Abstract
A DNA binding protein, BldR, was identified in the crenarchaeon Sulfolobus solfataricus as a protein 5- to 10-fold more abundant in cells grown in the presence of toxic aldehydes; it binds to regulatory sequences located upstream of an alcohol dehydrogenase gene (Sso2536). BldR is homologous to bacterial representatives of the MarR (multiple antibiotic resistance) family of transcriptional regulators that mediate response to multiple environmental stresses. Transcriptional analysis revealed that the bldR gene was transcribed in a bicistronic unit composed of the genes encoding the transcriptional regulator (Sso1352) and a putative multidrug transporter (Sso1351) upstream. By homology to bacterial counterparts, the bicistron was named the mar-like operon. The level of mar-like operon expression was found to be increased at least 10-fold in response to chemical stress by aromatic aldehydes. Under the same growth conditions, similar enhanced in vivo levels of Sso2536 gene transcript were also measured. The gene encoding BldR was expressed in E. coli, and the recombinant protein was purified to homogeneity. DNA binding assays demonstrated that the protein is indeed a transcription factor able to recognize site specifically both the Sso2536 and mar-like promoters at sites containing palindromic consensus sequences. Benzaldehyde, the substrate of ADHSs, stimulates DNA binding of BldR at both promoters. The role of BldR in the auto-activation as well as in the regulation of the Sso2536 gene, together with results of increased operon and gene expression under conditions of exposure to aromatic aldehydes, indicates a novel coordinate regulatory mechanism in cell defense against stress by aromatic compounds.
A chimeric nature of the transcription machinery with eukaryote-like basal factors and bacterium-like regulative components has been found in all members of the domain Archaea (50, 60). In fact, in most cases, homologs of bacterial regulators function in the context of the archaeal basal transcriptional apparatus, which resembles that of Eukarya (8, 26). Transcription initiation is mediated by a single RNA polymerase (RNAP) and two general transcription factors, TATA element binding protein (TBP) and transcription factor B (TFB), a homologue of the transcription factor TFIIB, which binds to the B recognition element (BRE) and determines the directionality of the transcription (7). The complex containing RNAP, TBP, and TFB is sufficient to initiate transcription in cell-free systems (31, 49), although an additional factor, transcription factor E, is required to increase transcription from some promoters (6, 29).
A few homologs of eukaryal transcriptional regulators (33) and unique archaeal regulators (57) as well as non-sequence-specific DNA binding proteins (5, 30) have been investigated for their contribution to the regulation of archaeal genes.
Homologs of bacterial transcriptional regulators are more common in Archaea, and representatives that have been studied experimentally include the Lrp homologs (13) that can function as autorepressors and/or activators of both their own and different genes. Binding sites for the Lrp-like factors can be located upstream of the TATA box, sometimes overlapping it, or downstream and encompassing the transcriptional start site (4, 44, 45, 47). Depending on the position, activation or inhibition of the different steps of the transcriptional initiation process has been established (48, 53).
However, how bacterium-like transcriptional regulators positively regulate the eukaryote-like basal transcription apparatus still remains an intriguing question. To date, the few studies reported highlight at least two different molecular mechanisms of transcriptional activation in Archaea (32, 45, 53).
The role of repressor and the interaction with specific effectors has been more thoroughly investigated in cases such as, for example, the expression control of the genes in the operon involved in nitrogen fixation in the euryarchaeon Methanococcus maripaludis (36) and in the operon of trehalose/maltose ABC transport in the crenarchaeon Thermococcus litoralis (34, 35). The negative control of transcription generally involves DNA binding proteins that bind or release target promoter DNAs in response to signaling ligands and modulate transcription by competing for the binding sites of TBP/TFB or RNAP (26).
We have chosen the gene encoding an alcohol dehydrogenase (ADH) in the archaeon Sulfolobus solfataricus, adhSs (14, 27), annotated on the genome as Sso2536 (52), as a model of transcriptional regulation in Archaea. Multiple ADHs have been found in single members of the three domains of life as generally encoded by distinct genes (10), their expression being regulated at both the transcriptional and posttranscriptional levels (24, 56). In S. solfataricus, besides Sso2536, 12 additional open reading frames (ORFs) encoding putative ADHs have been identified and annotated (adh-1 to -13). All but adh-9 are actively expressed, as demonstrated by proteome analyses (17, 18). The presence of multiple ADH ORFs indicates a metabolic relevance of alcohol metabolism by this organism (16). Nevertheless, recent investigations have attributed an important role to ADHs in defense against different forms of stresses, such as those arising in extreme environments (20).
In a previous study, we have demonstrated that Sso2536 gene expression is transcriptionally up-regulated in response to the specific substrate of the enzyme, benzaldehyde, when added to the growth medium (15). This molecule is able to block cell growth at concentrations higher than 2 mM. Further studies have proposed that this response could be mediated by a sequence-specific DNA binding protein that is increased intracellularly when cells are exposed to the stress source. This protein, named Bald, was purified to homogeneity for its ability to bind the 5′ flanking region of the Sso2536 gene at a specific regulatory sequence made of two adjacent inverted repeats localized immediately upstream of the BRE sequence (25). Moreover, the 5′ flanking region of the Sso2536 gene has been demonstrated to drive the heterologous expression of a bacillar reporter gene in S. solfataricus, maintaining in vivo inducibility by benzaldehyde in gene fusion (19).
Prokaryotic transcriptional regulators involved in the catabolism of aromatic compounds frequently belong to the MarR family, also including transcription factors involved in the modulation of multiple response to other toxic molecules, such as antibiotics (2). The main feature of this family is the winged helix DNA binding motif. Although the majority of the members of this family have been characterized as transcriptional repressors, examples of transcriptional activators (BadR) have also been described (22, 40). In Escherichia coli and other bacteria, the genes encoding MarR and its homologs are generally located in operons and share similar overall genetic organizations. As an example, the MexR protein isolated from Pseudomonas aeruginosa is a repressor of the MexAB-OprM operon, which encodes a tripartite multidrug efflux system (1). In general, the mar loci mediate a global stress response involving a large network of genes (3, 39, 51), and the same operon contributes to ethanol tolerance in engineered ethanologenic E. coli cells (28).
Phenolic ligands have been shown to regulate the expression of specific genes in both Eukarya and Bacteria (59). In most cases, like that of the mesophilic bacterium Deinococcus radiodurans, this influence can be exerted through MarR homologs by affecting their interaction with the target promoter regions (58). A very recent study reported on the three-dimensional structure of an archaeal homologue of the MarR repressor, but no information on protein function has been yet provided (41).
In this study, we extended our investigation on the regulative roles of the Bald protein, which we renamed BldR, in the response to stress caused by aromatic aldehydes. Sequence analysis revealed that BldR is homologous to members of the MarR family. Transcriptional and translational analysis demonstrated that BldR is positively autoregulated by the addition of aromatic aldehydes to the growth medium; an analogous trend was observed also for Sso2536 gene expression; as in the cases of other MarR representatives, BldR up-regulation involved also the cotranscribed gene encoding a putative drug efflux transporter, Sso1351 (37). Recombinant BldR was able to bind to both its own and Sso2536 gene regulatory sequences.
MATERIALS AND METHODS
S. solfataricus cultivation and preparation of crude extracts and total RNA.
S. solfataricus P2 was grown aerobically at 82°C in 100 ml of DSM 182 medium containing Brock's salts supplemented with 0.1% (wt/vol) yeast extract and 0.1% (wt/vol) Casamino Acids (12) and buffered at pH 3.5. In some cases, benzaldehyde, cynnamaldehyde, and veratrylaldehyde were added to final concentrations of 1 mM, 0.35 mM, and 1 mM, respectively, after dilution of an exponentially growing culture up to an A600 of 0.08 optical density (OD) units. Cells were grown to mid-logarithmic phase, corresponding to about 0.3 OD600 units, and harvested by centrifugation at 4,000 × g for 10 min. Crude extracts and total RNA were prepared, following previously reported procedures (15, 25).
Northern blot analysis.
RNAs (10 μg) extracted from cells grown under different conditions were electrophoretically separated in 1.5% agarose gel containing 10% formaldehyde and transferred to nylon filters (Amersham Biosciences). Hybridization was carried out as described by Cannio et al. (15), alternately using the Sso1352, Sso1351, Sso2536, and Sso7d genes as probes. Signals were visualized by autoradiography and quantified with a densitometric analysis using a Personal Fx phosphorimager and Quantity One software (Bio-Rad).
PCR amplification of the mar-like promoter region.
The region upstream of the ORF Sso1351 defined as the mar-like promoter was amplified with the primer pair Marfw (5′-CTATTGGATCGATGGGTTGC-3′) and Marrv (5′-GGCAACCCATTTGTAATGCATA-3′) by PCR amplification on S. solfataricus P2 genomic DNA and cloned in the pUC19 vector. The Marfw primer anneals starting at position −271 with respect to the translation start codon of the Sso1351 gene and contains the recognition sequence for the enzyme ClaI (underlined). Marrv anneals downstream in the proximity of the ATG start codon and contains a recognition sequence for the enzyme NsiI (underlined). The insertion and correct sequence of the PCR product were verified by DNA sequencing.
Primer extension assay.
To determine the first transcribed nucleotide, total RNA extracted from cells grown in the presence or in the absence of benzaldehyde and harvested at 0.3 OD600 units was subjected to primer extension analysis as described in Limauro et al. (38), using the primer 5′-GGTACTGAAATGAGGTAAAGGGG-3′, annealing from position +160 to position +139 in the Sso1351 gene. The same primer was used to produce a sequence ladder by using a Sequenase version 2.0 sequencing kit (Amersham) according to the manufacturer instructions to locate the products on 6% urea-polyacrylamide gel.
Heterologous expression of Sso1352 and purification of the recombinant protein.
The gene encoding Sso1352 from S. solfataricus P2 was amplified by PCR from genomic DNA, using Pfx DNA polymerase and the oligonucleotides Baldup (5′CAAAAAATAGATGAAAAACTCCAATTAA3′) and Balddw (5′-CAT TAC ATT GGG ATC CCT AGT CC-3′). Baldup was the phosphorylated 5′ primer starting with the second Sso1352 translation codon, while the Balddw primer contained the stop codon and introduced the restriction site PstI (underlined in the sequence). Amplified fragments were purified, digested with appropriate restriction enzymes, and cloned in the NcoI-filled/PstI-digested pTrc99A vector. The sequence of the cloned fragment was shown to be identical to the original annotated sequence available on the S. solfataricus P2 genome (http://www-archbac.u-psud.fr/projects/sulfolobus/). Homology comparison and multiple alignment of the BldR protein were performed using the BLAST and ClustalW programs, respectively, available on the Internet.
E. coli RB791 cells (11) transformed with pTrcBldR were grown in 1 liter of LB medium containing ampicillin (100 μg/ml). When the culture reached an A600 of 1.0 OD units, protein expression was induced by the addition of 1 mM isopropyl-1-thio-β-d-galactopyranoside and the bacterial culture grown for additional 16 h. Cells were harvested by centrifugation, and pellets were lysed by sonication in 30 ml of lysis buffer (50 mM Tris-HCl [pH 7.0], 1 mM phenylmethylsulfonyl fluoride) in an ultrasonic liquid processor (Heat system Ultrasonic Inc.). The lysate was centrifuged at 30,000 × g for 60 min (SW41 rotor; Beckman). The supernatant was heated to 70°C for 5 min, and denatured proteins were precipitated by centrifugation at 20,000 × g for 20 min at 4°C.
The supernatant was loaded onto a Resource S column (1 ml; Amersham Biosciences) connected to a fast-performance liquid chromatography system and preequilibrated in 50 mM Tris-HCl (pH 7.0) (buffer A). After a washing step with buffer A, the elution was carried out with 20 ml of a KCl gradient (0 to 0.4 M). Fractions were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to detect the BldR protein. These fractions were pooled, concentrated by ultrafiltration using a YM10 membrane (Millipore), dialyzed against buffer A, and loaded onto a heparin column (5 ml; HiTrap heparin; Amersham Biosciences) preequilibrated in the same buffer. After a washing step with buffer A, the elution was carried out with 40 ml of a KCl gradient (0 to 0.8 M). Fractions containing the BldR protein were pooled, concentrated, dialyzed, and stored at 4°C.
Analytical methods for protein characterization.
Protein concentration was determined by the method of Bradford (9), using the Bio-Rad protein staining assay and bovine serum albumin as the standard.
Protein homogeneity was estimated by SDS-PAGE (12.5% [wt/vol] gels). To determine the native molecular mass of BldR, the purified protein was applied to an analytical Superdex PC75 column (0.3 cm × 3.2 cm) connected to the AKTA Explorer system (Amersham Biosciences) and equilibrated in buffer B (50 mM Tris-HCl, pH 8, 0.2 M KCl). The column was calibrated using a set of gel filtration markers (low range; Amersham), including bovine serum albumin (67.0 kDa), ovalbumin (43.0 kDa), chymotrypsinogen A (25.0 kDa), and RNase A (13.7 kDa).
The molecular mass of the protein was also estimated using electrospray mass spectra recorded on a Bio-Q triple quadrupole instrument (Micromass, Thermofinnigan, San Jose, CA).
Western blot analysis.
Extracts prepared from cells grown under different conditions (10 μg) were subjected to SDS-PAGE, electroblotted onto polyvinylidene difluoride membranes, and detected immunologically using rabbit polyclonal antisera raised against the BldR protein (Igtech, Paestum, Salerno, Italy). Antigen-antibody interactions were detected with horseradish peroxidase-conjugated secondary antibodies and an enhanced chemiluminescence kit (Amersham Bioscience). To determine the relative abundances of the BldR protein in extracts prepared from cells grown in the presence of the different aldehydes, aliquots of the extracts were run on SDS-PAGE gel together with increasing amounts of purified recombinant BldR before being subjected to the same procedure. The relative amount of BldR was quantified using a Gel-Doc phosphorimager and Quantity One software (Bio-Rad).
EMSA.
Binding of BldR protein to target DNA sequences was measured by an electrophoretic mobility shift assay (EMSA) using different DNA probes. DNA containing the Sso2536 promoter was purified in gel after digestion with EcoRI endonuclease from the pUC18Ssadh promoter plasmid (15); DNA containing the mar-like promoter region was recovered by digestion of the recombinant pUC19 plasmid with NsiI-ClaI. DNA containing the SOD promoter was excised by digestion of a recombinant pUC18 plasmid with SspI/NcoI. All fragments were purified by native PAGE and radiolabeled with [α-32P]dATP by a Klenow fill-in reaction. The double-stranded oligonucleotide PAL was obtained by annealing the complementary sequences FPAL (5′-TAATGCTATTACGTTATATAACCCCGGG-3′) and RPAL (5′-CCCGGGGTTATTTTTAATTAA-3′) as already described (25). A typical reaction mixture (in a final volume of 15 μl) contained 15,000 cpm (0.2 ng) of radiolabeled fragments or oligonucleotides, 1 μg poly(dI-dC), and 2 μM BldR in binding buffer (25 mM Tris HCl, pH 8, 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 5% glycerol).
To test the effect of the aromatic aldehydes (cynnamaldehyde, veratrylaldehyde, and benzaldehyde), the benzyl alcohol, or the benzoic acid on the properties of BldR binding to the Sso2536 and mar-like promoters, an EMSA was performed, preincubating 0.8 μM BldR with the aromatic compounds in the following concentrations: 0.08, 0.12, 0.16, 0.2, 0.4, 1.6, 3, and 10 μM. Binding reactions and electrophoresis were carried out following the procedure previously described (25).
DNase I footprinting.
Probes containing the promoter regions of the Sso2536 gene and the mar-like operon (Sso1351-1352) were produced by PCR using a combination of an unlabeled primer and a second primer end labeled with T4 polynucleotide kinase and [γ-32P]ATP. The Sso2536 probe was prepared as described by Fiorentino et al. (25), while the DNA fragment containing the mar-like promoter was amplified using the Marfw primer (5′-CTATTGGATCGATGGGTTGC-3′) and the labeled foot pUC19p primer (5′-CGGCCAGTGAATTCGAGCT-3′), designed based on the mar-like promoter and pUC19 sequences, respectively. Labeled primers were used to generate a dideoxynucleotide sequence ladder with the Promega f-Mol DNA sequencing system. The labeled PCR products (about 40 nM) were incubated with 0.5 to 2 μg of pure BldR at 60°C in binding buffer (see above) and digested with 0.5 units of DNase I for 1 min at 60°C. Subsequent steps were performed as described by Bell and Jackson (4).
RESULTS
Identification of the BldR transcription factor.
In a previous study, Bald, a 16-kDa DNA binding protein able to bind a specific regulatory sequence in the Sso2536 promoter, was identified and purified (25). Inspection of the S. solfataricus genomic sequence in search of ORFs encoding transcription factors with similar molecular weights and putative involvement in xenobiotic response revealed the presence of a putative transcriptional regulator (Sso1352) belonging to the MarR family (59). The translated Sso1352 polypeptide is composed of 144 amino acids, with a predicted molecular mass of 16485.02 and a basic pI of 10.18. Sequence alignment with the BlastP program revealed identity with bacterial transcriptional regulators belonging to the MarR family and with archaeal proteins of still-unknown functions. Among the proteins characterized biochemically, the most significant similarity scores were obtained in multiple sequence alignment with SlyA (42.6% similarity, 19.6% identity) from Salmonella enterica serovar Typhimurium (23), MarR (47.3% similarity, 16.9% identity) from E. coli (2), and HucR (35.2% similarity, 15.4% identity) from Deinococcus radiodurans (58). Six fundamental residues were found to be identical in the C-terminal half of the protein; of these, four occur within the conserved beta sheet and turn elements of MarR, namely, the “wing” motif and one in the helix immediately adjacent to this region (Fig. 1).
FIG. 1.
Multiple alignment of BldR and characterized MarR family members. Proteins are BldR (Sso1352) from S. solfataricus, SlyA from Salmonella enterica serovar Typhimurium, PecS from Erwinia chrysantemi, MarR from E. coli, HucR from Deinococcus radiodurans, and MexR from Pseudomonas aeruginosa. The alignment was generated using Clustal W, and the numbering is based upon the HucR sequence. The winged helix-turn-helix DNA binding motif predicted for BldR is also represented: alfa-helices are gray bars, beta sheets are black arrows, and turns are black lines.
Analysis of the DNA sequence in the 5′ flanking region of ORF1352 did not reveal any consensus involved in transcriptional activity, namely, no stretch resembling an archaeal A/T-rich box (TATA box) and/or a TFB binding site. The Sso1352 gene is adjacent to a gene, Sso1351, in a tandem array organization: the TAG stop codon of Sso1351 immediately precedes the ATG start codon of Sso1352 (Fig. 2D). Intriguingly, the Sso1351 protein is annotated on the genome sequence of S. solfataricus as a permease involved in drug export; the search for structural motifs performed with the PFAM (Protein Families database of Alignments and Hidden Markov Models) program, available at the website www.sanger.ac.uk, revealed that the protein shows the typical transmembrane array of the permeases belonging to the major facilitator superfamily; these proteins are known to play a crucial role in the removal of toxic compounds, such as antibiotics, organic solvents, lipophilic anionic ligands, and, more interestingly, aromatic compounds (46).
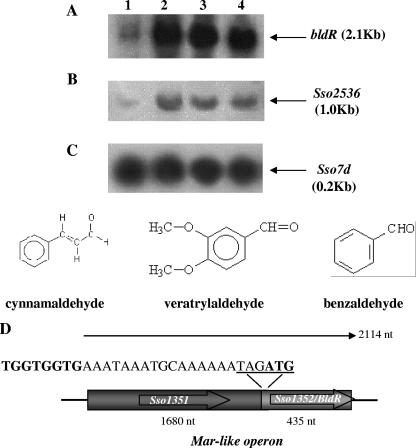
FIG. 2.
Northern blot analysis of the mar-like operon and Sso2536 mRNAs. Total RNA was prepared from S. solfataricus cells grown in the presence of different aromatic aldehydes and harvested in exponential phase. Lane 1, untreated control cells; lane 2, cells grown in the presence of 0.35 mM cynnamaldehyde; lane 3, 1 mM veratrylaldehyde; lane 4, 1 mM benzaldehyde. The filters were probed with the Sso1352 (A), Sso2536 (B), and Sso7d (C) genes. (D) Structure of the mar-like operon. BldR and the putative drug efflux permease are encoded by Sso1352 and Sso1351, respectively, and are transcribed in a single polycistronic mRNA as indicated. The stop/start codons of the ORFs are underlined in the sequence. The putative ribosome binding site(s) upstream of the Sso1352 gene is in bold. nt, nucleotides.
Transcriptional analysis of BldR.
Northern blot experiments were performed to analyze the transcription of the bldR gene in cells grown in the presence of different aromatic aldehydes; its expression profile was also compared to that of the Sso2536 gene (Fig. 2). All the aldehydes employed in this study had effect on cell growth, being toxic at concentrations of 2 mM, 2 mM, and 1 mM for benzaldehyde (15), veratrylaldehyde, and cynnamaldehyde, respectively (Fig. 2). When cells were cultivated in the presence of 1 mM benzaldehyde, 1 mM veratrylaldehyde, or 0.35 mM cynnamaldehyde, the growth rate was delayed but not arrested; the doubling times calculated in the exponential growth phase changed from 6 h for the nontreated cells to 10 h for the drug-exposed ones.
bldR mRNA revealed a single hybridization band under all conditions tested and the molecular size of the transcript, when referred to molecular weight RNA standards, was calculated to be about 2.1 kb, i.e., significantly larger than the expected length of a monocistronic transcript of the bldR gene (432 nucleotides). The size of Sso1351 located upstream of the bldR gene is 1,680 bp and matches the extra value measured for the message detected. In fact, identical hybridization signals were revealed when RNA was probed with the putative permease coding sequence (data not shown). Therefore, these results demonstrated that the two genes are transcribed in a bicistronic operon, which was defined “mar-like” by analogy to the bacterial counterparts (37, 39). This gene ordering was recently reported to be present also in other Archaea, like Sulfolobus tokodaii, Sulfolobus acidocaldarius, Thermoplasma acidophilum, Methanosarcina barkeri, and Picrophilus torridus (41).
Furthermore, as shown in Fig. 2A, in vivo levels of mar-like mRNA were significantly increased at the log phase of growth as a consequence of aldehyde supplementation to the medium.
Densitometric analysis evidenced that the levels of mRNA were increased about 10-fold in the presence of all the aldehydes with respect to nontreated cells. The results indicate a role for the aromatic aldehydes in determining increased expression levels of the mar-like operon. Interestingly, the profile of the specific Sso2536 mRNA followed a similar trend, indicating that under these growing conditions, the mar-like and Sso2536 gene expressions were both positively regulated (Fig. 2B). The amounts of total cellular RNA were comparable for all the samples, as indicated by hybridization of the same filter with the Sso7d gene (21) (Fig. 2C).
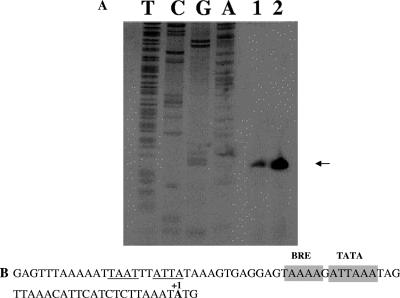
To determine the transcriptional start site of the mar-like operon, we performed a primer extension analysis on RNAs prepared from nontreated and benzaldehyde-exposed cells. The results depicted in Fig. 3 confirmed the relative increase in the mRNA upon benzaldehyde induction. Moreover, in both samples the transcription start site coincides with the first nucleotide of the translation initiation codon (Fig. 3). This result confirms a previous hypothesis that the first genes of an operon are often leaderless (54, 55). Also, as expected for leaderless transcripts, the putative TATA box (ATTAAA) is centered at position −27 relative to the transcription/translation start site and shows 84% conservation with respect to the consensus sequence ([T/C]TTA[T/A]A). A stretch of four A's corresponding to the BRE signal was also identified as centered at position −33 (7).
FIG. 3.
Primer extension and sequence analysis of the mar-like promoter region. Total RNA was prepared from cells grown in the presence or absence of benzaldehyde and harvested in exponential growth phase. (A) Primer extended products were separated by electrophoresis under denaturing conditions alongside sequencing reactions with the same primer. (B) Promoter sequence. The mapped transcription/translation start site (+1) is in bold. The TBP and TFB binding sites are indicated by boxed regions, and tandem inverted repeats are underlined.
Heterologous expression of BldR.
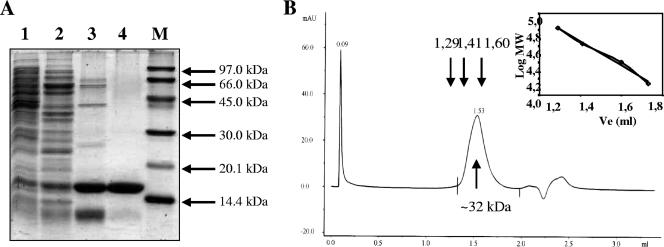
BldR was expressed in E. coli as a soluble protein and was purified to homogeneity, using the same chromatographic steps described for the purification of the native protein inducible by benzaldehyde in S. solfataricus (25). Prior to cation exchange and heparin chromatographies, the cell extract was subjected to a thermal treatment at 65°C for 10 min (Fig. 4A). Fractions containing active protein were identified by an EMSA using the same regulative sequences used to assay the native BldR protein (25). From a 1-liter culture of E. coli, it was possible to recover about 4 mg of pure BldR. Electrospray mass spectrometry confirmed the molecular mass of the recombinant polypeptide, and gel filtration experiments allowed the dimeric state in the solution of BldR to be assessed (Fig. 4B). This finding is consistent with crystallographic and biochemical analysis of MarR homologs shown to form homodimers (36).
FIG. 4.
Purification and gel filtration analysis of recombinant BldR. (A) Coomassie brilliant blue-stained SDS-PAGE gels of the protein samples from E. coli transformed with Sso1352 after each purification step. Lane 1, crude extract; lane 2, heat-treated cell extract; lane 3, fraction from Resource S chromatography; lane 4, fraction from Heparin chromatography. M, molecular mass markers. (B) Elution profile of the purified protein from gel filtration on a superdex PC75 column. Arrows indicate the elution volumes of the protein standards in the relative calibration of the column. MW, molecular mass (kDa).
Circular dichroism spectra were recorded at 25°C to determine the secondary structure composition of BldR. The content in alfa-helix (42%), beta-sheet (13%), turns (16%), and random coil (29%), calculated by the method of Young, matched the parameters predicted on the basis of the mere primary structure.
Expression analysis of BldR.
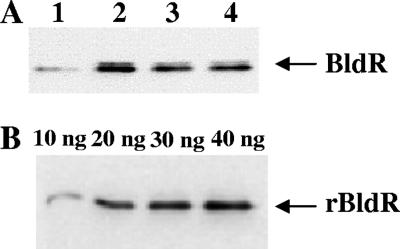
The variation in protein abundance of BldR was qualitatively estimated by Western blot analysis performed on S. solfataricus extracts derived from cells growing in media with or without aldehydes, using rabbit polyclonal antibodies raised against recombinant BldR. The accumulation of the protein followed an increase similar to that observed in the transcriptional analysis. In fact, enhancement in the BldR levels was measured depending on the added aldehyde at exponential growth phase (Fig. 5A). The BldR representativity in S. solfataricus cell extracts was determined by densitometric analysis of immunoblots containing known amounts of recombinant BldR protein as the reference (Fig. 5B). The intensity of the chemiluminescent signal obtained from the standards was linear in the range considered. BldR represented about the 0.01% of the total proteins in cells grown in basal medium, and this value increased 5- to 10-fold in induced cells.
FIG. 5.
Detection of BldR by Western blot analysis. S. solfataricus protein extracts were prepared from cells grown in the presence of different aromatic aldehydes and harvested in exponential phase, as for Northern analysis. (A) Untreated control cells (lane 1) and cells grown in the presence of 0.35 mM cynnamaldehyde (lane 2), 1 mM veratrylaldehyde (lane 3), and 1 mM benzaldehyde (lane 4). (B) Western blot analysis was performed on known amounts (indicated on the top) of purified recombinant protein (rBldR) as a reference for quantitative determination of the native BldR protein in S. solfataricus cell extracts.
Therefore, since the observed variation of protein and cognate RNA abundances match in actively growing cells, we conclude that the response to toxic aromatic aldehydes occurs at the level of transcriptional regulation control of the mar-like operon.
Binding of BldR to DNA.
DNA binding activity of BldR was investigated by mobility shift assays of suitable DNA fragments contained in the 5′ flanking regions of either the Sso2536 gene or the mar-like operon.
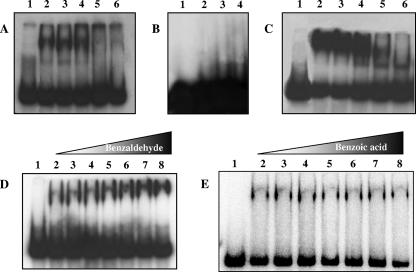
In particular, to test the affinity of BldR for the Sso2536 regulatory sequences, a 160-bp EcoRI-NcoI restriction fragment of the Sso2536 promoter located at positions −152 to +10 with respect to the transcription/translation start site was used in the binding assays (25). To assess BldR for autoregulation, an NsiI-EcoRI fragment located upstream of the mar-like operon at positions −271 to +21 was employed. BldR binds to both labeled DNA fragments site specifically (Fig. 6A and C). Eight micromolar was the concentration of BldR able to completely shift the DNA. Unlabeled specific DNA fragments abolished gel retardation when added at the ratio of 400:1, whereas a nonspecific competitor DNA produced no effect even at a 2,000-fold excess. Furthermore, under the same stringent conditions, the protein failed to associate to a DNA fragment containing a 220-bp 5′ flanking region of a superoxide dismutase (Sso0316) gene, used as a negative control for binding specificity (Fig. 6B). These results demonstrated a specific recognition by BldR of both the Sso2536 and the mar-like promoters. The dissociation constant of the BldR-promoter interaction was calculated by incubating increasing amounts of protein (0.2 to 25 μM) with the regulatory regions and analyzing the intensity of the shifted complex by signal-to-noise densitometric scanning. The dissociation constant, defined as the protein concentration at which 50% of protein is bound, was calculated using Graph Pad Prism software. The values of 0.8 μM and 1.0 μM for the Sso2536 and mar-like promoters, respectively (average values from three independent experiments), indicated that BldR binds to both promoters with comparable affinities.
FIG. 6.
Binding of recombinant BldR to the promoter regions of Sso2536 and the mar-like operon. EMSAs of the −152/+10 region of the Sso2536 gene (A) and of the −271/+21 region of the mar-like operon (C) were performed both in the absence (lanes 2) and in the presence (lanes 3 to 6) of two different excess amounts of unlabeled unspecific (Sso10b coding sequence; lanes 3 and 4) or specific (lanes 5 and 6) DNAs, using 2 μM protein. The specificity of the binding was tested with three different protein amounts (5, 10, and 20 μM) on the S. solfataricus superoxide dismutase gene promoter (B, lanes 2, 3, and 4). Shown are the effects of increasing concentrations of benzaldehyde (D) or benzoic acid (E) on the mobility of the mar-like promoter with BldR. Lanes 2 to 9, 0.8 μM BldR and 0, 0.08, 0.16, 0.2, 0.4, 1.6, and 10 μM aromatic compounds. Similar results were obtained with the Sso2536 promoter. Lanes 1 in all panels indicate free labeled DNAs.
Since members of the MarR family are phenolic sensors, we tested benzaldehyde, veratrylaldehyde, and cynnamaldehyde as potential ligands of BldR, incubating the protein with the Sso2536 and Sso1351 promoters in the presence of different amounts of aromatic aldehydes. Figure 6D shows results obtained with benzaldehyde, and similar results were obtained for the other two compounds. As shown in the figure, the aldehyde facilitated the binding of the protein to its target promoters when used at equimolar concentrations. Under the same conditions, neither benzoic acid (Fig. 6E) nor benzyl alcohol (not shown) was able to influence the binding of BldR to the Sso2536 and Sso1351 promoter regions.
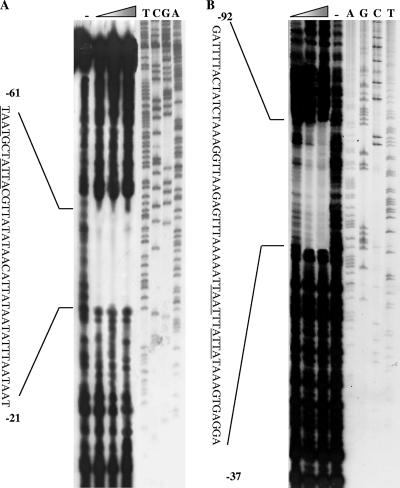
DNase I footprinting was carried out on both Sso2536 and mar-like promoter sequences to define the specific binding sites of BldR. BldR protected the regions of positions −61 to −21 and −92 to −37 (with respect to the transcription start site) on both DNAs from DNase I digestion (Fig. 7). This region encompasses, as expected, the palindromic sites at the Sso2536 promoter (25). Sequence alignment of the protected regions evidenced a conserved inverted repeat (TAATNATTA, where N indicates two or three nucleotides) located on both promoters at the same distance upstream of the TATA box.
FIG. 7.
BldR binding sites at the promoters of the Sso2536 gene and the mar-like operon. DNase I footprinting analyses of BldR were performed at the nontemplate strand of both the Sso2536 gene (A) and the mar-like operon (B). DNase I footprint analyses were performed using 0.0, 0.5, 1.0, and 2.0 μg of purified recombinant BldR. DNA fragments were analyzed in parallel with a sequencing reaction (relative lanes are indicated by the corresponding nucleotide positions on the top) by denaturing gel electrophoresis. The positions of the BldR footprints are indicated on the nucleotide sequences relative to the transcription start site.
These results strongly indicate that benzaldehyde could be the natural ligand of BldR and confirm a regulatory system in which Sso2536 gene and marR-like operon expression are responsive to levels of the substrate of the ADH enzyme via binding of BldR to their promoters.
DISCUSSION
In this study, we investigated the regulatory strategy adopted by the crenarchaeon Sulfolobus solfataricus for the cellular response to stress caused by aromatic aldehydes. Biochemical methods as well as regulation studies of the genes and proteins involved were used to survey cellular responses against the addition of the exogenous drugs. These toxic compounds were chosen since they can derive from the biodegradation of polymers (42), such as plant lignin, which can be found in the natural environment of S. solfataricus, but they can also originate from environmental pollutants, herbicides, and pesticides.
The gene product of the ORF Sso2536 had been demonstrated to be an atypical ADH (ADHSs) since it is sensibly more efficient in the catalytic reduction of aldehydes than in the oxidation of the alcoholic counterparts (27). Moreover, it is highly specific toward aromatic rather than aliphatic aldehydes and the expression of the encoding gene is indeed induced by benzaldehyde (15). In a previous study, a protein named Bald was purified for its ability to bind specifically to the Sso2536 regulatory sequences and demonstrated to increase intracellularly upon exposure to the toxic benzaldehyde (14). For this reason, the protein was postulated to be the transcription factor responsive to xenobiotic agents in the defense mechanisms involving ADH. The properties determined for the native BldR protein helped in the identification of its coding sequence on the genome of S. solfataricus. In fact, the in silico screening of sequences encoding putative transcription factors selected only one ORF (Sso1352) sharing with good confidence features coincident with those determined at both the structural and functional levels for the native protein.
Interestingly, multiple sequence alignment of BldR revealed conservation with the prototype of the family MarR in the DNA binding domain that is a winged helix-turn-helix motif. The physiological role of the members of this family can be classified into three general categories: (i) regulation of response to environmental stress, (ii) regulation of virulence factors, and (iii) regulation of aromatic catabolic pathways (59).
The majority of the genes encoding MarR homologs, in particular, and regulators containing the HTH motif, in general, are part of a gene cluster containing the gene(s) under their regulation. In particular, the adjacent genes regulated by these transcription factors code for multisubstrate efflux pumps that contribute to multidrug resistance and are cotranscribed as polycistronic transcriptional units (46).
This typical gene array is conserved also for Sso1352, as evidenced by the analysis of its genetic environment; Sso1352 overlaps to an upstream ORF, Sso1351, coding for a multidrug efflux permease. Similar orderings of the genes were also found in other Archaea, including S. tokodaii and S. acidocaldarius, but the functions of the gene products were not established (41). Alignment of the 5′ flanking region of Sso1351 with its hortolog from S. tokodaii ST1709 (not shown) allowed the identification of an AT-rich conserved sequence (TTAAAAATTAA) located in both promoters at the same distance relative to the putative TATA boxes.
The transcriptional analysis of S. solfataricus confirmed the prediction that the two genes are cotranscribed and showed that their expression increased in actively dividing cells in response to the addition of aromatic aldehydes to the medium in a fashion identical to the trend observed for the specific induction of Sso2536 gene transcription.
The recombinant BldR protein was demonstrated to be a dimer and to bind site specifically to both its own and Sso2536 promoters in delimited and specific regions. Winged helix proteins from the MarR family typically bind as dimers to inverted repeats located upstream of the transcription start site, but to date, they have been characterized only from bacterial sources (59). The analysis of the DNase I footprints evidenced an extended protection. The sequence of this region included the inverted repeat TAATNATTA (n = 2 and 3 for the mar-like and Sso2536 promoters, respectively) on both promoters at the same small distance from the TATA box and matched the consensus sequence already identified during the isolation of native BldR. The wide extension of the protection is not unusual for archaeal transcriptional regulators, and it has been suggested that it could be due to multiple adjacent sites with different binding affinities. However, the location of a binding site in this region can account for an activator role for BldR since it could act by enhancing the recruitment of TBP or TFB or by stabilizing their binding to DNA. This has been recently demonstrated for the regulatory protein Sta1 from S. solfataricus, which is able to activate transcription from viral promoters (32).
As for most of the genes in the MarR family, the binding of BldR to its own promoter would induce auto-activation and in turn increase in the coexpressed drug export permease levels. Moreover, the binding of BldR to the Sso2536 promoter would stimulate the gene transcription, the accumulation of the ADH enzyme, and hence the enzyme-catalyzed conversion of aldehydes to alcohols. This mechanism could be the strategy adopted by S. solfataricus to reduce the aromatic aldehyde concentration inside the cells.
This drug-triggered response mode is consistent with the observation that reduction of aldehydes to their corresponding alcohols mitigates the toxicity of these compounds. In fact, alcohols can be accumulated intracellularly with minor damaging effect before being extruded by efflux pumps and/or metabolized via fission by aromatic ring cleavage enzymes (43). This prompt but finely regulated response represents, in our opinion, the effective and productive strategy for detoxification against naturally occurring aromatic aldehydes in S. solfataricus.
By inspection of the S. tokodaii genome at http://www.genome.ad.jp, among 10 putative adh genes, a hortolog of SSo2536, ST2577 (identified using the KEGG orthology database), was found. Very interestingly, the gene contained, at specific positions, the same regulative sequences of the ST1709-1710 promoter (not shown). This corroborates evidence that a similar detoxification strategy involving MarR-like family members could be adopted by other Sulfolobales.
BldR is the first example of a MarR family member to be functionally characterized as a positive transcriptional regulator in archaea. Similarly to other bacterial counterparts, it could respond to the effector molecule by direct binding. This study also further supports a specific involvement of ADHSs in stress response to aromatic compounds rather than in aldehyde/alcohol metabolism (16).
In this field, more-detailed time course studies using proteomics and/or interaction analysis among cellular regulators may help in clarifying the interplay between the stress response, the synthesis of efflux pumps, and the induction of catabolic pathways as well as their connections at the molecular level.
Acknowledgments
This work was supported in part by research grants from Ministero dell'Università e della Ricerca Scientifica (MIUR, PRIN 2004). Support from the Regional Center of Competence (CRdC ATIBB, Regione Campania, Naples, Italy) is also gratefully acknowledged.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Adewoye, L., A. Sutherland, R. Srikumar, and K. Poole. 2002. The mexR repressor of the mexAB-oprM multidrug efflux operon in Pseudomonas aeruginosa: characterization of mutations compromising activity. J. Bacteriol. 184:4308-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, S. D., and S. P. Jackson. 2000. Mechanism of autoregulation by an archaeal transcriptional repressor. J. Biol. Chem. 275:31624-31629. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. D., C. H. Botting, B. N. Wardleworth, S. P. Jackson, and M. F. White. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148-151. [DOI] [PubMed] [Google Scholar]
- 6.Bell, S. D., A. B. Brinkman, J. van der Oost, and S. P. Jackson. 2001. The archaeal TFIIEalpha homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, S. D., P. L. Kosa, P. B. Sigler, and S. P. Jackson. 1999. Orientation of the transcription preinitiation complex in archaea. Proc. Natl. Acad. Sci. USA 96:13662-13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, S. D., C. P. Magill, and S. P. Jackson. 2001. Basal and regulated transcription in Archaea. Biochem. Soc. Trans. 29:392-395. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Branden, C. I., H. Jornvall, H. Eklund, and B. Furugren. 1975. Alcohol dehydrogenase, p. 103-190. In P. D. Boyer(ed.), The enzymes, 3rd ed. Academic Press, Orlando, FL.
- 11.Brent, R., and M. Ptashne. 1981. Mechanism of action of the lexA gene product. Proc. Natl. Acad. Sci. USA 78:4204-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brock, T. D., K. M. Brock, R. T. Belly, and R. L. Weiss. 1972. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84:54-68. [DOI] [PubMed] [Google Scholar]
- 13.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannio, R., G. Fiorentino, P. Carpinelli, M. Rossi, and S. Bartolucci. 1996. Cloning and overexpression in Escherichia coli of the genes encoding NAD-dependent alcohol dehydrogenase from two Sulfolobus species. J. Bacteriol. 178:301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannio, R., G. Fiorentino, M. Rossi, and S. Bartolucci. 1999. The alcohol dehydrogenase gene: distribution among Sulfolobales and regulation in Sulfolobus solfataricus. FEMS Microbiol. Lett. 170:31-39. [DOI] [PubMed] [Google Scholar]
- 16.Chong, P. K., A. M. Burja, H. Radianingtyas, A. Fazeli, and P. C. Wright. 2007. Proteome analysis of Sulfolobus solfataricus P2 propanol metabolism. J. Proteome Res. 6:1430-1439. [DOI] [PubMed] [Google Scholar]
- 17.Chong, P. K., and P. C. Wright. 2005. Identification and characterization of the Sulfolobus solfataricus P2 proteome. J. Proteome Res. 4:1789-1798. [DOI] [PubMed] [Google Scholar]
- 18.Chong, P. K., A. M. Burja, H. Radianingtyas, A. Fazeli, and P. C. Wright. 2007. Translational and transcriptional analysis of Sulfolobus solfataricus P2 to provide insights into alcohol and ketone utilisation. Proteomics 7:424-435. [DOI] [PubMed] [Google Scholar]
- 19.Contursi, P., R. Cannio, S. Prato, G. Fiorentino, M. Rossi, and S. Bartolucci. 2003. Development of a genetic system for hyperthermophilic Archaea: expression of a moderate thermophilic bacterial alcohol dehydrogenase gene in Sulfolobus solfataricus. FEMS Microbiol. Lett. 218:115-120. [DOI] [PubMed] [Google Scholar]
- 20.Echave, P., J. Tamarit, E. Cabiscol, and J. Ros. 2003. Novel antioxidant role of alcohol dehydrogenase E from Escherichia coli. J. Biol. Chem. 278:30193-30198. [DOI] [PubMed] [Google Scholar]
- 21.Edmondson, S. P., and J. W. Shriver. 2001. DNA binding proteins Sac7d and Sso7d from Sulfolobus. Methods Enzymol. 334:129-145. [DOI] [PubMed] [Google Scholar]
- 22.Egland, P. G., and C. S. Harwood. 1999. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an Fnr family member. J. Bacteriol. 181:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellison, D. W., and V. L. Miller. 2006. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9:153-159. [DOI] [PubMed] [Google Scholar]
- 24.Felenbok, B., M. Flipphi, and I. Nikolaev. 2001. Ethanol catabolism in Aspergillus nidulans: a model system for studying gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 69:149-204. [DOI] [PubMed] [Google Scholar]
- 25.Fiorentino, G., R. Cannio, M. Rossi, and S. Bartolucci. 2003. Transcriptional regulation of the gene encoding an alcohol dehydrogenase in the archaeon Sulfolobus solfataricus involves multiple factors and control elements. J. Bacteriol. 185:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiduschek, E. P., and M. Ouhammouch. 2005. Archaeal transcription and its regulators. Mol. Microbiol. 56:1397-1407. [DOI] [PubMed] [Google Scholar]
- 27.Giordano, A., R. Cannio, F. La Cara, S. Bartolucci, M. Rossi, and C. A. Raia. 1999. Asn249Tyr substitution at the coenzyme binding domain activates Sulfolobus solfataricus alcohol dehydrogenase and increases its thermal stability. Biochemistry 38:3043-3054. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez, R., H. Tao, J. E. Purvis, S. W. York, K. T. Shanmugam, and L. O. Ingram. 2003. Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: comparison of KO11 (parent) to LY01 (resistant mutant). Biotechnol. Prog. 19:612-623. [DOI] [PubMed] [Google Scholar]
- 29.Hanzelka, B. L., T. J. Darcy, and J. N. Reeve. 2001. TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEα. J. Bacteriol. 183:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinicke, I., J. Muller, M. Pittelkow, and A. Klein. 2004. Mutational analysis of genes encoding chromatin proteins in the archaeon Methanococcus voltae indicates their involvement in the regulation of gene expression. Mol. Genet. Genomics 272:76-87. [DOI] [PubMed] [Google Scholar]
- 31.Hudepohl, U., W. D. Reiter, and W. Zillig. 1990. In vitro transcription of two rRNA genes of the archaebacterium Sulfolobus sp. B12 indicates a factor requirement for specific initiation. Proc. Natl. Acad. Sci. USA 87:5851-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kessler, A., G. Sezonov, J. I. Guijarro, N. Desnoues, T. Rose, M. Delepierre, S. D. Bell, and D. Prangishvili. 2006. A novel archaeal regulatory protein, Sta1, activates transcription from viral promoters. Nucleic Acids Res. 34:4837-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruger, K., T. Hermann, V. Armbruster, and F. Pfeifer. 1998. The transcriptional activator GvpE for the halobacterial gas vesicle genes resembles a basic region leucine-zipper regulatory protein. J. Mol. Biol. 279:761-771. [DOI] [PubMed] [Google Scholar]
- 34.Lee, S. J., A. Engelmann, R. Horlacher, Q. Qu, G. Vierke, C. Hebbeln, M. Thomm, and W. Boos. 2003. TrmB, a sugar-specific transcriptional regulator of the trehalose/maltose ABC transporter from the hyperthermophilic archaeon Thermococcus litoralis. J. Biol. Chem. 278:983-990. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. J., C. Moulakakis, S. M. Koning, W. Hausner, M. Thomm, and W. Boos. 2005. TrmB, a sugar sensing regulator of ABC transporter genes in Pyrococcus furiosus exhibits dual promoter specificity and is controlled by different inducers. Mol. Microbiol. 57:1797-1807. [DOI] [PubMed] [Google Scholar]
- 36.Lie, T. J., G. E. Wood, and J. A. Leigh. 2005. Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J. Biol. Chem. 280:5236-5241. [DOI] [PubMed] [Google Scholar]
- 37.Lim, D., K. Poole, and N. Strynadka. 2002. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J. Biol. Chem. 277:29253-29259. [DOI] [PubMed] [Google Scholar]
- 38.Limauro, D., A. Falciatore, A. L. Basso, G. Forlani, and M. De Felice. 1996. Proline biosynthesis in Streptococcus thermophilus: characterization of the proBA operon and its products. Microbiology 142:3275-3282. [DOI] [PubMed] [Google Scholar]
- 39.Martin, R. G., and J. L. Rosner. 2004. Transcriptional and translational regulation of the marRAB multiple antibiotic resistance operon in Escherichia coli. Mol. Microbiol. 53:183-191. [DOI] [PubMed] [Google Scholar]
- 40.Martin, R. G., W. K. Gillette, S. Rhee, and J. L. Rosner. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431-441. [DOI] [PubMed] [Google Scholar]
- 41.Miyazono, K. I., M. Tsujimura, Y. Kawarabayasi, and M. Tanokura. 2007. Crystal structure of an archaeal homologue of multidrug resistance repressor protein, EmrR, from hyperthermophilic archaea Sulfolobus tokodaii strain 7. Proteins 67:1138-1146. [DOI] [PubMed] [Google Scholar]
- 42.Nishimura, M., O. Ooi, and J. Davies. 2006. Isolation and characterization of Streptomyces sp. NL15-2K capable of degrading lignin-related aromatic compounds. Biosci. Bioeng. 102:124-127. [DOI] [PubMed] [Google Scholar]
- 43.Notomista, E., A. Lahm, A. Di Donato, and A. Tramontano. 2003. Evolution of bacterial and archaeal multicomponent monooxygenases. J. Mol. Evol. 56:435-445. [DOI] [PubMed] [Google Scholar]
- 44.Ouhammouch, M., and E. P. Geiduschek. 2005. An expanding family of archaeal transcriptional activators. Proc. Natl. Acad. Sci. USA 102:15423-15428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouhammouch, M., G. E. Langham, W. Hausner, A. J. Simpson, N. M. El-Sayed, and E. P. Geiduschek. 2005. Promoter architecture and response to a positive regulator of archaeal transcription. Mol. Microbiol. 56:625-637. [DOI] [PubMed] [Google Scholar]
- 46.Paulsen, I. T. 2003. Multidrug efflux pumps and resistance: regulation and evolution. Curr. Opin. Microbiol. 6:446-451. [DOI] [PubMed] [Google Scholar]
- 47.Peeters, E., T. L. Thia-Toong, D. Gigot, D. Maes, and D. Charlier. 2004. Ss-LrpB, a novel Lrp-like regulator of Sulfolobus solfataricus P2, binds cooperatively to three conserved targets in its own control region. Mol. Microbiol. 54:321-336. [DOI] [PubMed] [Google Scholar]
- 48.Peeters, E., R. Willaert, D. Maes, and D. Charlier. 2006. Ss-LrpB from Sulfolobus solfataricus condenses about 100 base pairs of its own operator DNA into globular nucleoprotein complexes. J. Biol. Chem. 281:11721-11728. [DOI] [PubMed] [Google Scholar]
- 49.Qureshi, S. A., and S. P. Jackson. 1998. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell 1:389-400. [DOI] [PubMed] [Google Scholar]
- 50.Reeve, J. N. 2003. Archaeal chromatin and transcription. Mol. Microbiol. 48:587-598. [DOI] [PubMed] [Google Scholar]
- 51.Schneiders, T., and S. B. Levy. 2006. MarA-mediated transcriptional repression of the rob promoter. J. Biol. Chem. 281:10049-10055. [DOI] [PubMed] [Google Scholar]
- 52.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thaw, P., S. E. Sedelnikova, T. Muranova, S. Wiese, S. Ayora, J. C. Alonso, A. B. Brinkman, J. Akerboom, J. van der Oost, and J. B. Rafferty. 2006. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 34:1439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tolstrup, N., C. W. Sensen, R. A. Garrett, and I. G. Clausen. 2000. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles 4:175-179. [DOI] [PubMed] [Google Scholar]
- 55.Torarinsson, E., H. P. Klenk, and R. A. Garrett. 2005. Divergent transcriptional and translational signals in Archaea. Environ. Microbiol. 7:47-54. [DOI] [PubMed] [Google Scholar]
- 56.Tussey, L., and M. R. Felder. 1989. Tissue-specific genetic variation in the level of mouse alcohol dehydrogenase is controlled transcriptionally in kidney and posttranscriptionally in liver. Proc. Natl. Acad. Sci. USA 86:5903-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vierke, G., A. Engelmann, C. Hebbeln, and M. Thomm. 2003. A novel archaeal transcriptional regulator of heat shock response. J. Biol. Chem. 278:18-26. [DOI] [PubMed] [Google Scholar]
- 58.Wilkinson, S. P., and A. Grove. 2004. HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J. Biol. Chem. 279:51442-51450. [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson, S. P., and A. Grove. 2006. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Biol. 8:51-62. [PubMed] [Google Scholar]
- 60.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]