Abstract
The Bacillus subtilis spoIIIA locus encodes eight proteins, SpoIIIAA to SpoIIIAH, which are expressed in the mother cell during endospore formation and which are essential for the activation of σG in the forespore. Complementation studies indicated that this locus may be transcribed from two promoters, one promoter upstream from the first gene and possibly a second unidentified promoter within the locus. Fragments of the spoIIIA locus were expressed at an ectopic site to complement the sporulation-defective phenotype of a spoIIIAH deletion, and we determined that complementation required a fragment of DNA that extended into spoIIIAF. To confirm that there was a promoter located in spoIIIAF, we constructed transcriptional fusions to lacZ and found strong sporulation-induced promoter activity. Primer extension assays were used to determine the transcription start site, and point mutations introduced into the −10 and −35 regions of the promoter reduced its activity. This promoter is transcribed by σE-RNA polymerase and is repressed by SpoIIID. Therefore, we concluded that the spoIIIA locus is transcribed from two promoters, one at the start of the locus (P1spoIIIA) and the other within the locus (P2spoIIIA). Based on Campbell integrations and reverse transcription-PCR analysis of the P2spoIIIA region, we determined that P2spoIIIA is sufficient for transcription of spoIIIAG and spoIIIAH. Inactivation of P2spoIIIA blocked spore formation, indicating that P2spoIIIA is essential for expression of spoIIIAG and spoIIIAH. The P2spoIIIA activity is twice the P1spoIIIA activity; therefore, larger amounts of SpoIIIAG and SpoIIIAH than of proteins encoded at the upstream end of the locus may be required.
In response to nutrient depletion, Bacillus subtilis can respond by differentiating into an endospore, which can remain dormant until nutrients become available to stimulate its germination. Soon after the onset of endospore formation, the cell divides into two morphologically distinct cells, the larger mother cell and the smaller forespore. During sporulation, 383 genes are expressed in the mother cell (8), while 143 different genes are expressed in the forespore (23). The cell type-specific patterns, as well as the temporal patterns of gene expression, are governed largely by the appearance and activation of four RNA polymerase sigma factors (for reviews, see references 9, 12, and 17). The activity of each of these sigma factors is tightly regulated for two purposes, thereby coupling the activation of gene expression to the completion of morphological landmarks and synchronizing the developmental programs of gene expression in each of the two cell types. For example, activation of σG in the forespore requires σE-directed gene expression in the mother cell.
Transcription of the spoIIIA locus by σE is required for σG activation (13, 15). The spoIIIA locus, which is conserved throughout spore-forming bacteria, encodes eight proteins, SpoIIIAA to SpoIIIAH, each of which is required for σG activation (15). Seven of the eight products, SpoIIIAB to SpoIIIAH, are predicted to be membrane associated (4, 20, 22); SpoIIIAA shows homology to AAA-type ATPases, but the other seven proteins show homology only to their orthologs in other sporulating bacteria. SpoIIIAH interacts with the forespore-expressed protein SpoIIQ via the large extracellular C-terminal domains (1, 6, 18). The two proteins make contact first at the septum between the mother cell and the forespore and then migrate with the mother cell membrane as the forespore is engulfed (1, 2). It is not known how this complex contributes to σG activation or what roles the other spoIIIA-encoded proteins play.
Transcription of the spoIIIA locus is initiated at a σE-dependent promoter located immediately upstream of spoIIIAA (13). Transcription from this promoter is repressed by the DNA binding protein SpoIIID (13) and possibly to a lesser extent by GerR (8). Transcription from the gene immediately preceding the spoIIIA locus, yqhV, may also read into the spoIIIA locus since there is no obvious transcription terminator between the two genes. Since yqhV transcription is also σE dependent and repressed by GerR, this may account for the effect of GerR on spoIIIA transcription (8).
A number of operons found in both gram-positive and gram-negative bacteria are transcribed from more than one promoter. In some instances, these promoters are expressed under different conditions, and their activities are controlled by different RNA polymerase sigma factors or DNA binding proteins (3, 21). The use of alternative promoters can also allow different levels of expression of different genes in the locus in response to different signals (16, 24).
Two observations led us to suspect that a second promoter located within the spoIIIA locus may play a role in its expression. A transposon insertion into the extreme 5′ end of spoIIIAA does not prevent expression of SpoIIIAH, as determined by Western blotting (1). We also noted that when SpoIIIAH was directly expressed from the promoter located upstream from spoIIIAA (P1spoIIIA) at an ectopic locus, this construct did not complement the sporulation defect of a spoIIIAH mutant strain. Moreover, a second putative promoter within the locus had been proposed previously (19a). Therefore, we began a search for the promoter responsible for spoIIIAH transcription. We found a promoter (P2spoIIIA) that is located in spoIIIAF and is necessary and sufficient for expression of spoIIIAG and spoIIIAH.
MATERIALS AND METHODS
Bacterial strains and culture media.
The strains and plasmids used in this study are shown in Tables 1 and 2. Routine microbiological manipulations and procedures were carried out as described by Cutting and Harwood (5). The concentrations of antibiotics used for selection in Luria broth (LB) and Difco sporulation medium (DSM) were as follows: 100 μg/ml ampicillin, 5 μg/ml chloramphenicol, 10 μg/ml kanamycin, 100 μg/ml spectinomycin, and 20 μg/ml tetracycline. Cultures were grown in LB, and sporulation was induced by nutrient exhaustion in DSM. Competent cells were prepared and transformed using the Spizizen method (5).
TABLE 1.
Plasmids used in this study
| Plasmid | Genotype | Reference or source |
|---|---|---|
| pDG1661 | 10 | |
| pDG1662 | 10 | |
| pDG784 | 9 | |
| pDG1515 | 9 | |
| pDG1726 | 9 | |
| pMS38 | Lab stock | |
| pLitmus28 | NEB | |
| pCG123 | pDG1662 + spoIIIAH | This study |
| pCG124 | pDG1662 + spoIIIAGH | This study |
| pCG125 | pDG1662 + spoIIIAFGH | This study |
| pCG126 | pDG1662 + spoIIIA′EFGH (bp 1010 to 1215 of spoIIIAE) | This study |
| pCG129 | pDG1661 + spoIIIAH | This study |
| pCG130 | pDG1661 + spoIIIAGH | This study |
| pCG131 | pDG1661 + spoIIIAFGH | This study |
| pCG132 | pDG1661 + spoIIIA′EFGH | This study |
| pCG134 | pDG1661 + P1spoIIIA | This study |
| pCG135 | pDG1661 + spoIIIAFGH (bp 1 to 42 of spoIIIAH) | This study |
| pCG136 | pDG1661 + spoIIIAFG (bp 1 to 42 of spoIIIAG) | This study |
| pCG137 | pDG1661 + spoIIIAF | This study |
| pCG138 | pDG1661 + spoIIIAF (200 bp 3′ of spoIIIAF deleted) | This study |
| pCG139 | pDG1661 + spoIIIAF (400 bp 3′ of spoIIIAF deleted) | This study |
| pCG140 | pDG1661 + spoIIIA′EFGH (bp 1 to 42 spoIIIAH) | This study |
| pCG141 | pDG1661 + spoIIIA′EFG (bp 1 to 42 spoIIIAG) | This study |
| pCG142 | pDG1661 + spoIIIA′EF | This study |
| pCG143 | pDG1661 + spoIIIA′EF (200 bp 3′ of spoIIIAF deleted)a | This study |
| pCG144 | pDG1661 + spoIIIA′EF (400 bp 3′ of spoIIIAF deleted) | This study |
| pCG145 | pDG1661 + spoIIIA′EF (600 bp 3′ of spoIIIAF deleted) | This study |
| pCG146 | pDG1661 + spoIIIA′E | This study |
| pCG149 | pDG1661 + spoIIIAF-lacZ (100 bp 5′ of spoIIIAF deleted) | This study |
| pCG150 | pDG1661 + spoIIIAF-lacZ (200 bp 5′ of spoIIIAF deleted)b | This study |
| pCG151 | pDG1661 + spoIIIAF-lacZ (300 bp 5′ of spoIIIAF deleted) | This study |
| pCG152 | pMS38 + spoIIIAF | This study |
| pCG153 | pMS38 + spoIIIAE | This study |
| pCG154 | pMS38 + spoIIIAD | This study |
| pCG155 | pMS38 + spoIIIAC | This study |
| pCG156 | pMS38 + spoIIIAB | This study |
| pCG162 | pDG1726 ΔgerR | This study |
| pCG163 | pDG784 ΔspoIIID | This study |
| pCG164 | pDG1726 ΔsigE | This study |
| pCG165 | pDG1515 ΔsigK | This study |
| pCG166 | pLitmus28 + spoIIIAFGH | This study |
| pCG167 | pDG1661 + PsspE (bp −165 to 14) | This study |
| pCG169 | pLitmus28 + spoIIIAFGH −30 C-to-T mutation | This study |
| pCG170 | pLitmus28 + spoIIIAFGH −12/−11 CA-to-GT mutation | This study |
| pCG179 | pDG1661 + P2spoIIIA −30 C-to-T mutation | This study |
| pCG180 | pDG1661 + P2spoIIIA −12/−11 CA-to-GT mutation | This study |
| pCG184 | pDG1661 + spoIIIAFGH −12/−11 CA-to-GT mutation | This study |
| pCG185 | pDG1662 + spoIIIAFGH −12/−11 CA-to-GT mutation | This study |
| pCG202 | pDG784 + spoIIIAFGH −12/−11 CA-to-GT mutation | This study |
This site was used as 3′ end for all remaining experiments.
The fragment of DNA from base 200 to base 415 of spoIIIAF is referred to as P2spoIIIA.
TABLE 2.
B. subtilis strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JH642 | trpC2 pheA1 | J. Hoch |
| AOB114 | MB24 gerE::kan | Lab stock |
| RL2045 | ΔspoIIIAG | P. Stragier |
| RL2046 | ΔspoIIIAH | P. Stragier |
| CGB125 | amyE::spoIIIAH | This study |
| CGB126 | amyE::spoIIIAGH | This study |
| CGB127 | amyE::spoIIIAFGH | This study |
| CGB128 | amyE::spoIII′AEFGH | This study |
| CGB129 | RL2046 amyE::spoIIIAH | This study |
| CGB130 | RL2046 amyE::spoIIIAGH | This study |
| CGB131 | RL2046 amyE::spoIIIAFGH | This study |
| CGB132 | RL2046 amyE::spoIII′AEFGH | This study |
| CGB133 | RL2045 amyE::spoIIIAH | This study |
| CGB134 | RL2045 amyE::spoIIIAGH | This study |
| CGB135 | RL2045 amyE::spoIIIAFGH | This study |
| CGB136 | RL2045 amyE::spoIIIA′EFGH | This study |
| CGB142 | amyE::spoIIIAH-lacZ | This study |
| CGB143 | amyE::spoIIIAGH-lacZ | This study |
| CGB144 | amyE::spoIIIAFGH-lacZ | This study |
| CGB145 | amyE::spoIIIA′EFGH-lacZ (last 200 bp of spoIIIAE) | This study |
| CGB147 | amyE::P1spoIIIA-lacZ | This study |
| CGB158 | amyE::spoIIIAFGH-lacZ (42 bp of spoIIIAH) | This study |
| CGB159 | amyE::spoIIIAFG-lacZ (42 bp of spoIIIAG) | This study |
| CGB160 | amyE::spoIIIAF-lacZ | This study |
| CGB161 | amyE::spoIIIAF-lacZ (200 bp at 3′ end of spoIIIAF deleted) | This study |
| CGB162 | amyE::spoIIIAF-lacZ (400 bp at 3′ end of spoIIIAF deleted) | This study |
| CGB163 | amyE::spoIIIA′EFGH-lacZ (42 bp of spoIIIAH) | This study |
| CGB164 | amyE::spoIIIA′EFG-lacZ (42 bp of spoIIIAG) | This study |
| CGB165 | amyE::spoIIIA′EF-lacZ (200 bp at 3′ end of spoIIIAF deleted)a | This study |
| CGB166 | amyE::spoIIIA′EF-lacZ (400 bp at 3′ end of spoIIIAF deleted) | This study |
| CGB167 | amyE::spoIIIA′EF-lacZ (600 bp at 3′ end of spoIIIAF deleted) | This study |
| CGB168 | amyE::spoIIIA′E-lacZ (last 200 bp of spoIIIAE) | This study |
| CGB172 | amyE::spoIIIAF-lacZ (100 bp at 5′ end of spoIIIAF deleted) | This study |
| CGB173 | amyE::spoIIIAF-lacZ (200 bp at 5′ end of spoIIIAF deleted)b | This study |
| CGB174 | amyE::spoIIIAF-lacZ (300 bp at 5′ end of spoIIIAF deleted) | This study |
| CGB175 | spoIIIAF Campbell | This study |
| CGB176 | spoIIIAE Campbell | This study |
| CGB177 | spoIIIAD Campbell | This study |
| CGB178 | spoIIIAC Campbell | This study |
| CGB179 | spoIIIAB Campbell | This study |
| CGB187 | gerR::spec amyE::P1spoIIIA | This study |
| CGB188 | spoIIID::kan amyE::P1spoIIIA | This study |
| CGB189 | sigE::spec amyE::P1spoIIIA | This study |
| CGB190 | sigK::tet amyE::P1spoIIIA | This study |
| CGB191 | gerE::kan amyE::P1spoIIIA | This study |
| CGB192 | gerR::spec amyE::P2spoIIIA | This study |
| CGB193 | spoIIID::kan amyE::P2spoIIIA | This study |
| CGB194 | sigE::spec amyE::P2spoIIIA | This study |
| CGB195 | sigK::tet amyE::P2spoIIIA | This study |
| CGB196 | gerE::kan amyE::P2spoIIIA | This study |
| CGB197 | amyE::PsspE-lacZ | This study |
| CGB198 | CGB175 amyE::PsspE-lacZ | This study |
| CGB202 | amyE::P2spoIIIA-lacZ (−30 C-to-T mutation) | This study |
| CGB203 | amyE::P2spoIIIA-lacZ (−12/−11 CA-to-GT mutation) | This study |
| CGB206 | amyE::spoIIIAFGH-lacZ (−12/−11 CA-to-GT mutation) | This study |
| CGB207 | amyE::spoIIIAFGH (−12/−11 CA-to-GT mutation) | This study |
| CGB208 | RL2045 amyE::spoIIIAFGH (−12/−11 CA-to-GT mutation) | This study |
| CGB209 | RL2046 amyE::spoIIIAFGH (−12/−11 CA-to-GT mutation) | This study |
| CGB225 | spoIIIAFGH (−12/−11 CA-to-GT mutation) | This study |
| CGB226 | CGB197 spoIIIAFGH (−12/−11 CA-to-GT mutation) | This study |
| CGB227 | CGB225 amyE::spoIIIAFGH-lacZ | This study |
This site was used as the 3′ end for all remaining experiments.
The fragment of DNA from base 200 to base 415 of spoIIIAF is referred to as P2spoIIIA.
To construct the plasmids used for complementation of the sporulation-defective phenotype of RL2045 (JH642 ΔspoIIIAG) and RL2046 (JH642 ΔspoIIIAH), fragments of DNA corresponding to spoIIIAH, spoIIIAGH, spoIIIAFGH and spoIIIA′EFGH were amplified from JH642 chromosomal DNA, cloned into pDG1662 (11) digested with BamHI and EcoRI, and transformed into competent JH642, RL2045, and RL2046 with selection for chloramphenicol resistance and screening for spectinomycin sensitivity to confirm double crossovers into the amyE locus.
In order to more positively identify which region of DNA contained the putative promoter, we amplified the sections of DNA described above and cloned them into EcoRI- and BamHI-digested pDG1661 (11), which contains a promoterless lacZ gene fused to the ribosome binding site of spoVG, and then we transformed the resulting plasmids into competent JH642, with selection for chloramphenicol resistance and screening for spectinomycin sensitivity to confirm double crossovers into the amyE locus. To compare the known spoIIIA operon promoter P1spoIIIA (upstream of spoIIIAA) to P2spoIIIA, we amplified DNA from position −246 to position 7 relative to the P1spoIIIA transcription start site and cloned it into pDG1661 as described above.
Since we wanted to define the P2spoIIIA promoter region more narrowly, we made sequential deletions from the 5′ and 3′ ends of the spoIIIAF gene. First, we kept the 5′ end constant either at the second codon of spoIIIAF or at base 1016 of spoIIIAE, and the 3′ end was moved inward from the last codon of spoIIIAF to the second codon of spoIIIAF in 200-bp intervals. DNA fragments resulting from the PCR amplifications were cloned into pDG1661 digested with EcoRI and BamHI and transformed into competent JH642 as described above. Once a suitable 3′ end was established, as determined by β-galactosidase activity, we constructed and transformed pDG1661-based plasmids as described above that had DNA fragments with successive 100-bp deletions from the second codon of spoIIIAF.
The following double-crossover knockouts of potential P2spoIIIA regulators were constructed in early and late stages of mother cell transcription: (i) σE and regulators SpoIIID and GerR and (ii) σK and a regulator GerE. Approximately 500 bp of 5′- and 3′-flanking DNA of each gene was amplified from chromosomal DNA and cloned into vectors designed for gene replacement. First gerR and sigE 5′ homology regions were cloned into EcoRV- and EcoRI-digested pDG1726 (10) (spectinomycin), and then the 3′ homology regions were cloned into the resulting plasmids with BamHI and SalI. The spoIIID 5′ homology region was cloned into EcoRI- and BamHI-digested pDG784 (10) (kanamycin), and the 3′ region was cloned into the resulting plasmid with PstI and SphI. Homology regions for sigK were cloned first into the EagI and BamHI sites of pDG1515 (10) (tetracycline) and then into the EcoRI and HindIII sites of the resulting plasmid. Each of the final plasmids was linearized with PstI or ScaI and transformed into competent CGB147 and CGB173 with selection for the appropriate antibiotic marker. Double crossovers were confirmed by PCR analysis. The gerE knockout was constructed by transforming competent CGB147 and CGB173 with chromosomal DNA isolated from laboratory stock strain AOB114 (MB24 gerE::kan) and selecting for kanamycin resistance.
To introduce point mutations into the −35 and −10 regions of P2spoIIIA and into both potential alternative start codons for SpoIIIAG, we designed two complementary QuikChange oligonucleotides per desired mutation and performed QuikChange mutagenic PCR on pCG166 as described below. After sequencing to ensure the presence of the desired mutation, we subcloned (i) the previously defined promoter region into pDG1661 containing the separate −30 and −12/−11 mutations using EcoRI and BamHI and (ii) the entire spoIIIAFGH fragment containing the −12/−11 point mutations into both pDG1661 and pDG1662 using BamHI and EcoRI. The four resulting plasmids were transformed into competent JH642 (or RL2045 and RL2046 for pDG1662 derivatives) with selection for chloramphenicol resistance and screening for spectinomycin sensitivity to confirm double crossovers into the amyE gene.
Plasmids for Campbell integration into spoIIIAF, spoIIIAE, spoIIIAD, spoIIIAC, and spoIIIAB were constructed by amplifying each entire gene from chromosomal DNA along with its ribosome binding site and a few bases downstream of the stop codon and cloning the DNA into pMS38 digested with EcoRV and BamHI. The resulting plasmids were transformed into competent JH642 with selection for chloramphenicol resistance. The presence of single crossovers was confirmed by amplification of the ampicillin resistance gene on the pMS38 vector backbone from chromosomal DNA.
The promoter region of sspE, beginning 130 bp upstream of the −35 region and extending 5 bases past its ribosome binding site, was amplified from chromosomal DNA and cloned into pDG1661 digested with BamHI and HindIII. The plasmid was transformed into competent JH642 and CGB175 with selection for chloramphenicol resistance and screening for spectinomycin sensitivity to confirm double crossovers into the amyE gene.
In order to isolate a strain with the two base pair substitutions in the −10 region of P2spoIIIA at the spoIIIA chromosomal locus, we cloned the spoIIIAFGH DNA fragment harboring the −12/−11 CA-to-GT P2spoIIIA mutation from pCG170 into pDG784 (10) that had been cleaved with EcoRI and BamHI. In the second step, an approximately 500-bp fragment of DNA from the region immediately downstream of spoIIIAH was amplified and cloned into the pDG784 derivative between the PstI and SphI sites. The resulting plasmid was used to transform competent JH642 and CGB197 to kanamycin resistance. PCR amplification and DNA sequencing of the transformants were used to show that a double crossover resulted in replacement of the wild-type copy of P2spoIIIA with the mutated P2spoIIIA, resulting in strains CGB225 and CGB226, respectively. To test for complementation of the sporulation defect of CGB225, we isolated a derivative of CGB225 in which spoIIIAFGH was inserted at amyE.
Sporulation efficiency test.
In order to determine the sporulation efficiency of B. subtilis strains, 5-ml cultures were incubated in DSM (supplemented with appropriate antibiotics for Campbell insertion strains) for approximately 48 h at 37°C (for 24 h at 37°C for point mutants). Aliquots of each culture were heated at 80°C for 10 min, serially diluted, and plated on LB agar (with appropriate antibiotics for Campbell strains). Unheated aliquots were also serially diluted and plated. Colonies were counted after 16 h of incubation at 37°C.
β-Galactosidase assays.
Overnight LB cultures (incubated at 30°C for point mutants and at 37°C for other strains) were diluted 1:100 in fresh DSM with 5 μg/ml chloramphenicol (100 μg/ml spectinomycin for CGB197 and its derivatives) and allowed to grow for 2 h at 37°C. Two 300-μl aliquots were collected every 0.5 h until 6 h after the onset of sporulation (usually 8 to 8.5 h); the first aliquot was used to measure the optical density at 600 nm, while the second aliquot was spun down and the cell pellet was stored at −80°C until it was assayed for β-galactosidase activity (5).
RNA isolation.
RNA was isolated from sporulating B. subtilis strains 4 h after the onset of sporulation using the Epicenter MasterPure RNA purification kit protocol (catalog no. MCR85102; Epicenter Biotechnologies), with the following modifications: 2 ml of RNAprotect bacterial reagent (catalog no. 76506; QIAGEN) was added to 1 ml of a culture and incubated 5 min at room temperature before cells were harvested. Six hundred microliters of tissue and cell lysis solution was added to the cell pellets, which were resuspended by vortexing. The cell suspension was placed in a tube containing lysing matrix B (catalog no. 6911-100; MP Biomedical) and then lysed in a Bio101 Fastprep FP120 machine (MP Biomedical) three times at speed 6 for 45 s. The glass beads were spun down by centrifugation at 16,100 × g for 2 min, and 300 μl of supernatant was transferred to a clean tube. Proteinase K was added as described in the kit protocol instructions, and the protocol was followed until the DNase I digestion step (step C). At this point, we incubated samples for 30 min after DNase I addition and repeated the entire step C to ensure that all DNA was removed. RNA was quantitated by spectrophotometric analysis at 260 nm.
Primer extension.
Thirty picomoles of oligonucleotide primer lacZ-rev2 (Table 3) (100 pmol/μl) was end labeled with [γ-32P]ATP (Amersham Biosciences) according to the protocol for an Epicenter SequiTherm EXCEL II DNA sequencing kit (catalog no. SEM79020; Epicenter Biotechnologies), except that we incubated the reaction mixture for 60 min at 37°C and terminated the reaction by incubation at 70°C for 10 min. Labeled primer was purified over a MicroSpin G25 column (Amersham Biosciences) as described in the product manual. Six picomoles of purified labeled primer was added to 1.7 μg of total RNA from CGB173, CGB193, and CGB194. cDNA synthesis was performed according to the Invitrogen ThermoScript reverse transcription (RT)-PCR system (catalog no. 11146-024; Invitrogen) protocol using a cDNA synthesis temperature of 55°C.
TABLE 3.
Oligonucleotide primers used for PCR, sequencing, and mutagenesisa
| Primer | Sequence (5′ to 3′) |
|---|---|
| spoIIIAH-rev-EcoRI | CACACACAGAATTCTTTAGCGGGCTTTTTTCCCTCATTCTTA |
| spoIIIAH-for-BamHI | CACACACAGGATCCCTTAAAAAACAAACCGTTTGGCTATT |
| spoIIIAG-for-BamHI | CACACACAGGATCCAATAAAAACGGATTATGGAATGTA |
| spoIIIAF-for-BamHI | CACACACAGGATCCAGTTTTTTAACGGAATGGCTTACC |
| spoIIIAE-for-BamHI | CACACACAGGATCCGTAATCAGCGCCTCTTTACT |
| spoIIIAH-rev-BamHI | CACACACAGGATCCTTTAGCGGGCTTTTTTCCCTCATTC |
| spoIIIAH-for-EcoRI | CACACACAGAATTCCTTAAAAAACAAACCGTTTGGCTATT |
| spoIIIAG-for-EcoRI | CACACACAGAATTCAATAAAAACGGATTATGGAATGTA |
| spoIIIAF-for-EcoRI | CACACACAGAATTCAGTTTTTTAACGGAATGGCTTACC |
| spoIIIAE-for-EcoRI | CACACACAGAATTCGTAATCAGCGCCTCTTTACT |
| spoIIIAH-rev-fuse-BamHI | CACACACAGGATCCTAAGACTGAGCATTGTTAATAGCCAAA |
| spoIIIAG-rev-fuse-BamHI | CACACACAGGATCCTAAGAGACTGCTTTTTCAATACATTCCA |
| spoIIIAF-rev-BamHIA | CACACACAGGATCCTTTCATTGCCGACACTCTC |
| spoIIIAF-rev-BamHIB | CACACACAGGATCCTGGCCATATACACGCTGATTGTTTT |
| spoIIIAF-rev-BamHIC | CACACACACAGGATCCTCTCTGACTGCCCGTTTTT |
| spoIIIAE-rev-BamHI | CACACACAGGATCCATGAGAGACACAATGGCGAGA |
| spoIIIAF-for-EcoRIA | CACACACAGAATTCATGGTAGTCAGCCTGCTCTTGATT |
| spoIIIAF-for-EcoRIB | CACACACAGAATTCAAACGGGCAGTCAGAGTCT |
| spoIIIAF-for-EcoRIC | CACACACAGAATTCAATGGCTGTCCAACTAAA |
| lacZ-rev2 | ATCTTACGTCAGTAACTTCCACAGT |
| PspoIIIA-for-EcoRI | CACACACAGAATTCACGGCAGCAATTGTCATGCTTGTGA |
| PspoIIIA-rev-BamHI | CACACACAGGATCCATTGGCTTCTTTAAAATGTATGATGTGAG |
| spoIIIAF-for-EcoRV | CACACACAGATATCTCACGATGATGATGAAATGAAGGA |
| spoIIIAF-rev-BamHI | CACACACAGGATCCGAAGAGACTGCTTTTTCAATACAT |
| spoIIIAE-for-EcoRV | CACACACAGATATCTGTCATAACCGAAAGGAGGCGGTA |
| spoIIIAE-rev-BamHI | CACACACAGGATCCACAACACGAATGGTGGTAAGC |
| spoIIIAD-for-EcoRV | CACACACAGATATCAGCTGTGTTTTTATTCCAAGGATAGG |
| spoIIIAD-rev-BamHI | CACACACAGGATCCTTCAATCTACCGCCTCCTTT |
| spoIIIAC-for-EcoRV | CACACACAGATATCGATGTAAACGTGAGGGGAGCAA |
| spoIIIAC-rev-BamHI | CACACACAGGATCCAATGTCAATCTGCAAGCCCCCCTAT |
| spoIIIAB-for-EcoRV | CACACACAGATATCAAATTTATGACAAAGACGGAAATGTG |
| spoIIIAB-rev-BamHI | CACACACAGGATCCTTTATTTTTGCTCCCCTCACGTTA |
| gerR-for-del-EcoRV | CACACACAGATATCCTGAACAAGAAAAAGGAGCTGCTC |
| gerR-rev-del-EcoRI | CACACACAGAATTCAGTCCAAGCATCTTGTCTTGTAATGGT |
| gerR-for-del-BamHI | CACACACAGGATCCATGAAAAAAGCGGCTCAAGAA |
| gerR-rev-del-SalI | CACACACAGTCGACTCAAAGAATACGGCATTCAGGA |
| spoIIID-for-del-EcoRI | CACACACAGAATTCGAGTCATTGGTCCGATCGTA |
| spoIIID-rev-del-BamHI | CACACACAGGATCCTGTTCGCTCTTTGATGTAATCGTGCA |
| spoIIID-for-del-PstI | CACACACACTGCAGGAAGGAGAGCCTGTTCAGCAAT |
| spoIIID-rev-del-SphI | CACACACAGCATGCTTTGAGAACAGGCCTTTTACA |
| sigE-for-del-EcoRV | CACACACAGATATCGTCAGCGTGCAGGCAGATT |
| sigE-rev-del-EcoRI | CACACACAGAATTCCAGCAGCTTATACCAGAGGTGCGTCAA |
| sigE-for-del-BamHI | CACACACAGGATCCAAAGAGTTCAACAAAATGGTG |
| sigE-rev-del-SalI | CACACACAGTCGACTTGTAAGCGATGTCCC |
| sigK-for-del-EagI | CACACACACGGCCGACAATAAACAGCACTCTGGTACC |
| sigK-rev-del-BamHI | CACACACAGGATCCAACAAAGCCGAGCGCTGCGAAAA |
| sigK-for-del-EcoRI | CACACACAGAATTCATCTTCAAGAGTTAAGTTATCGCACCGAT |
| sigK-rev-del-HindIII | CACACACAAAGCTTGGTTTGGTTAGATGCGGAAAATGC |
| AF35for | CAAAAAAAATAGAAACACAAGCTTCCCAGCG |
| AF35rev | CGCTGGGAAGCTTGTGTTTCTATTTTTTTTG |
| AF10for | CAAGCTTCCCAGCGCGGTTATATTCTAGAAGAAATGG |
| AF10rev | CCATTTCTTCTAGAATATAACCGCGCTGGGAAGCTTG |
| sspE-for-HindIII | CACACACAAAGCTTACGCATGGTCGAAATTAAAGAC |
| sspE-rev-BamHI | CACACACAGGATCCTGTTATCACCTCCACGGTCA |
Restriction enzyme sites are indicated by bold type, and base changes in QuikChange primers are indicated by underlining.
For sequencing, 1.5 pmol of purified labeled primer was used along with 50 fmol of pCG150 according to the Epicenter SequiTherm EXCEL II DNA sequencing kit (catalog no. SEM79020; Epicenter Biotechnologies) protocol, except that during cycle sequencing we added a 30-s annealing step at 55°C. Before electrophoresis on a 6% polyacrylamide gel with 6 M urea, loading buffer was added to all samples and the samples were heated at 90°C for 10 min. Samples were electrophoresed in 1× Tris-borate-EDTA, (pH 8.3) at 65 W for 2 to 2.5 h or until the xylene cyanol had just run off the gel. The gel was dried and exposed overnight to an Amersham Biosciences phosphor screen.
Site-directed mutagenesis.
Point mutations were introduced as described in the manual for a Stratagene QuikChange site-directed mutagenesis kit (catalog no. 200518; Stratagene) with the following PCR cycle parameters: one cycle of 95°C for 1 min, followed by 18 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 7 min and a final extension step of 10 min at 68°C. One microliter of DpnI was added to each reaction mixture, and the tubes were incubated at 37°C for 1 to 2 h. Five to seven microliters of each reaction mixture was then transformed into One Shot Top10 chemically competent cells (Invitrogen) as described in the product manual. The entire transformation reaction mixture was plated on LB plates containing 100 μg/ml ampicillin. Individual colonies were grown in liquid culture, and plasmid DNA was isolated (QIAGEN QIAprep spin miniprep kit) and sequenced to ensure that the desired mutation was present.
RT-PCR.
RNA was prepared from wild-type B. subtilis JH642 at 4 h after the onset of sporulation as described above and used to make cDNA as described above, except that primer spoIIIAF-rev-BamHIB was used. PCR was then performed using primers spoIIIAF-for-EcoRIA and spoIIIAF-for-EcoRIC with spoIIIAF-rev-BamHIB and Platinum Taq high-fidelity DNA polymerase (Invitrogen) for 30 cycles according to the ThermoScript RT-PCR system protocol (Invitrogen). PCR products were electrophoresed on a 1.5% agarose gel. A negative control containing no reverse transcriptase was included for each primer set.
RESULTS
Second promoter within the spoIIIA locus.
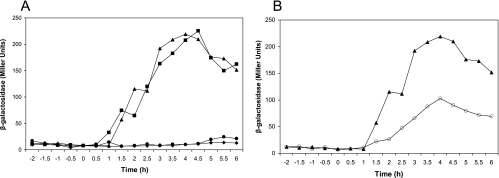
To search for the promoter that drives expression of spoIIIAG and spoIIIAH independent of the upstream spoIIIA genes, we tested the ability of fragments of the spoIIIA operon inserted into the chromosome at amyE to complement the sporulation-defective phenotypes of RL2045 and RL2046, which have in-frame nonpolar deletions of spoIIIAG and spoIIIAH, respectively. Only the fragments of DNA which contained DNA extending from spoIIIAF through spoIIIAH were able to complement mutations in spoIIIAG or spoIIIAH (Table 4). Since a region in spoIIIAF was required for complementation of spoIIIAH and spoIIIAG mutants, we hypothesized that a promoter is located within spoIIIAF. To test this hypothesis, the same DNA fragments of the spoIIIA locus were transcriptionally fused to lacZ and inserted into the amyE locus. We then assayed β-galactosidase activity during growth and sporulation. In agreement with the complementation experiments, we found that only the fragments of DNA that extended into spoIIIAF were able to drive production of β-galactosidase (Fig. 1A). In these strains, β-galactosidase activity began to accumulate about 1.5 h after the onset of sporulation, and maximum activity was reached approximately 2.5 h later (Fig. 1A). This temporal pattern of β-galactosidase activity was similar to that observed when P1spoIIIA was fused to lacZ (Fig. 1B). However, the promoter within spoIIIAF, P2spoIIIA, produced twice as much β-galactosidase activity as P1spoIIIA (Fig. 1B).
TABLE 4.
Complementation of ΔspoIIIAG and ΔspoIIIAH by spoIIIA fragments
| DNA fragment at amyE | ΔspoIIIAGa
|
ΔspoIIIAHb
|
||
|---|---|---|---|---|
| No. of heat-resistant spores formed/ml | Complementation (% relative to wild type) | No. of heat-resistant spores formed/ml | Complementation (% relative to wild type) | |
| spoIIIAH | 1.1 × 104 | 0.02 | 2.2 × 104 | 0.03 |
| spoIIIAGH | 2.9 × 104 | 0.05 | 3.9 × 103 | 0.002 |
| spoIIIAFGH | 1.3 × 108 | 99 | 1.1 × 108 | 97 |
| spoIIIA′EFGH | 1.25 × 108 | 98 | 1.04 × 108 | 96 |
| spoIIIAFGH −12/−11c | 4.5 × 104 | 0.07 | 1.7 × 104 | 0.025 |
| P1spoIIIA-spoIIIAHd | NDe | ND | 6.3 × 104 | 0.09 |
spoIIIA fragments in RL2045 (ΔspoIIIAG) and strains CGB129, CGB130, CGB131, and CGB32.
spoIIIA fragments in RL2046 (ΔspoIIIAH) and strains CGB133, CGB134, CGB135, and CGB136.
spoIIIAFGH fragment with base substitutions at positions −12 and −11 of P2spoIIIA (strains CGB208 and CGB209).
Fusion in only RL2046. spoIIIAH was directly expressed from P1spoIIIA at amyE.
ND, not determined.
FIG. 1.
Expression of spoIIIA-lacZ fusions. (A) β-Galactosidase accumulation in cultures of strains containing various regions of the spoIIIA locus, including spoIIIAH (⧫), spoIIIAGH (•), spoIIIAFGH (▴), and spoIIIA′EFGH (▪), transcriptionally fused to lacZ. (B) Comparison of P1spoIIIA and P2spoIIIA activities. β-Galactosidase activity was measured in strains containing P1spoIIIA (○) and spoIIIAFGH, which contains P2spoIIIA (▴), transcriptionally fused to lacZ. The data are the averages of three replicates. The x axis indicates the time (in hours) before and after the onset of sporulation (zero time).
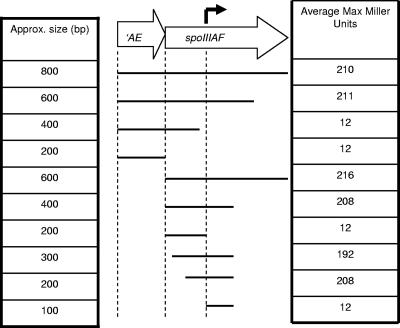
To more precisely locate P2spoIIIA, we examined the effect of sequential deletions from the 3′ end of spoIIIAF on β-galactosidase activity (Fig. 2). A fragment that included the first 415 bp of spoIIIAF directed transcription of lacZ and produced as much β-galactosidase activity as the 800-bp fragment. Sequential deletions were then made from the 5′ end of spoIIIAF, and we found that a fragment containing only about 200 bp of spoIIIAF directed expression of lacZ at the same level as the 800-bp fragment (Fig. 2). Therefore, the new promoter P2spoIIIA was located between bp 200 and 415 of spoIIIAF.
FIG. 2.
Deletion analysis of the P2spoIIIA region. The large arrows indicate the region of the spoIIIA locus that contains P2spoIIIA. The solid lines show the regions of the spoIIIA locus (approximate sizes are indicated on the left) that were transcriptionally fused to lacZ for β-galactosidase assays. The maximum accumulation of β-galactosidase for strains containing the fusion is indicated on the right (averages of three replicates).
σE directs transcription from P2spoIIIA.
SpoIIIAH accumulates in the mother cell (1), and transcriptional array data indicate that its transcription is dependent on the mother cell-specific sigma factor σE (8, 19). Therefore, we examined the effects of mutations in genes responsible for both early and late mother cell gene expression on the activity of P2spoIIIA. The early transcriptional regulators GerR and SpoIIID and secondary sigma factor σE and the late transcription factor GerE and secondary sigma factor σK were inactivated by insertion of antibiotic cassettes. The effects of these mutations on expression of P1spoIIIA-lacZ and P2spoIIIA-lacZ were monitored during sporulation. Both promoters were found to be regulated by the same factors. σE was solely responsible for transcription of the promoters since very little β-galactosidase accumulated in the sigE mutant, and SpoIIID was responsible for repression of the promoters; in fact, in the absence of SpoIIID, twice as much β-galactosidase was produced from P2spoIIIA (Fig. 3 and data not shown). There were no significant effects on β-galactosidase activity when GerR, GerE, or σK was absent (Fig. 3).
FIG. 3.
Regulation of P2spoIIIA: expression of a P2spoIIIA-lacZ fusion in the wild type (▴) and mutant strains, including gerR (▪), spoIIID (⧫), sigE (•), gerE (□), and sigK (○) mutants. The data are averages of three replicates.
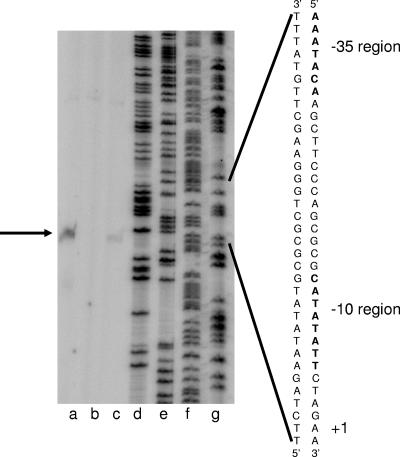
We used primer extension to determine the transcription start site of P2spoIIIA. RNA was harvested 4 h after the onset of sporulation from cultures of wild-type, ΔsigE, and ΔspoIIID strains harboring the P2spoIIIA-lacZ fusion at amyE. We found that the 5′ end of the major transcription product mapped to bp 289 of spoIIIAF (Fig. 4). The effects on P2spoIIIA transcription of the mutations in sigE, which encodes σE, and spoIIID were the same as those seen in the β-galactosidase assays (Fig. 4); essentially, there was no transcript in the sigE deletion strain and there was about twice as much transcript in the spoIIID deletion strain.
FIG. 4.
Primer extension of analysis P2spoIIIA. Total RNA was isolated 4 h after the onset of sporulation from three B. subtilis strains (ΔspoIIID [lane a], ΔsigE [lane b], and wild type [lane c]) harboring a P2spoIIIA-lacZ fusion at amyE. The primer extension product and putative transcription start point are indicated by an arrow on the left and by +1 on the right. The −35 and −10 regions are also indicated to the right of the DNA sequence of the expanded region. Lanes d, e, f, and g contained the dideoxy sequencing reactions for G, A, T, and C, respectively; the same primer was used for sequencing and for primer extension.
Inspection of the sequence upstream from the putative P2spoIIIA start point of transcription revealed the sequences 5′AAATACA and 5′CATATATT (Fig. 5) in the −35 and −10 regions, respectively, that are similar to the consensus sequences in the −35 and −10 regions of σE-dependent promoters (7), especially in the most highly conserved sequence in the −35 region (ATA) and all of the −10 region. Since transcription from P2spoIIIA was dependent on σE (Fig. 3 and 4), we expected that the highly conserved −10 and −35 region sequences would play important roles in promoter activity if the 5′ end of the transcript indicated in our primer extension assays represented the actual start point of transcription. Therefore, we examined the effects of point mutations in the conserved sequences. The critical T residue in the −35 region (position −30) was changed to the nonconsensus base C, and the CA nucleotide pair in the −10 region (positions −12 and −11) was mutated to the nonconsensus nucleotide pair GT (Fig. 5). Both of these mutations caused complete inactivation of P2spoIIIA (data not shown).
FIG. 5.
Sequence of the P2spoIIIA region. The nontranscribed strand encompassing P2spoIIIA from B. subtilis (B. s.), Oceanobacillus iheyensis (O. i.), Bacillus cereus (B. c.), Bacillus thuringiensis (B. t.), Bacillus anthracis Ames (B. a. A.), Bacillus licheniformis (B. l.), and Geobacillus thermodenitrificans (G. t.) is shown. The −35 and −10 regions are indicated by boldface type. The start point of transcription is indicated by +1, and positions −30 and −12 are indicated above the sequence. The underlined sequence is one potential SpoIIID binding site in which five of eight base pairs match the consensus sequence (8).
The effects of mutations in the −10 and −35 regions of P2spoIIIA demonstrated that this promoter was responsible for the σE-dependent expression of the lacZ fusions described above, but we also tested whether this promoter was responsible for the expression of spoIIIAG and spoIIIAH at the amyE locus in the complementation experiments described above. We inserted the spoIIIAFGH fragment containing the −10 point mutations into the amyE locus and tested its ability to complement the spoIIIAG and spoIIIAH deletion mutants. We found that the mutant allele did not complement the sporulation defect in these strains (Table 4). Therefore, P2spoIIIA is required for expression of spoIIIAG and spoIIIAH in these strains.
P1spoIIIA plays no role in spoIIIAG and spoIIIAH expression.
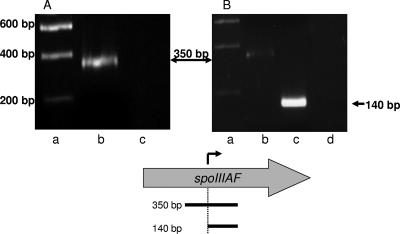
All of our results are consistent with the hypothesis that σE directs transcription from the P2spoIIIA promoter and that this transcription probably is sufficient for spoIIIAG and spoIIIAH expression. P2spoIIIA is located within spoIIIAF; therefore, it seemed likely that transcription initiated at P1spoIIIA, which is required for spoIIIAF expression, would continue through P2spoIIIA and into spoIIIAG and spoIIIAH. We used RT-PCR to determine whether this read-through occurred. RNA was isolated from wild-type strain JH642 4 h after the onset of sporulation, and RT-PCR was performed for two regions near P2spoIIIA: a 350-bp region that extended 180 bp upstream of P2spoIIIA to 150 bp downstream and a 140-bp region immediately downstream of P2spoIIIA. After 30 cycles, the PCR products were electrophoresed on a 1.5% agarose gel. We detected the 350-bp product indicative of the read-through transcript (Fig. 6A). The 140-bp RT-PCR product that resulted from transcription from both P1spoIIIA and P2spoIIIA was much more abundant than the 350-bp product that represented transcription from only P1spoIIIA (Fig. 6B).
FIG. 6.
RT-PCR analysis of transcripts in the P2spoIIIA region. RT-PCR was performed using two different forward primers in order to compare the amounts of transcript generated by P1spoIIIA alone and by P1spoIIIA and P2spoIIIA together. Each panel shows the ethidium bromide-stained DNA after electrophoresis in agarose. Lane a contained molecular weight standards in both panels. Lane b in both panels contained the 350-bp RT-PCR product of the transcript from P1spoIIIA, whereas lane c in panel B contained the 140-bp RT-PCR product from transcripts originating from both P1spoIIIA and P2spoIIIA. Lane c in panel A and lane d in panel B contained the products from control reactions in which no reverse transcriptase was added. Below panels A and B is a map of spoIIIAF. The arrow above spoIIIAF and the dotted vertical line indicate the P2spoIIIA start point of transcription. The horizontal lines below spoIIIAF indicate the 350- and 140-bp cDNA amplified from the P1spoIIIA and P2spoIIIA transcripts, respectively.
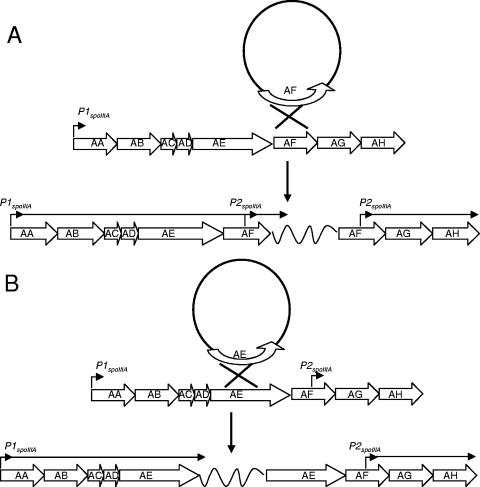
Although our complementation studies, in which we expressed spoIIIAG and spoIIIAH at an ectopic location, indicated that P2spoIIIA was necessary and sufficient for the expression of spoIIIAG and spoIIIAH, the RT-PCR results showing that transcripts from P1spoIIIA read into spoIIIAG raised the possibility, which was unlikely, that transcription originating from P1spoIIIA of spoIIIAG and spoIIIAH at the spoIIIA locus may enhance their expression and thus affect sporulation. Therefore, we isolated strains with Campbell-type insertions that would separate spoIIIAG and spoIIIAH from P1spoIIIA (Fig. 7A). A strain harboring the insertion in spoIIIAF, CGB175, exhibited almost wild-type levels of sporulation (99% of the wild-type levels). We also examined the effect of this Campbell-type insertion on expression of a σG-dependent sspE-lacZ fusion and found no effect on its expression, indicating that there were no subtle effects on σG activation (data not shown). As a control for these experiments, we also isolated a strain with an insertion in spoIIIAE, which resulted in separation of P1spoIIIA from spoIIIAF and downstream genes (Fig. 7B). This insertion reduced sporulation to 0.05% of that seen with a wild-type strain, indicating that the inserted sequences blocked transcription. Evidently, transcripts from P1spoIIIA read into spoIIIAG and spoIIIAH since they traverse P2spoIIIA; however, they are not necessary to support sporulation. We also used the Campbell insertion technique to search for other potential promoters in the spoIIIA locus, but strains bearing such insertions in spoIIIAD, spoIIIAC, and spoIIIAB were sporulation defective (the sporulation efficiencies were 0.03 to 0.07% of the wild-type sporulation efficiencies), indicating that there are no other promoters (data not shown). This, along with our previous complementation results (Fig. 1A and B), indicated that transcription of spoIIIAG and spoIIIAH from P2spoIIIA alone is sufficient for sporulation.
FIG. 7.
Separation of spoIIIAG and spoIIIAH from P1spoIIIA: models of the Campbell-type integration of plasmids into the spoIIIA locus. (A) Model in which a nonreplicating circular plasmid carrying spoIIIAF integrates into the spoIIIA locus by a single homologous recombination event. A horizontal arrow indicates that transcription from P1spoIIIA proceeds uninterrupted through spoIIIAF, while spoIIIAG and spoIIIAH are transcribed from P2spoIIIA. (B) Model in which a nonreplicating circular plasmid carrying spoIIIAE integrates into the spoIIIA locus by a single homologous recombination event. A horizontal arrow indicates that transcription from P1spoIIIA is interrupted by plasmid sequences (wavy line) before entering spoIIIAF, resulting in a sporulation-defective phenotype. The intervening plasmid DNA is represented by a wavy line in both panels and is not to scale.
P2spoIIIA is essential for sporulation.
The results of complementation tests indicated that transcription of spoIIIAG and spoIIIAH from P2spoIIIA is sufficient when these genes are located at the ectopic amyE locus. At the chromosomal spoIIIA locus, transcription of spoIIIAH from P1spoIIIA is not essential (1), so we predicted that transcription of both spoIIIAG and spoIIIAH from P2spoIIIA at the spoIIIA locus is essential for sporulation. Therefore, we isolated a strain (CGB225) containing two base pair substitutions at positions −12 and −11 of P2spoIIIA within the spoIIIA locus. These mutations resulted in a sporulation-deficient phenotype (0.025 to 0.7% of the wild-type sporulation level) and also reduced the expression of a σG-dependent sspE-lacZ fusion (CGB226) to levels that were less than 30% of the wild-type levels (data not shown). We noted that the mutations in P2spoIIIA resulted in a change in the coding region of spoIIIAF by producing an alanine-to-glycine substitution in SpoIIIAF. However, this amino acid substitution was not responsible for the sporulation-defective phenotype since spore formation was complemented to wild-type levels by expression of spoIIIAG and spoIIIAH from the P2spoIIIA promoter at amyE in strain CGB227, a derivative of CGB225.
DISCUSSION
Our results indicate that the spoIIIA locus, which encodes eight genes, is transcribed from two promoters, P1spoIIIA, which is located at the start of the locus, and P2spoIIIA, which is located within the open reading frame of spoIIIAF. P2spoIIIA is required for transcription of spoIIIAG and spoIIIAH. RT-PCR analysis showed that transcripts from P1spoIIIA probably read through spoIIIAG and spoIIIAH, but the P1spoIIIA-dependent transcripts may not produce enough spoIIIAG and spoIIIAH products. This conclusion follows from our observations that P2spoIIIA-lacZ was expressed at twice the levels of P1spoIIIA-lacZ (Fig. 1B) and that expression of a P1spoIIIA-spoIIIAH fusion was not sufficient to complement the sporulation defect of a mutant spoIIIAH strain (Table 4). Furthermore, expression of spoIIIAG and spoIIIAH from P2spoIIIA is sufficient to support sporulation, since P2spoIIIA-driven expression of spoIIIAG and spoIIIAH complements from an ectopic location (Table 4), and an insertion physically separating spoIIIAG and spoIIIAH from P1spoIIIA had no detectable effect on sporulation efficiency or on the timing of σG activation (data not shown). Moreover, mutation of P2spoIIIA at the spoIIIA locus caused a sporulation-defective phenotype that could be complemented by expression of spoIIIAG and spoIIIAH at the amyE locus.
There are many examples of complex operons containing more than one promoter in both gram-positive and gram-negative bacteria (3, 14, 16, 24). However, the most common reason for having more than one promoter for an operon is to ensure its expression under different growth conditions or requirements. The promoters of these operons are usually recognized by different sigma factors or may require other types of different transcription factors for expression. Therefore, a potential explanation for the necessity of a second promoter within the spoIIIA locus could be that P1spoIIIA and P2spoIIIA are differentially regulated. However, this possibility is unlikely because transcription from both promoters appears to be directly dependent on σE and the activities of both promoters are repressed by SpoIIID. We found at least two potential SpoIIID binding sites centered at position −22 relative to the P2spoIIIA transcription start site (bp 301 of spoIIIAF) (Fig. 5) and at position 11 (bp 268 of spoIIIAF) (not shown). These conserved sequences and the elevated expression from P2spoIIIA that we observed in the spoIIID mutant (Fig. 3 and 4) are consistent with a model in which SpoIIID directly represses P2spoIIIA activity, as is the case for P1spoIIIA (8, 13). Therefore, it is unlikely that the two promoters in the spoIIIA locus are differentially regulated; rather, it is likely that the primary purpose of P2spoIIIA has to do only with increasing the expression of the last two genes of the locus.
Although the reason that a second promoter in the locus, P2spoIIIA, is required is not entirely clear, this promoter may be a conserved feature within the spoIIIA locus of other spore formers (Fig. 5). The hypothesis that the second promoter in the locus, P2spoIIIA, is required because higher levels of SpoIIIAG and SpoIIIAH than of the upstream spoIIIA products are required for sporulation led us to speculate that SpoIIIAG and SpoIIIAH play different types of roles in sporulation than the other members of the locus. This idea is supported by topology predictions of SpoIIIAG and SpoIIIAH that show that these proteins are more similar to one another than to any other protein encoded by the spoIIIA locus (4, 20, 22). SpoIIIAH is known to recruit other proteins involved in intercellular signaling to the septum separating the mother cell and forespore (6). SpoIIIAH has also been postulated to help drive the engulfment of the forespore (2). The SpoIIIA proteins encoded by upstream genes are also essential for spore formation and, like SpoIIIAH and SpoIIIAG, are required for activation of σG (15). However, these proteins, SpoIIIAA to SpoIIIAF, may act catalytically or at least at lower stoichiometries than SpoIIIAG and SpoIIIAH.
Acknowledgments
We gratefully acknowledge Adriano Henriques, Bill Shafer, and Gordon Churchward for their suggestions on this work.
This work was supported by Public Health Service grant GM54395 from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Blaylock, B., X. Jiang, A. Rubio, C. P. Moran, Jr., and K. Pogliano. 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 18:2916-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broder, D. H., and K. Pogliano. 2006. Forespore engulfment mediated by a ratchet-like mechanism. Cell 126:917-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter, H. L., L. F. Wang, R. H. Doi, and C. P. Moran, Jr. 1988. rpoD operon promoter used by sigma H-RNA polymerase in Bacillus subtilis. J. Bacteriol. 170:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in procariotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 5.Cutting, S. M., and C. Harwood. 1990. Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 6.Doan, T., K. A. Marquis, and D. Z. Rudner. 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 55:1767-1781. [DOI] [PubMed] [Google Scholar]
- 7.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 8.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 10.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 11.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 12.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Illing, N., and J. Errington. 1991. The spoIIIA operon of Bacillus subtilis defines a new temporal class of mother-cell-specific sporulation genes under the control of the sigma E form of RNA polymerase. Mol. Microbiol. 5:1927-1940. [DOI] [PubMed] [Google Scholar]
- 14.Kajitani, M., and A. Ishihama. 1984. Promoter selectivity of Escherichia coli RNA polymerase. Differential stringent control of the multiple promoters from ribosomal RNA and protein operons. J. Biol. Chem. 259:1951-1957. [PubMed] [Google Scholar]
- 15.Kellner, E. M., A. Decatur, and C. P. Moran, Jr. 1996. Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol. Microbiol. 21:913-924. [DOI] [PubMed] [Google Scholar]
- 16.Maxson, M. E., and A. J. Darwin. 2006. Multiple promoters control expression of the Yersinia enterocolitica phage-shock-protein A (pspA) operon. Microbiology 152:1001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579-586. [DOI] [PubMed] [Google Scholar]
- 18.Rubio, A., and K. Pogliano. 2004. Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis. EMBO J. 23:1636-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steil, L., M. Serrano, A. O. Henriques, and U. Volker. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 215:399-420. [DOI] [PubMed] [Google Scholar]
- 19a.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 20.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 21.Unniraman, S., M. Chatterji, and V. Nagaraja. 2002. DNA gyrase genes in Mycobacterium tuberculosis: a single operon driven by multiple promoters. J. Bacteriol. 184:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Heijne, G. 1992. Membrane protein structure prediction: hydrophobicity analysis and the ‘Positive Inside’ rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 23.Wang, S. T., B. Setlow, E. M. Conlon, J. L. Lyon, D. Imamura, T. Sato, P. Setlow, R. Losick, and P. Eichenberger. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16-37. [DOI] [PubMed] [Google Scholar]
- 24.Yang, B., and T. J. Larson. 1998. Multiple promoters are responsible for transcription of the glpEGR operon of Escherichia coli K-12. Biochim. Biophys. Acta 1396:114-126. [DOI] [PubMed] [Google Scholar]