8-fold), Cu(II) (11.8-fold), Ni(II) (5.5-fold), Pb(II) (4.2-fold), and Zn(II) (22.8-fold); the CzcP P1-type ATPase was classified in the family of lead, cadmium, and zinc efflux P-type ATPases (24, 28). Quantitative PCR revealed induction of czcP (28; D. Nies, personal communication) by Zn(II), Cd(II), and Cu(II), in agreement with the microarray data.
czcJ has two paralogs, orf-167 (proposed designation, mmrQ, for “multiple metal response”) and copQ. These three genes encoding proteins having functions that are still unknown with a signal peptide were highly induced (between 3- and 77-fold) in the presence of all the metals that we tested except Hg(II). The corresponding proteins have a common motif, RDXXTDG, repeated three times. The upstream regions of czcJ and mmrQ are very similar, suggesting that they have a common promoter.
(b) New putative gene in the pbr operon.
Only one ORF, pbrB/C, codes for functional PbrB and PbrC. An additional gene, pbrU (likely comprised of orf-125 and orf-130), encoding a putative permease belonging to the major facilitator family (MFS1) and predicted to be located in the inner membrane, should be included in the pbr cluster (although it is knocked out as described above). Examination of the upstream sequence showed the presence of a promoter (pbrUP; GTGAATTCATTCGAGCGCCCGATTTAAC) that is very similar to the promoters of pbrA and pbrR (GTGTATTCATCTCGCGTTGCCGATTTAAC) (identical residues are underlined).
The complete pbr cluster (except pbrR) was up-regulated in the presence of Pb(II) (with major induction of pbrA [7.3-fold]) and Cd(II) (see Table S2 in the supplemental material). The truncated pbrU gene had the same expression pattern as the rest of the pbr cluster, with induction by Pb(II) and Cd(II).
(c) Tn4380 and its mer operon.
The tnpA gene (encoding a transposase) is induced 2.15-fold by Cd(II), whereas tnpR (encoding a resolvase) is induced by Zn(II) (2.4-fold), Cd(II) (6.89-fold), Pb(II) (4.38-fold), and Hg(II) (2.41-fold). urf-2 (putative diguanylate phosphodiesterase gene) was induced when preparations were exposed to Cd(II) (2.73-fold) and Pb(II) (2.75-fold). The expression of the merRTPADE genes is discussed above.
On pMOL30, an incomplete mer cluster (merRTP) was discerned (Fig. 1b) (the region 65 to 67 kb from parA). Its genes encoded proteins that showed between 40 and 50% protein similarity with their Tn4378 and Tn4380 counterparts. They were not induced under any of the experimental conditions and likely are nonfunctional. We suggest that these genes are a remnant of the rearrangements, having generated the czc-pbr-mer genomic island.
(d) Silent and defective ncc-nre cluster.
No Ni(II) resistance was shown to be carried by plasmid pMOL30. The presence of a frameshift covering 28 bp into nccB suggests that the ncc genes probably are not functional. These genes were not induced by heavy metals, confirming previous observations (31).
(e) How extended is the cop region?
The 19 cop genes of pMOL30 are highly induced by Cu(II) (from 2.2- to 21.6-fold), confirming previous data (27). They are also induced by Zn(II), Cd(II), and Ni(II), while Pb(II) induces copF, copR1, copS1, copK, copM, and copT. Microarray data also indicate that the cluster includes some of the neighboring genes (especially outside copE) (see Table S2 in the supplemental material). The larger cluster of copper response genes would contain 33 genes, from copV to ubiE (encoding a putative methyltransferase), including the cop, gtr-2, and sil genes (Fig. 1b). The cop and sil genes encode the efflux and resistance mechanisms required for detoxifying the cytoplasm and periplasm. Equivalents of the silCBA genes were found on the megaplasmid of R. solanacearum (24). Proteomic data revealed that SilB and SilC were overexpressed in the presence of Cu(II) and Ag(I) (24).
The gtr-2 cluster is required for lipopolysaccharide exposition on the outer membrane (20).
(iv) Response of pMOL30 hypothetical and unknown genes to metal exposure.
Besides the genes involved in heavy metal resistance, 85 genes were up-regulated more than twofold, and 32 of them were up-regulated more than threefold. In the latter group, the functions of 17 genes are unknown, and 10 of them are unique to C. metallidurans according to the current databases (Fig. 3) [e.g., orf-231, located in the island flanked by ISRme10 and its putative remnant, is up-regulated when preparations are treated with Zn(II) (8.3-fold), Cd(II) (2.1-fold), Cu(II) (5.6-fold), and Ni(II) (6.8-fold) (Fig. 1b)].
DISCUSSION
Our annotation of plasmids pMOL28 and pMOL30 confirmed all previously known heavy metal resistance clusters (24), identified a few additional ORFs within these clusters, and described plasmid maintenance and transfer genes, as well as IS elements. However, the function of most of the newly described genes remains hypothetical or is unknown. Although these findings may suggest that these ORFs do not have any real function and therefore might be considered evolutionary remnants, microarray data has shed more light on their possible role because they are over- or underexpressed in the presence of heavy metals.
Plasmid pMOL28 shares a common set of genes with plasmids pSym of R. taiwanensis and pHG1 of R. eutropha H16 (Fig. 2). This common set contains orthologs for basic plasmid functions in a ∼90-kb backbone indicative of a common origin. This situation resembles the genesis of IncPβ plasmids, which also contain a conserved region involved in plasmid maintenance next to a variety of functional modules concerned with resistance to antibiotics, mercury, or xenobiotics or combinations thereof (8, 41). Plasmid pMOL30, in contrast, belongs to another plasmid family; parA, parB, and some other closely linked genes of pMOL30 are highly similar to B. vietnamiensis G4 equivalents.
Besides the determinants for metal resistance, other plasmid-borne genes were strongly induced by heavy metals, and some of these genes are involved in conjugative transfer (such as pMOL28 trbN, trbI, trbG, and trbJ), transposition (Tn4378 and Tn4380), and membrane maintenance. Of particular interest for the latter function are the putative glycosyltransferase genes that are situated between known metal resistance loci and that appear to be organized as they are in Shigella flexneri (20). Their products may play a role in maintaining the integrity of the cell wall. Thus, the induction of the gtr gene clusters in the presence of high concentrations of heavy metals suggests that such exposure may heighten the demands for membrane biogenesis and restoration of the outer membrane lipopolysaccharides.
Some truncated IS elements also were overexpressed during exposure to heavy metals. The fact that the metal-induced partial IS elements encode only the DNA-binding domain opens the possibility that their corresponding proteins could have evolved to serve another function unrelated to gene mobility but possibly related to regulation. Interestingly, other researchers suggested that some eukaryotic mobile genetic elements might have evolved in such a way (44).
An intriguing feature of the microarray data was the multiple-metal responses exhibited by several genes belonging to the cop, cnr, mer, and pbr loci. These genes inevitably were induced by the expected substrate and also by other metals. Most notably, mer genes were activated in response to Cd(II) and Pb(II) and cop genes were activated in response to Zn(II), Cd(II), and Ni(II), while the expression of the cnr genes rose in response to Cu(II) and Cd(II), usually by the same order of magnitude irrespective of which metal was tested. These observations contrast with the more specific metal responses that previously were noted with gene fusions (biosensors) (6, 7, 32, 42, 43). The responses of these gene fusions, made with pMOL28 and pMOL30 as various in vivo and in vitro constructions, mostly relied on bioluminescence. Indeed, they were elicited mostly by the metallic substrates of the corresponding resistance proteins (6, 7, 32, 42, 43). This apparent contradiction may mainly reflect the differences in the timing of the responses to metal induction; thus, the luminescent responses of the gene fusions are observed only after a couple of hours, while the results for quantitative PCR or microarrays correspond to a 30-min pulse. Nonetheless, data reported for cnr genes and cnr-lac fusions (32) are consistent with the concept of a multiple-metal response [e.g., up-regulation of cnr genes in the presence of Cu(II), although this metal is not a substrate of cnr] and experimental detection after short exposure times (10 min) (32), followed by a more specific response [up-regulation in the presence of Ni(II) or Co(II), which are the main substrates of cnr] when cells are exposed to the metals for longer times (32).
Thus, multiple-metal responses could be transient phases in global resistance to heavy metals, with an early stage of multimetallic up-regulation of the metal resistance genes followed by a more substrate-specific response directed towards a particular metal. These apparent different phases involved in the response and resistance to heavy metals need to be investigated further by detailed kinetic studies during exposure. Recently, such an analysis was performed with copper-induced Pseudomonas aeruginosa cultures (40). For now, we hypothesize that such layered multiple responses would mean that the heavy metal resistance genes are controlled by various regulatory pathways with different regulators, each responding to several metals. The genes for these regulatory circuits could be located on the two plasmids, but they could also be located on the larger replicons.
Alternatively, highly specific sigma factors may be involved. Currently, 11 sigma factors have been recognized in C. metallidurans CH34, half of which are up- or down-regulated in the presence of heavy metals; furthermore, deleting five of these factors caused a decrease in heavy metal resistance (D. Nies, personal communication). The only sigma factor encoded on a plasmid (pMOL28), cnrH, is induced in the presence of Cu(II), Cd(II), Co(II), or Ni(II) (D. Nies, personal communication; this study). The cellular defense mechanisms of C. metallidurans against heavy metals might encompass several stages, implying a response to various signals, some of which are distinct from the substrates of the detoxification genes, with the corresponding genes located on the various replicons.
In both plasmids, mobile genetic elements may have participated in acquiring genes involved in heavy metal resistance. In pMOL28, the metal resistance island is flanked by inactivated IS elements belonging to the IS3 family (IS1071).
In pMOL30, the metal resistance genes are grouped in two putative islands separated by a small 13-kb region that contains some tra genes that might have belonged to the pMOL30 backbone before the acquisition of the “islands” (Fig. 1b) (region 130 to 140 kb from parA). These islands, as well as the pMOL28 island, do not seem to be mobile, which might reflect ancient acquisition.
For all the heavy metals tested, microarrays showed that the genes most up-regulated were located on the two plasmids. But many chromosomal genes and genes on the megaplasmid also are up-regulated. It would be interesting to compare these responses with the microarray data reported previously for Escherichia coli (19) and for P. aeruginosa (40) after exposure to copper. These microarray data (and likely the data for the CH34 chromosome and the megaplasmid) give an overview of possible microbial responses to moderate to high concentrations of heavy metals. pMOL28 and pMOL30 gene expression data probably describe the bacterial reaction to the most acute viable metallic stress for mesophilic heterotrophs growing at neutral pH. Classic metal resistance genes are only part of the gene arsenal on which bacterial survival depends. We expect that the transcriptomic data from this study will be a stepping stone for additional research on the unknown and hypothetical genes in the form of proteomics, mutagenesis, and phenotypic analyses.
Supplementary Material
Acknowledgments
Sequence analysis of the C. metallidurans genome was carried out by the Joint Genome Institute (JGI) under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program, by the University of California Lawrence Livermore National Laboratory under contract W-7405-Eng-48, by Lawrence Berkeley National Laboratory under contract DE-AC03-76SF00098, and by Los Alamos National Laboratory under contract W-7405-ENG-36. S.M. is a research assistant at SCK · CEN. D.V.D.L. and S.T. are supported by laboratory-directed research and development funds at the Brookhaven National Laboratory under contract with the U.S. Department of Energy and by the U.S. Department of Energy Biological and Environmental Research (ESRP) Program under contract DE-AC02-98CH10886. This work was partially supported by ESA and Federal Belgian Scientific Policy under PRODEX contracts 90037, 90094, and 90150.
We especially thank Alla Lapidus and her team at JGI. We thank Jacques Mahillon for suggestions about the data on mobile genetic elements, Ariane Toussaint and Raphael Leplae for critically reading the manuscript, Catherine Masson, Mehmet Berkman, and Jon Hobman for exchange of information, Claudine Medigue for access to MAGE tools, and Avril Woodhead for correcting the language. The skillful technical assistance of Albert Bossus, Arlette Michaux, Ann Provoost, and Werner Schoonjans is gratefully acknowledged.
Footnotes
Published ahead of print on 3 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Borremans, B., J. L. Hobman, A. Provoost, N. L. Brown, and D. van Der Lelie. 2001. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J. Bacteriol. 183:5651-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brim, H., M. Heyndrickx, P. de Vos, A. Wilmotte, D. Springael, H. Schlegel, and M. Mergeay. 1999. Amplified rDNA restriction analysis and further genotypic characterisation of metal-resistant soil bacteria and related facultative hydrogenotrophs. Syst. Appl. Microbiol. 22:258-268. [DOI] [PubMed] [Google Scholar]
- 3.Cha, J. S., and D. A. Cooksey. 1991. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. USA 88:8915-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, W. M., L. Moulin, C. Bontemps, P. Vandamme, G. Bena, and C. Boivin-Masson. 2003. Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J. Bacteriol. 185:7266-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collard, J. M., A. Provoost, S. Taghavi, and M. Mergeay. 1993. A new type of Alcaligenes eutrophus CH34 zinc resistance generated by mutations affecting regulation of the cnr cobalt-nickel resistance system. J. Bacteriol. 175:779-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbisier, P. 1997. Bacterial metal-lux biosensors for a rapid determination of the heavy metal bioavailability and toxicity in solid samples. Res. Microbiol. 148:534-536. [DOI] [PubMed] [Google Scholar]
- 7.Corbisier, P., D. van der Lelie, B. Borremans, A. Provoost, V. de Lorenzo, N. L. Brown, J. R. Lloyd, J. L. Hobman, E. Csoregi, G. Johansson, and B. Mattiasson. 1999. Whole cell- and protein-based biosensors for the detection of bioavailable heavy metals in environmental samples. Anal. Chim. Acta 387:235-244. [Google Scholar]
- 8.Dennis, J. J. 2005. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16:291-298. [DOI] [PubMed] [Google Scholar]
- 9.Diels, L., M. Faelen, M. Mergeay, and D. Nies. 1985. Mercury transposons from plasmids governing multiple resistance to heavy metals in Alcaligenes eutrophus CH34. Arch. Intern. Physiol. Biochem. 93:27-28. [Google Scholar]
- 10.Di Gioia, D., M. Peel, F. Fava, and R. C. Wyndham. 1998. Structures of homologous composite transposons carrying cbaABC genes from Europe and North America. Appl. Environ Microbiol. 64:1940-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong, Q., A. Sadouk, D. van der Lelie, S. Taghavi, A. Ferhat, J. M. Nuyten, B. Borremans, M. Mergeay, and A. Toussaint. 1992. Cloning and sequencing of IS1086, an Alcaligenes eutrophus insertion element related to IS30 and IS4351. J Bacteriol. 174:8133-8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feulner, G., J. A. Gray, J. A. Kirschman, A. F. Lehner, A. B. Sadosky, D. A. Vlazny, J. Zhang, S. Zhao, and C. W. Hill. 1990. Structure of the rhsA locus from Escherichia coli K-12 and comparison of rhsA with other members of the rhs multigene family. J. Bacteriol. 172:446-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich, B., C. Hogrefe, and H. G. Schlegel. 1981. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J. Bacteriol. 147:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghai, R., T. Hain, and T. Chakraborty. 2004. GenomeViz: visualizing microbial genomes. BMC Bioinformatics 5:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goris, J., P. De Vos, T. Coenye, B. Hoste, D. Janssens, H. Brim, L. Diels, M. Mergeay, K. Kersters, and P. Vandamme. 2001. Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int. J. Syst. Evol. Microbiol. 51:1773-1782. [DOI] [PubMed] [Google Scholar]
- 16.Hill, C. W., C. H. Sandt, and D. A. Vlazny. 1994. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol. Microbiol. 12:865-871. [DOI] [PubMed] [Google Scholar]
- 17.Hiniker, A., J. F. Collet, and J. C. Bardwell. 2005. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J. Biol. Chem. 280:33785-33791. [DOI] [PubMed] [Google Scholar]
- 18.Juhnke, S., N. Peitzsch, N. Hubener, C. Grosse, and D. H. Nies. 2002. New genes involved in chromate resistance in Ralstonia metallidurans strain CH34. Arch. Microbiol. 179:15-25. [DOI] [PubMed] [Google Scholar]
- 19.Kershaw, C. J., N. L. Brown, C. Constantinidou, M. D. Patel, and J. L. Hobman. 2005. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 151:1187-1198. [DOI] [PubMed] [Google Scholar]
- 20.Korres, H., M. Mavris, R. Morona, P. A. Manning, and N. K. Verma. 2005. Topological analysis of GtrA and GtrB proteins encoded by the serotype-converting cassette of Shigella flexneri. Biochem. Biophys. Res. Commun. 328:1252-1260. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. M., G. Grass, C. Rensing, S. R. Barrett, C. J. Yates, J. V. Stoyanov, and N. L. Brown. 2002. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 295:616-620. [DOI] [PubMed] [Google Scholar]
- 22.Liesegang, H., K. Lemke, R. A. Siddiqui, and H. G. Schlegel. 1993. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J. Bacteriol. 175:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mergeay, M., C. Houba, and J. Gerits. 1978. Extrachromosomal inheritance controlling resistance to cadmium, cobalt, copper and zinc ions: evidence from curing in a Pseudomonas. Arch. Int. Physiol. Biochim. 86:440-442. [PubMed] [Google Scholar]
- 24.Mergeay, M., S. Monchy, T. Vallaeys, V. Auquier, A. Benotmane, P. Bertin, S. Taghavi, J. Dunn, D. van der Lelie, and R. Wattiez. 2003. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27:385-410. [DOI] [PubMed] [Google Scholar]
- 25.Mergeay, M., D. Nies, H. G. Schlegel, J. Gerits, P. Charles, and F. Van Gijsegem. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merryweather, A., C. E. Rees, N. M. Smith, and B. M. Wilkins. 1986. Role of sog polypeptides specified by plasmid ColIb-P9 and their transfer between conjugating bacteria. EMBO J. 5:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monchy, S., M. A. Benotmane, R. Wattiez, S. van Aelst, V. Auquier, B. Borremans, M. Mergeay, S. Taghavi, D. van der Lelie, and T. Vallaeys. 2006. Transcriptomic and proteomic analyses of the pMOL30-encoded copper resistance in Cupriavidus metallidurans strain CH34. Microbiology 152:1765-1776. [DOI] [PubMed] [Google Scholar]
- 28.Monchy, S., T. Vallaeys, A. Bossus, and M. Mergeay. 2006. Metal efflux P1-ATPase genes of Cupriavidus metallidurans CH34: a transcriptomic approach. Int. J. Environ. Anal. Chem. 86:677-692. [Google Scholar]
- 29.Nakatsu, C., J. Ng, R. Singh, N. Straus, and C. Wyndham. 1991. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc. Natl. Acad. Sci. USA 88:8312-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nies, D., M. Mergeay, B. Friedrich, and H. G. Schlegel. 1987. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J. Bacteriol. 169:4865-4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 32.Nies, D. H., G. Rehbein, T. Hoffmann, C. Baumann, and C. Grosse. 2006. Paralogs of genes encoding metal resistance proteins in Cupriavidus metallidurans strain CH34. J. Mol. Microbiol. Biotechnol. 11:82-93. [DOI] [PubMed] [Google Scholar]
- 33.Rees, C. E., and B. M. Wilkins. 1989. Transfer of Tra proteins into the recipient cell during bacterial conjugation mediated by plasmid ColIb-P9. J. Bacteriol. 171:3152-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakai, D., and T. Komano. 2002. Genes required for plasmid R64 thin-pilus biogenesis: identification and localization of products of the pilK, pilM, pilO, pilP, pilR, and pilT genes. J. Bacteriol. 184:444-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 36.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies. Arch. Mikrobiol. 38:209-222. [PubMed] [Google Scholar]
- 37.Schluter, A., H. Heuer, R. Szczepanowski, S. M. Poler, S. Schneiker, A. Puhler, and E. M. Top. 2005. Plasmid pB8 is closely related to the prototype IncP-1beta plasmid R751 but transfers poorly to Escherichia coli and carries a new transposon encoding a small multidrug resistance efflux protein. Plasmid 54:135-148. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz, E., A. Henne, R. Cramm, T. Eitinger, B. Friedrich, and G. Gottschalk. 2003. Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H(2)-based lithoautotrophy and anaerobiosis. J. Mol. Biol. 332:369-383. [DOI] [PubMed] [Google Scholar]
- 39.Taghavi, S., A. Provoost, M. Mergeay, and D. van der Lelie. 1996. Identification of a partition and replication region in the Alcaligenes eutrophus megaplasmid pMOL28. Mol. Gen. Genet. 250:169-179. [DOI] [PubMed] [Google Scholar]
- 40.Teitzel, G. M., A. Geddie, S. K. De Long, M. J. Kirisits, M. Whiteley, and M. R. Parsek. 2006. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 188:7242-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPbeta plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 42.Tibazarwa, C., S. Wuertz, M. Mergeay, L. Wyns, and D. van Der Lelie. 2000. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J. Bacteriol. 182:1399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Lelie, D., P. Corbisier, W. Baeyens, S. Wuertz, L. Diels, and M. Mergeay. 1994. The use of biosensors for environmental monitoring. Res. Microbiol. 145:67-74. [DOI] [PubMed] [Google Scholar]
- 44.Volff, J. N. 2006. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28:913-922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.