Abstract
Methanosarcina acetivorans, a strictly anaerobic methane-producing species belonging to the domain Archaea, contains a gene cluster annotated with homologs encoding oxidative stress proteins. One of the genes (MA3736) is annotated as a gene encoding an uncharacterized carboxymuconolactone decarboxylase, an enzyme required for aerobic growth with aromatic compounds by species in the domain Bacteria. Methane-producing species are not known to utilize aromatic compounds, suggesting that MA3736 is incorrectly annotated. The product of MA3736, overproduced in Escherichia coli, had protein disulfide reductase activity dependent on a C67XXC70 motif not found in carboxymuconolactone decarboxylase. We propose that MA3736 be renamed mdrA (methanosarcina disulfide reductase). Further, unlike carboxymuconolactone decarboxylase, MdrA contained an Fe-S cluster. Binding of the Fe-S cluster was dependent on essential cysteines C67 and C70, while cysteines C39 and C107 were not required. Loss of the Fe-S cluster resulted in conversion of MdrA from an inactive hexamer to a trimer with protein disulfide reductase activity. The data suggest that MdrA is the prototype of a previously unrecognized protein disulfide reductase family which contains an intermolecular Fe-S cluster that controls oligomerization as a mechanism to regulate protein disulfide reductase activity.
The oxidative stress defense mechanisms utilized by prokaryotes of the domain Bacteria are well understood (61). Considerably less is known about these mechanisms in members of the domain Archaea, including the strictly anaerobic methane-producing archaea (methanoarchaea). It has been documented that Methanosarcina and Methanobrevibacter species are aerotolerant (34, 38). Methanosarcina barkeri survives exposure to air and resumes growth immediately after a return to anaerobiosis (20, 67), suggesting that it mounts a substantial defense against oxidative stress. An iron superoxide dismutase and catalase have been characterized from M. barkeri (7, 58). Recently, an iron-sulfur flavoprotein (Isf) from Methanosarcina thermophila was shown to reduce O2 and H2O2 to water (13). The sequenced genomes of Methanosarcina species (14, 22) contain homologs of genes encoding superoxide reductase and rubrerythrin, proteins unique to anaerobes that reduce superoxide and hydrogen peroxide, respectively, and have been characterized from other strict anaerobes (12, 25, 31, 46, 64). The genome annotations also include homologs of genes encoding flavoprotein A (FprA), which reduces O2 to water (56).
RC-IMRE50 is an uncultured methanoarchaeon closely related to Methanosarcina species and is a representative of the rice cluster I (RC-I) methanoarchaea, which are the predominant methanoarchaea in the rice rhizosphere (11, 16). The RC-IMRE50 group is the primary contributor to methane emissions from rice fields, which are estimated to contribute 10 to 25% of the global methane emissions to the atmosphere (17). The recent sequencing of the RC-IMRE50 genome revealed genes encoding homologs of antioxidant enzymes, including superoxide dismutase, superoxide reductase, catalase, rubrerythrin, FprA, and peroxiredoxins. Thus, it has been suggested that aerotolerance is a key component of the competitive superiority of RC-IMRE50, allowing survival during transient oxic conditions associated with life in the rhizosphere (17). The genome of Methanosarcina acetivorans, a marine methanoarchaeon phylogenetically related to RC-IMRE50 (16), also contains homologs of genes encoding antioxidant enzymes similar to those found in RC-IMRE50 (17, 22), suggesting that M. acetivorans can also survive transient oxic conditions found in the kelp bed sediment from which it was isolated (60). To date, attempts to obtain RC-I organisms in pure culture have not been successful. M. acetivorans has a robust genetic system (49, 66), making this organism an attractive model for studying the specific function of the annotated antioxidant genes and for discovering additional genes important for aerotolerance of Methanosarcina and related species, including RC-IMRE50.
Here we show that the genome of M. acetivorans contains a 10-gene transcriptional unit annotated with homologs of genes encoding superoxide reductase, FprA, and Isf. MA3736 in the cotranscribed gene cluster is annotated as a gene encoding carboxymuconolactone decarboxylase (CMD), an enzyme essential in aerobic species in the domain Bacteria utilizing aromatic compounds as growth substrates (18, 52). Methanogens are strictly anaerobic, and none are known to metabolize aromatic compounds for growth (68), suggesting that MA3736 is annotated incorrectly. We overproduced the MA3736 product in Escherichia coli and found that the purified product had protein disulfide reductase activity dependent on a CXXC motif typical of protein disulfide reductases. Unexpectedly, the MA3736 product was found to contain an Fe-S cluster(s) with binding also dependent on the CXXC motif. Loss of the Fe-S cluster(s) was necessary for protein disulfide reductase activity. We propose that MA3736 is distinct from CMD and should be renamed mdrA (methanosarcina disulfide reductase).
MATERIALS AND METHODS
RT-PCR analysis.
Sequence information for M. acetivorans, Methanosarcina mazei, and M. barkeri was obtained from The Institute for Genomic Research (http://www.tigr.org), and sequence information for Methanococcoides burtonii was obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Total RNA was isolated from methanol-grown M. acetivorans, and reverse transcription (RT)-PCR analysis of the gene cluster containing MA4664 and MA3734 to MA3743 (designated the MA4664/MA3734-MA3743 cluster) was performed as described previously (43). The primer sequences used are listed in Table S1 in the supplemental material.
Cloning, expression, and purification of MdrA.
The gene encoding MdrA was amplified from M. acetivorans genomic DNA by PCR. The PCR-amplified DNA fragment was cloned into the pTYB12 vector from an IMPACT T7 kit (New England Biolabs), generating plasmid pDJL200. pDJL200 contains the chitin-binding domain (CBD)-intein-MdrA fusion.
The CBD-intein-MdrA fusion was overproduced in E. coli Rosetta (DE3)(pLacI) cells transformed with pDJL200. Cells were grown in Terrific broth at 37°C with shaking at 250 rpm until an optical density at 600 nm of 0.5 to 0.7 was reached, at which time the growth temperature was adjusted to 16°C. After 30 min the culture was induced with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) and then harvested by centrifugation 16 h after induction. All subsequent purification procedures were performed anaerobically using an anaerobic chamber (Coy Laboratory Products) containing an atmosphere of 95% N2 and 5% H2. Approximately 15 g (wet weight) of cells was suspended in 20 ml of 50 mM HEPES (pH 7.5) containing 300 mM NaCl and 2 mM benzamidine. The cells were lysed by two passages through a French pressure cell at 138 MPa. The lysate was centrifuged at 74,000 × g for 30 min at 4°C. The supernatant solution containing the CBD-intein-MdrA fusion protein was filtered (pore size, 0.45 μm) and applied at a flow rate of 0.5 ml/min to a column containing 20 ml of chitin bead resin (New England Biolabs). The column was then washed with 200 ml of 50 mM HEPES (pH 7.5) containing 300 mM NaCl and 1% Triton X-100 at a flow rate of 2 ml/min. MdrA was cleaved from the CBD by flushing the column with 60 ml of 50 mM HEPES (pH 7.5) containing 300 mM NaCl and 40 mM dithiothreitol (DTT), followed by incubation of the column for 16 h at room temperature. MdrA was then eluted from the column with 60 ml of 50 mM HEPES (pH 7.5) containing 300 mM NaCl. The elute was concentrated to 2.5 ml using a Vivacell concentrator with a 10,000-molecular-weight cutoff under a nitrogen flow inside the anaerobic chamber. The concentrated protein was desalted with 3.5 ml of 50 mM HEPES (pH 7.5) containing 300 mM NaCl using a PD-10 column (Amersham Biosciences). The purity of MdrA was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. MdrA purified using this method contained one additional histidine residue in the N terminus.
MdrA variants were generated by site-directed mutagenesis with primers listed in Table S1 in the supplemental material, using a QuickChange site-directed mutagenesis kit (Stratagene). Each variant protein was purified as described above for wild-type MdrA.
Protein concentrations were determined by the method of Bradford (6), using bovine serum albumin as a standard.
Enzyme assays.
The protein disulfide reductase activity of MdrA was determined using the turbidimetric assay for insulin disulfide reduction described by Holmgren (30). For determination of DTT-dependent activity, the assay mixture contained 0.4 ml (final volume) of 100 mM potassium phosphate (pH 7.0), 0.13 mM insulin, 1 mM EDTA, and 0 to 10 μM MdrA. The reaction was initiated by addition of 0.33 mM DTT and was performed at 21°C. The absorbance at 650 nm was plotted against time. Assays were done in an anaerobic chamber (Coy). Activity was expressed as the ratio of the slope of a linear part of the turbidity curve to the lag time (reported as ΔA650/min2, 10−5), as described previously (48, 57). The lipoamide-dependent insulin disulfide reduction activity of MdrA was assayed with an assay similar to the DTT-dependent assays using NADH, lipoamide, and bovine lipoamide dehydrogenase (8, 30). The typical assay was performed anaerobically, and the assay mixture contained 100 mM potassium phosphate (pH 7.0), 1 mM EDTA, 0.13 mM bovine insulin, 0.4 U of lipoamide dehydrogenase, 50 μM lipoamide, and 0 to 10 μM MdrA. The reaction was initiated by addition of 0.5 mM NADH, and turbidity was monitored at 650 nm.
Characterization of chromophore content.
The iron and acid-labile sulfide content of MdrA was determined as previously described (4, 65). UV-visible spectra of MdrA and variants were recorded with a Beckman DU-7400 spectrophotometer inside an anaerobic chamber (Coy). The putative Fe-S cluster was removed by anaerobic incubation of MdrA with dithionite and 20 mM EDTA in 50 mM HEPES (pH 7.5) containing 300 mM NaCl for 2 h at 25°C. The protein was then desalted with a PD-10 column equilibrated with 50 mM HEPES (pH 7.5) containing 300 mM NaCl. The resulting form of MdrA is referred to as apo-MdrA below.
Size exclusion chromatography.
Estimates of the native molecular masses of MdrA and variants of MdrA were based on elution from a Sephacryl Hiprep S-200 gel filtration fast protein liquid chromatography column (Amersham Biosciences) using an AKTA explorer (Pharmacia Biotech). The column was calibrated with the following proteins having known molecular masses: β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). The buffer used was 50 mM HEPES (pH 7.5) containing 150 mM NaCl and 10 mM DTT to provide reducing conditions. A flow rate of 0.5 ml min−1 was used. Samples containing 0.5 to 0.6 mM protein were loaded onto the column. To determine the effect of EDTA on the oligomeric state of wild-type MdrA and cysteine variants of MdrA, proteins were incubated with 10 mM EDTA under anaerobic conditions at 25°C for 30 min prior to injection onto the column with 10 mM EDTA in the elution buffer.
Construction of a phylogenetic tree.
Database searches and alignments were carried out using BLAST and CLUSTALX. The output was edited with the Alignment Editor of MEGA (v3.1) (37). A phylogenetic tree was constructed with the MEGA package using the neighbor-joining method, including 500 bootstrap replicates. The accession numbers for all protein sequences used for the phylogenetic analysis are as follows: M. acetivorans MA3736, gi: 19917805; M. mazei Goe1 MM0631, gi: 20905023; uncultured RC-I methanogenic archaeon RCIX2594, gi: 110622368; Thermus thermophilus HB8 TTHA0727, gi: 55772109; Rhodococcus sp. strain RHA1 RHA1_ro11235, gi: 110825601; Mycobacterium tuberculosis H37Rv Rv1767, gi: 2131035; Thermotoga maritima MSB8 TM1620, gi: 15644368; Rhodopseudomonas palustris BisB18 RPC_4301, gi: 90107787; Lactobacillus sakei 23K LSA1776, gi: 78611031; Thermoanaerobacter tengcongensis MB4(T) TTE0299, gi: 20515286; R. palustris BisB18 RPC444, gi: 90107930; Legionella pneumophila Philadelphia 1 lpg2349, gi: 52629670; Streptomyces coelicolor A3(2) SCO5031, gi: 9967658; M. tuberculosis H37Rv Rv2429, gi: 1666155; Caulobacter crescentus CB15 CC_3698, gi: 13425462; Myxococcus xanthus DK 1622 MXAN_1563, gi: 108465278; Brucella abortus 9-941 BruAb2_0523, gi: 62197643; Corynebacterium diphtheriae NCTC13129 DIP1419, gi: 38200266; Ralstonia eutropha JMP134 Reut_A1364, gi: 72118471; Nocardia farcinica IFM10152 nfa37900, gi: 54017268; Cytophaga hutchinsonii ATCC 33406 CHU_3759, gi: 110282806; Acinetobacter sp. strain ADP1 ACIAD1710, gi: 49530840; Methanobacterium thermoautotrophicum delta H MTH234, gi: 2621282; M. acetivorans C2A MA0409, gi: 19914189; Sulfolobus acidocaldarius DSM 639 Saci_1814, gi: 68568191; M. tuberculosis H37Rv Rv0771, gi: 1550649; Rhodococcus sp. strain RHA1 RHA1_ro01338, gi: 110817878; Pseudomonas putida KT2440 PP_1381, gi: 24982843; Burkholderia xenovorans LB400 Bxe_B0647, gi: 91692108; S. coelicolor A3(2) SCO6339, gi: 3367745; R. palustris CGA009 RPA4740, gi: 39651658; and Shewanella oneidensis MR-1 SO_0083, gi: 24345456.
RESULTS
Analysis of the MA4664/MA3734-MA3743 gene cluster.
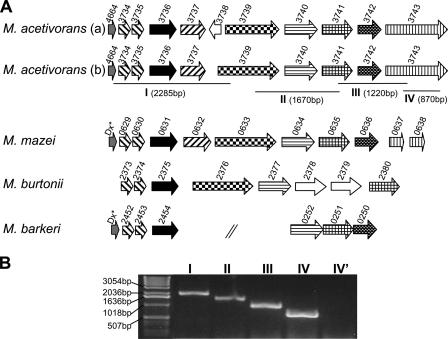
Similar to other Methanosarcina spp. (34), M. acetivorans can withstand prolonged exposure to atmospheric levels of O2 and resume growth once anaerobiosis is restored (data not shown), suggesting that this organism contains enzymes for protection from and/or repair of damage caused by reactive O2 species. Indeed, the MA4664/MA3734-MA3743 gene cluster (Fig. 1) contains homologs of genes encoding oxidative stress proteins that have been characterized from other strict anaerobes. This gene arrangement is similar to that of gene clusters in other sequenced Methanosarcina and related species (Fig. 1), suggesting that the gene products have an important function in these organisms. However, the original annotation of MA3739 appears to be incorrect, as the first 53 amino acids of the gene product are missing compared to the gene products of MM0633 and Mbur2376 (see Fig. S1 in the supplemental material). We propose that MA3739 starts at a codon that is within MA3738 previously annotated as divergently transcribed from MA3739, suggesting that MA3738 is not a functional open reading frame (Fig. 1). RT-PCR analysis of each intergenic region in the MA4664/MA3734-MA3743 gene cluster (data not shown) and across several genes (Fig. 1B) indicated that the genes are cotranscribed and further suggested that MA3738 is not a functional gene. Furthermore, the products of most of the genes (MA3735, MA3736, MA3737, MA3740, MA3741, MA3742, and MA3743) were detected at similar levels in CO-, acetate-, and methanol-grown cells by global proteomic analyses (41, 42), consistent with a physiological function for the encoded proteins.
FIG. 1.
Organization of the M. acetivorans MA4664/MA3734-MA3743 gene cluster and comparison to gene clusters in other Methanosarcina species. (A) The MA4664/MA3734-MA3743 gene organization shown in line a is the original annotation, and that shown in line b is the proposed annotation. MA4664/MA3734-MA3743 is compared to the following gene clusters from other sequenced methanogens: M. mazei Go1 MM0629 to MM0638, M. burtonii DSM 6242 Mbur2373 to Mbur2380, and M. barkeri strain Fusaro Mbar_A2452 to Mbar_A2454 and Mbar_A0252 to Mbar_A0250. The arrows indicate the gene direction and relative size and spacing. Homologous genes are indicated by the same pattern and are centered on MA3736 (indicated by the solid arrow). Genes indicated by asterisks in the M. mazei and M. barkeri gene clusters were missed in the original annotation and encode desulforedoxin (Dx) homologs similar to the MA4664 product. Mbur2378 and Mbur2379 encode homologs of flavodoxin and rubrerythrin, respectively. The genes in M. barkeri are not contiguous, as indicated by slashes. (B) RT-PCR analysis of the MA4664/MA3734-MA3743 gene cluster in M. acetivorans. Predicted RT-PCR products are indicated in panel A by lines under the genes and are labeled with roman numerals. Predicted RT-PCR product sizes are indicated in parentheses. The roman numerals above the gel lanes correspond to the predicted RT-PCR products. For lane IV′ the reaction was performed without reverse transcriptase.
In the MA4664/MA3734-MA3743 transcriptional unit, three of the gene products are annotated as proteins that directly reduce reactive O2 species. MA3737 is annotated as a gene encoding a class II superoxide reductase (see Fig. S2 in the supplemental material) (3). MA3740 is annotated as a gene encoding a homolog of Isf (see Fig. S3 in the supplemental material), and MA3743 is annotated as a gene encoding FprA (see Fig. S4 in the supplemental material); both of these gene products reduce O2 to H2O (13, 56). In addition, MA4664 is annotated as a gene encoding a homolog of desulforedoxin (see Fig. S5 in the supplemental material), the physiological electron donor to the class II superoxide reductase of Desulfovibrio gigas (3). A role for the remaining gene products in response to oxidative stress has not been documented.
In contrast to the annotation of other genes in the MA4664/MA3734-MA3743 transcriptional unit that could function in oxidative stress, MA3736 is annotated as a gene encoding an uncharacterized CMD homolog. CMD is an essential enzyme in aerobic species in the domain Bacteria that utilize aromatic compounds as growth substrates (18, 52). Methanogens are strictly anaerobic, and none are known to metabolize aromatic compounds for growth (68), suggesting that MA3736 is annotated incorrectly, which prompted an investigation of the physiological function of the protein previously shown to be present in CO-, acetate-, and methanol-grown cells of M. acetivorans (41, 42).
Purification of the MA3736 product and initial characterization.
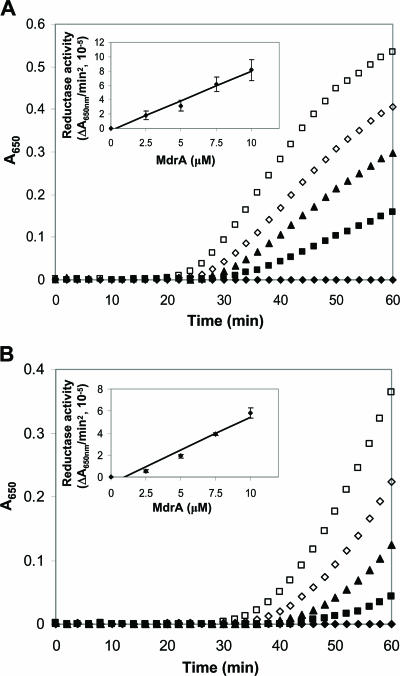
Unlike characterized CMD proteins, the deduced sequences of the MA3736 product and homologs (Fig. 1) contain a CXXC motif within a domain that has sequence identity (∼30%) to the active site domain of the prototypical AhpD protein from M. tuberculosis (Fig. 2). Although AhpD has alkylperoxide reductase activity, it functions primarily as a disulfide reductase, reducing the active site disulfide of AhpC, a peroxiredoxin (8, 28, 36). AhpD and AhpC are key components of the oxidative stress response in M. tuberculosis (8, 26). Thus, MA3736 was heterologously expressed, and the protein was anaerobically purified to test for AhpD-like activities. The protein was judged homogeneous by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, which also indicated that the subunit molecular mass was consistent with the calculated value, 12.9 kDa (data not shown). The purified MA3736 product was assayed for alkylperoxide reductase activity using DTT or a reducing system comprised of NADH, lipoamide, and lipoamide dehydrogenase, as previously described for AhpD (28, 36). No activity was detected under anaerobic conditions (data not shown), suggesting that the protein does not function as an alkylperoxide reductase. However, the MA3736 product exhibited both DTT- and lipoamide-dependent protein disulfide reductase activity as measured by the insulin turbidimetric assay (30) under anaerobic conditions (Fig. 3). No protein disulfide reductase activity was detected when the product was assayed aerobically. The DTT-dependent protein disulfide reductase activity of MdrA was approximately 20% of that measured for thioredoxin from E. coli (data not shown). Lipoamide-dependent activity was dependent on all three assay components (data not shown), suggesting that lipoamide directly reduces the oxidized MA3736 product, similar to AhpD (8). This is the first enzymatic activity determined for a product of genes annotated as genes encoding putative CMD enzymes with a CXXC motif. We propose that MA3736 encodes a protein distinct from CMD and should be renamed mdrA (methanosarcina disulfide reductase).
FIG. 2.
Alignment of amino acid sequences of MdrA homologs and AhpD from M. tuberculosis. Identical amino acid residues are indicated by asterisks. The active site cysteines of AhpD that are conserved in the MdrA homologs (C67 and C70 in MdrA) are indicated by solid arrowheads, and additional conserved cysteines (C39 and C107 in MdrA) not found in AhpD are indicated by open arrowheads. Sequences were aligned using CLUSTAL W. MdrA, M. acetivorans C2A; MM0631, M. mazei Go1; Mbar_A2454, M. barkeri strain Fusaro; Mbur2375, M. burtonii DSM 6242; AhpD, M. tuberculosis.
FIG. 3.
Protein disulfide reductase activity of MdrA as determined by the insulin turbidimetric method. (A) DTT-dependent protein disulfide reductase activity of MdrA. The assay was carried out by addition of 0.33 mM DTT in 100 mM potassium phosphate (pH 7.0) containing 0.13 mM bovine insulin in the absence of MdrA (⧫) and in the presence of different concentrations of MdrA, including 2.5 μM (▪), 5 μM (▴), 7.5 μM (□), and 10 μM (□). (B) Lipoamide-dependent protein disulfide reductase activity of MdrA. The assay was carried out by addition of 0.5 mM NADH in 100 mM potassium phosphate (pH 7.0) containing 0.13 mM bovine insulin, 0.05 mM lipoamide, and 0.4 U bovine lipoamide dehydrogenase in the absence of MdrA (⧫) and in the presence of different concentrations of MdrA, including 2.5 μM (▪), 5 μM (▴), 7.5 μM (⋄), and 10 μM (□). The insets show the linear dependence of the activity on the MdrA concentration.
It is unclear what protein(s) or cofactor(s) functions as an in vivo electron donor to MdrA. Reduced coenzyme F420, a universal electron carrier in methanogens, was ineffective as a direct electron donor (data not shown). NADPH-dependent thioredoxin reductase from E. coli also could not supply electrons to support MdrA protein disulfide reductase activity (data not shown). Reduction of AhpD in vivo is linked to metabolic enzymes of the tricarboxylic acid cycle in M. tuberculosis (8). Dihydrolipoamide succinyltransferase (SucB), a lipoamide-containing protein, is a reducing partner of AhpD. SucB is subsequently reduced by lipoamide dehydrogenase via NADH in vivo (8, 36). To examine the specificity of MdrA for the AhpD reducing partners, we assayed MdrA for disulfide reductase activity with the 5,5′-dithiobis(2-nitrobenzoic acid) assay developed by Bryk et al. (8), using purified SucB and lipoamide dehydrogenase from M. tuberculosis. MdrA could not substitute for AhpD in this assay (data not shown), suggesting that there are differences in the specificities of AhpD and MdrA for redox partners.
Analysis of MdrA cysteine variants.
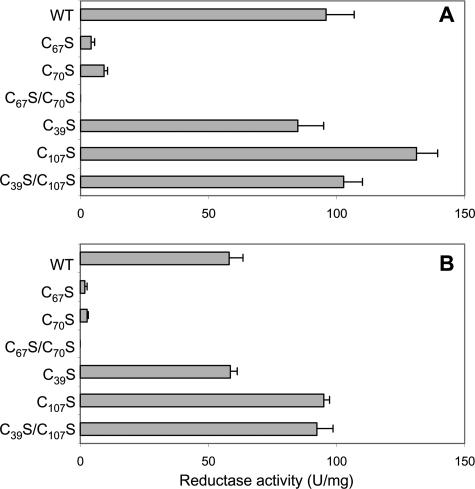
MdrA contains two additional conserved cysteine residues independent of the C67XXC70 motif; one is located in the N terminus (C39), while the second (C107) is located in the C terminus (Fig. 2). Protein disulfide reductases, including AhpD, thioredoxin, and glutaredoxin, possess redox-active cysteine residues within a CXXC motif (29). However, the redox-active cysteine residues in AhpC-like peroxiredoxins are located on opposite ends of the protein (15, 53), similar to the locations of C39 and C107 in MdrA. To determine which MdrA cysteines are functionally important for protein disulfide reductase activity, cysteine-to-serine variants, including single variants (C39S, C67S, C70S, and C107S) and double variants (C39S/C107S and C67S/C70S), were generated.
All of the MdrA variants were expressed and purified at levels similar to that of the wild type (data not shown). The C39S, C107S, and C39S/C107S variants retained wild-type levels of activity in the DTT- and lipoamide-dependent assays (Fig. 4). However, the C67S and C70S single variants exhibited only 3 to 9% of the wild-type MdrA activity in both assays (Fig. 4). In addition, the C67S/C70S double variant had no detectable activity in either assay (Fig. 4). These results indicate that C67 and C70 are required for protein disulfide reductase activity, consistent with a requirement for the CXXC motif in other characterized protein disulfide reductases (29).
FIG. 4.
Protein disulfide reductase activities of wild-type MdrA and cysteine variants. (A) DTT-dependent activity. (B) Lipoamide-dependent activity. Assays were performed as described in Materials and Methods. The values are reported as ΔA650/min2, 10−5. WT, wild type.
Detection of an Fe-S cluster in MdrA.
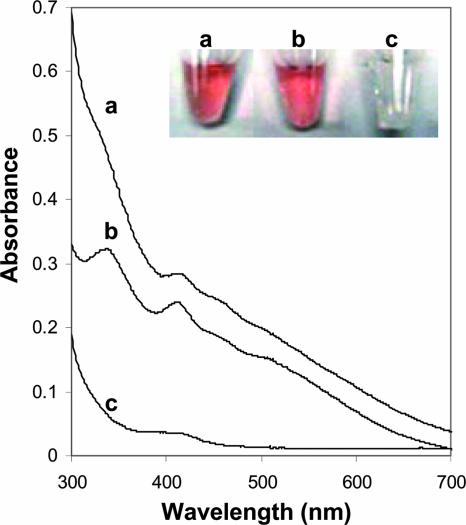
Unexpectedly, wild-type MdrA and the C39S/C107S variant were red-brown in color (Fig. 5), and iron and acid-labile sulfide were detected in both proteins (Table 1). The UV-visible spectrum of the C39S/C107S variant had absorbance peaks at 335, 412, 460, and 520 nm (Fig. 5). Similar spectral features were observed for the wild type, although the overall absorption was less (Fig. 5). These results suggest that MdrA contains an Fe-S cluster having an undetermined composition. The ratio of iron or acid-labile sulfide per monomer was less than unity for the wild type and the variant, which suggests either that the proteins do not contain a full complement of the Fe-S cluster(s) or that the cluster(s) is bound to more than one monomer. A double cysteine-to-alanine variant (C39A/C107A) was also red-brown in color and had a UV-visible absorption spectrum similar to that of the C39S/C107S variant (data not shown). These results suggest that residues at positions 39 and 107 are not essential for Fe-S cluster binding. However, the C67S/C70S variant was colorless and lacked spectral features of the wild type and the C39S/C107S variant (Fig. 5). Further, the levels of iron and acid-labile sulfide were below the limits of detection in the C67S/C70S variant (Table 1). Single-cysteine variants (C67S and C70S) were also colorless and lacked spectral features of the wild type and the C39S/C107S variant (data not shown). These results suggest not only that the active site cysteines function in protein disulfide reduction but also that both of these residues play a role in ligation of the Fe-S cluster(s).
FIG. 5.
UV-visible spectra of wild-type MdrA and variants. Line a, wild-type MdrA (400 μM); line b, C39S/C107S (200 μM); line c, C67S/C70S (400 μM). The inset shows vials containing the protein solutions.
TABLE 1.
Analysis of iron and acid-labile sulfide in wild-type MdrA and cysteine variants
| Protein | Amt of iron (nmol/ nmol of MdrA monomer) | Amt of sulfide (nmol/ nmol of MdrA monomer) |
|---|---|---|
| MdrAa | 0.37 ± 0.05 | 0.15 ± 0.02 |
| C39S/C107S | 0.44 ± 0.06 | 0.23 ± 0.02 |
| C67S/C70S | BDLb | BDL |
| apo-MdrAc | BDL | BDL |
As-purified MdrA.
BDL, below the detection limit (0.01 nmol).
As-purified MdrA pretreated with EDTA.
Effect of the Fe-S cluster on protein disulfide reductase activity and the oligomeric state of MdrA.
As residues Cys67 and Cys70 appear to be necessary for protein disulfide reductase activity and binding of an Fe-S cluster, the effect of the presence or absence of the Fe-S cluster on protein disulfide reductase activity was determined. The presence of EDTA in the assay mixture was necessary for activity with purified MdrA, unless MdrA was pretreated with EDTA (Table 2), in which case iron or acid-labile sulfide was undetectable (apo-MdrA) (Table 1). These results suggest that loss of the Fe-S cluster(s) is required for protein disulfide reductase activity.
TABLE 2.
Comparison of as-purified MdrA and apo-MdrA protein disulfide reductase activities
| Protein | DTT-dependent activity (U/mg)a
|
Lipoamide-dependent activity (U/mg)a
|
||
|---|---|---|---|---|
| With EDTA | Without EDTA | With EDTA | Without EDTA | |
| MdrA | 95 ± 11 | BDLb | 58 ± 6 | BDL |
| apo-MdrA | 80 ± 10 | 72 ± 15 | 50 ± 4 | 52 ± 9 |
Assays were performed as described in Materials and Methods with 10 μM protein with or without 1 mM EDTA in the assay mixture. The values are reported as ΔA650/min2, 10−5.
BDL, below the detection limit.
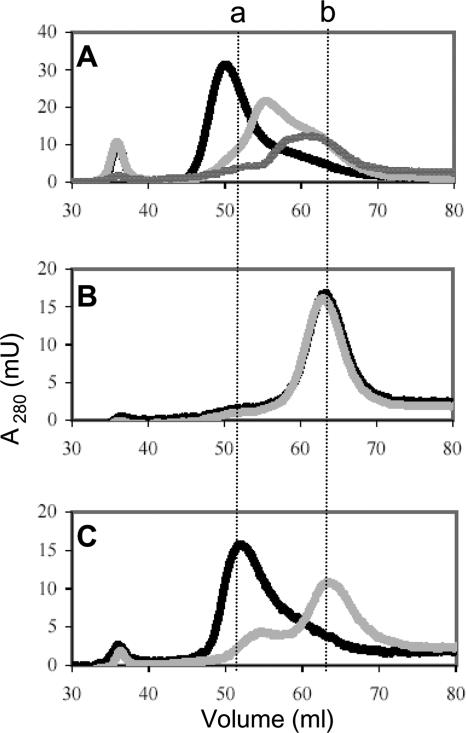
Four cysteines typically coordinate Fe-S clusters. However, only Cys67 and Cys70 appear to be required for Fe-S cluster binding in MdrA, suggesting that the cluster has ligands other than cysteine or that MdrA contains an intermolecular cluster coordinated by cysteines from more than one monomer. Indeed, the disulfide oxidoreductase glutaredoxin 2 (Grx2) from humans contains an intermolecular bridging [2Fe-2S] cluster that has been shown to regulate disulfide reductase activity (44). Thus, the effect of loss of the Fe-S cluster on the oligomeric state of wild-type MdrA and the cysteine variants was determined by size exclusion chromatography (Fig. 6). The elution profile of as-purified wild-type MdrA was consistent with the profile of a hexamer (Fig. 6A). The C39S/C107S variant elution profile was similar to that of the wild type, which was also consistent with the profile of a hexamer (Fig. 6C). However, the C67S/C70S variant migrated as a trimer (Fig. 6B). Inclusion of EDTA in the buffers used in size exclusion chromatography of as-purified wild-type MdrA resulted in a mixture of smaller oligomers of MdrA, including a trimer (Fig. 6A). A similar elution profile was obtained for MdrA pretreated with EDTA (apo-MdrA) even though EDTA was not included in the buffers (Fig. 6A and Table 1). The C39S/C107S variation had a similar effect, as the protein migrated primarily as a trimer when it was eluted in the presence of EDTA (Fig. 6C). However, the C67S/C70S variant continued to migrate as a trimer when it was eluted in the presence of EDTA (Fig. 6B). These results demonstrate the importance of Cys67 and Cys70 in modulating the oligomeric state of MdrA. Thus, Cys67 and Cys70 may coordinate an intermolecular bridging Fe-S cluster(s) between trimers to form a hexamer, and loss of the Fe-S cluster converts the enzyme to a trimer that is active.
FIG. 6.
Effect of EDTA on the oligomeric state of MdrA and cysteine variants as analyzed by size exclusion chromatography. (A) Elution profiles of wild-type MdrA as purified (black line) and with EDTA (light gray line) and of apo-MdrA (dark gray line). (B) Elution profiles of the C67S/C70S variant as purified (black line) and with EDTA (gray line). (C) Elution profiles of the C39S/C107S variant as purified (black line) and with EDTA (gray line). Dashed vertical line a indicates the volume corresponding to the hexameric form of MdrA, and dashed vertical line b indicates the volume corresponding to the trimeric form of MdrA. Hexameric and trimeric volumes were calculated based on a standard curve generated with molecular mass standards (data not shown).
Phylogenetic analyses.
The finding that MdrA is a protein disulfide reductase prompted an investigation of the databases to determine the extent to which proteins encoded by genes annotated as CMD genes contain the CXXC motif and have the potential to be MdrA-like protein disulfide reductases. A BLAST search of all nonredundant databases was performed with the protein sequence of MdrA as the query. A survey of the returned sequences revealed 189 putative proteins that contained a CXXC motif and had between 22 and 84% identity to MdrA, suggesting that putative homologs are widespread. The analysis was further extended to understand the relatedness of MdrA and putative homologs to prototypical CMD and AhpD. A BLAST search with prototypical CMD (PcaC from Acinetobactor sp. strain ADP1 [24]) as the query revealed 212 putative proteins without a CXXC motif that had ≥24% identity to PcaC. A BLAST search with prototypical AhpD from M. tuberculosis (28) as a query revealed 113 putative proteins with a CXXC motif that had ≥26% identity to AhpD. To elucidate the phylogeny of MdrA and CXXC-containing and non-CXXC-containing putative CMD and AhpD proteins, 32 sequences were selected from the first 50 sequences retrieved from each BLAST search. The selections were based on previously characterized proteins and uncharacterized proteins from physiologically and phylogenetically diverse organisms. These sequences were aligned, and a phylogenetic tree was constructed (Fig. 7). The non-CXXC-containing sequences from both Bacteria and Archaea group together (cluster III), including the prototypical CMD (PcaC) from Acinetobactor sp. strain ADP1. The CXXC-containing sequences display a dichotomy. Cluster II contains MdrA and various sequences from Bacteria and Archaea, whereas cluster I contains AhpD from M. tuberculosis (8), S. coelicolor (27), L. pneumophila (39) and sequences from other Bacteria. The phylogenetic analyses indicate that MdrA is distinct from both prototypical CMD and AhpD, suggesting that MdrA is the prototype of a new family. The phylogenetic analyses further suggest that there is wide distribution of CMD-related, MdrA-related, and AhpD-related enzymes among diverse prokaryotes. Two non-CXXC-containing proteins from methanogens, encoded by MTH234 from M. thermoautotrophicum and by MA0409 from M. acetivorans, group in cluster III with prototypical CMD (Fig. 7). Methanogens are strictly anaerobic, and none are known to metabolize aromatic compounds, indicating that MTH234 and MA0409 most likely do not function as CMD or as MdrA but may have an unknown function.
FIG. 7.
Phylogenetic tree of selected CMD-, MdrA-, and AhpD-related sequences. The phylogenetic tree was constructed using the neighbor-joining method. The scale bar indicates the average number of amino acid substitutions per site. Prototypical functionally analyzed CMD and AhpD, as well as MdrA, are in bold type. Cluster I contains AhpD-related proteins, and cluster II contains MdrA-related proteins. Cluster I and II proteins contain a CXXC motif, with the exception of TTHA0727 from T. thermophilus, which contains an SXXC motif (indicated by an asterisk). Cluster III contains prototypical CMD-related proteins that do not contain a CXXC motif.
DISCUSSION
A major challenge in the postgenomic era is avoiding the perpetuation of incorrect annotations. Resolution of this growing problem rests on biochemical and molecular biology experimental approaches for validation of questionable annotations, as discussed recently (63). The resolution of incorrect annotations often leads to discovery of function and protein families, as is the case reported here for MA3736 (encoding MdrA) from M. acetivorans. MA3736 was originally annotated as a gene encoding an uncharacterized CMD homolog, but the results presented here support the conclusion that MdrA is a protein disulfide reductase with the potential to function in the oxidative stress response of M. acetivorans and related species, including RC-IMRE50.
A role for CMD in the physiology of M. acetivorans is highly improbable as methanogens are strictly anaerobic and none are known to metabolize aromatic compounds for growth (68). Therefore, although MdrA shares some sequence identity (<30%) with CMD enzymes, such as PcaC from Acinetobacter sp. strain ADP1 (24), MdrA most likely does not function as previously annotated. Instead, MdrA was shown to contain an Fe-S cluster and to have protein disulfide reductase activity dependent on a CXXC motif that is not found in characterized CMD proteins (18, 45, 52) but is essential in other characterized protein disulfide reductases, including AhpD (8, 29, 36). Further, phylogenetic analyses indicate that MdrA is distinct from both CMD and AhpD, suggesting that MdrA is the prototype of a new family.
The active site domain of MdrA and AhpD also has identity to that of sestrins (9), proteins that play a role in peroxide signaling pathways in higher eukaryotic organisms, including humans. Analogous to AhpD, sestrin 2 catalyzes the reduction of a peroxiredoxin. However, sestrins contain only the proximal cysteine of the essential CXXC motif of AhpD and MdrA. Sestrins are not disulfide reductases but instead function as cysteine sulfinyl reductases, reducing overoxidized peroxiredoxins to modulate peroxide signaling and antioxidant defense (9). Therefore, MdrA and homologs found in ancient methanoarchaea may provide an evolutionary link not only to the structurally related proteins AhpD and CMD but also to sestrins.
The data presented here suggest that the CXXC-containing domain is important for oligomerization of MdrA and control of activity. Purified wild-type MdrA and the C39S/C107S variant are hexamers, while the C67S/C70S variant is a trimer. Oligomerization of MdrA also appears to be dependent on Fe-S cluster binding. Although additional characterization to identify the type of Fe-S cluster was beyond the scope of this study, the UV-visible spectrum and extrapolation of the amount of iron (nanomoles per nanomole of hexamer: 2.22 ± 0.30 for the wild type and 2.64 ± 0.36 for the C39S/C107S variant) are consistent with the hypothesis that wild-type MdrA and the C39S/C107S variant contain one [2Fe-2S] cluster per hexamer, while the Fe-S cluster is absent in the trimeric C67S/C70S variant. In addition, the protein disulfide reductase activity of wild-type MdrA was dependent on loss of the cluster, and addition of EDTA to wild-type MdrA and the C39S/C107S variant resulted in a change from a hexamer to primarily a trimer. Taken together, these results suggest that oligomerization of MdrA trimers is Fe-S cluster dependent and that Cys67 and Cys70 are important for Fe-S cluster binding.
The first disulfide reductase shown to contain a regulatory Fe-S cluster, [2Fe-2S], is Grx2 (44). Recently, a poplar glutaredoxin (Grx-C1) was also shown to contain a subunit-bridging [2Fe-2S] cluster (19, 55). The [2Fe-2S] cluster in Grx2 and Grx-C1 is coordinated by the N-terminal active site cysteine of two monomers and two noncovalently bound molecules of glutathione (5, 55). Dimeric holo-Grx2 and holo-Grx-C1 are inactive as disulfide oxidoreductases, similar to hexameric, [Fe-S]-containing MdrA. Loss of the [2Fe-2S] cluster results in activation of Grx2 and Grx-C1. In MdrA, the active site cysteines (Cys67 and Cys70) also appear to be necessary for Fe-S cluster binding, suggesting functional similarity to Grx2 and Grx-C1. Grx2 also contains two additional cysteine residues that are outside the active site cysteines and are postulated to play a structural role (5, 32). It is unclear what role, if any, the two additional cysteine residues (Cys39 and Cys107) play in MdrA. However, most CXXC-containing CMD homologs do not contain the additional cysteine residues found in the Methanosarcina-related MdrA homologs.
Recently, WhiB4/Rv3681c from M. tuberculosis was shown to have protein disulfide reductase activity and to contain a labile Fe-S cluster hypothesized to regulate protein disulfide reductase activity (2). WhiB homologs have been shown to be important for survival and for the response to oxidative stress (23, 35). WhiB4 and MdrA share no overall sequence identity, as confirmed by the inability to align the amino acid sequences (62), indicating that WhiB and MdrA are members of distinct protein disulfide reductase families. WhiB proteins have four conserved cysteines, two of which are in a CXXC motif (59), similar to MdrA, suggesting that WhiB and MdrA may be functionally similar protein disulfide reductases. However, all four cysteines are important for coordinating an intramolecular Fe-S cluster in WhiB, while only the CXXC motif appears to be necessary for coordinating an intermolecular Fe-S cluster in MdrA. The Fe-S cluster(s) in MdrA may also serve as a sensor of oxidative stress, similar to the [2Fe-2S] cluster in Grx2 and the Fe-S cluster in WhiB. Thus, it appears that at least three distinct protein disulfide reductase families that employ an Fe-S cluster as a mechanism to regulate activity have evolved.
The gene encoding MdrA (MA3736) was shown to reside in a transcriptional unit with several putative oxidative stress genes, consistent with a role for MdrA in the oxidative stress response of M. acetivorans. MdrA (encoded by MA3736) and the products of most of the other genes (MA3735, MA3737, MA3740, MA3741, MA3742, and MA3743) were detected at similar levels in CO-, acetate-, and methanol-grown cells by global proteomic analyses (41, 42), consistent with a physiological function for these proteins. Conservation of the gene organization in other methanogen species also supports the hypothesis that these genes have a physiological role. Further sequence analysis suggested potential functions for two of the remaining gene products. MA3742 is annotated as a gene encoding a conserved hypothetical protein, which contains a conserved di-iron-binding motif (see Fig. S6 in the supplemental material), similar to bacterioferritin and rubrerythrin, which function in iron storage/detoxification and in reduction of hydrogen peroxide to water, respectively (10, 21, 51). MA3739 encodes a protein with five CXXCH heme-binding motifs (see Fig. S1 in the supplemental material), which suggests that this protein is a multiheme cytochrome c.
One potential function that can be postulated for MdrA is the repair of proteins in which disulfide bonds are formed by oxidation during exposure to O2. An intriguing alternative hypothesis is that MdrA functions in Fe-S cluster assembly or delivery, a process which is relatively unknown in methanoarchaea. Indeed, the genome of M. acetivorans does not encode complete homologs of known Fe-S cluster biosynthesis proteins (e.g., NifU, Nfu, and IscA) (33, 40). Further, the properties of MdrA are consistent with those of proteins known to function in Fe-S cluster assembly, such as the CXXC-containing Fe-S cluster scaffold SyNifU/Nfu, which also binds a bridging [2Fe-2S] cluster (40, 50), and other disulfide reductases (glutaredoxins) (1, 54). Thus, MdrA may also function in repair of Fe-S cluster proteins damaged during oxidative stress.
The genome of M. acetivorans contains six additional genes annotated as genes encoding CMD homologs with CXXC motifs, which is similar to the number found in other Methanosarcina-related species. Although the genes encoding these homologs are not clustered with genes encoding oxidative stress proteins, the results are consistent with the hypothesis that the homologs have a function similar to that of MdrA. The multiple MdrA homologs found in Methanosarcina-related species, including RC-IMRE50 (17), suggest that these proteins are physiologically important components of methanoarchaea that significantly contribute to global methane emissions and may further suggest a broader function, such as Fe-S cluster assembly or delivery. Thus, it is important to note that methanoarchaea appear to contain the greatest number of Fe-S proteins, as estimated by the abundance of the CX2CX2CX3C motif in proteins encoded in methanoarchaeon genomes (47). Further, it is estimated that of the methanoarchaea, Methanosarcina species contain the highest number of Fe-S proteins, which may reflect their metabolic diversity and large genome size. That Methanosarcina species contain the highest number of putative Fe-S proteins may also reflect a need for a high number of Fe-S cluster assembly and delivery proteins, consistent with the multiple copies of MdrA functioning in Fe-S cluster assembly or delivery. MdrA may function in repair of Fe-S cluster proteins damaged during oxidative stress, and homologs could function in general Fe-S cluster biosynthesis. We are currently investigating the ability of MdrA and homologs to function in Fe-S cluster assembly or delivery.
Conclusions.
Although the mdrA gene was originally annotated as a gene encoding an uncharacterized CMD homolog, the results presented here support the hypothesis that MdrA is a protein disulfide reductase. The report of the protein disulfide reductase activity of MdrA is the first report of an enzymatic activity for CXXC-containing putative CMD homologs, and the findings suggest that MdrA is the prototype of a family. MdrA was also shown to contain an Fe-S cluster(s) with the potential to play a regulatory role in protein disulfide reductase activity or to additionally function in Fe-S cluster assembly or delivery. The activity of MdrA and the organization of mdrA in a transcriptional unit with oxidative stress genes are consistent with a role in the oxidative stress response of M. acetivorans.
Supplementary Material
Acknowledgments
We thank Rusalana Bryk and Carl Nathan for providing AhpD, SucB, and Lpd from M. tuberculosis and Eric Patridge for assistance with phylogenetic analyses.
This work was supported by postdoctoral fellowship grants to D.J.L. from the NRC/NASA Astrobiology Institute (grant 0386600) and NIH (grant ES013114-02).
Footnotes
Published ahead of print on 3 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Achebach, S., Q. H. Tran, A. Vlamis-Gardikas, M. Mullner, A. Holmgren, and G. Unden. 2004. Stimulation of Fe-S cluster insertion into apoFNR by Escherichia coli glutaredoxins 1, 2 and 3 in vitro. FEBS Lett. 565:203-206. [DOI] [PubMed] [Google Scholar]
- 2.Alam, M. S., S. K. Garg, and P. Agrawal. 2007. Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: a [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol. Microbiol. 63:1414-1431. [DOI] [PubMed] [Google Scholar]
- 3.Auchere, F., S. R. Pauleta, P. Tavares, I. Moura, and J. J. Moura. 2006. Kinetics studies of the superoxide-mediated electron transfer reactions between rubredoxin-type proteins and superoxide reductases. J. Biol. Inorg. Chem. 11:433-444. [DOI] [PubMed] [Google Scholar]
- 4.Beinert, H. 1983. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131:373-378. [DOI] [PubMed] [Google Scholar]
- 5.Berndt, C., C. Hudemann, E. M. Hanschmann, R. Axelsson, A. Holmgren, and C. H. Lillig. 2007. How does iron-sulfur cluster coordination regulate the activity of human glutaredoxin 2? Antioxid. Redox Signal. 9:151-157. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brioukhanov, A., A. Netrusov, M. Sordel, R. K. Thauer, and S. Shima. 2000. Protection of Methanosarcina barkeri against oxidative stress: identification and characterization of an iron superoxide dismutase. Arch. Microbiol. 174:213-216. [DOI] [PubMed] [Google Scholar]
- 8.Bryk, R., C. D. Lima, H. Erdjument-Bromage, P. Tempst, and C. Nathan. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073-1077. [DOI] [PubMed] [Google Scholar]
- 9.Budanov, A. V., A. A. Sablina, E. Feinstein, E. V. Koonin, and P. M. Chumakov. 2004. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science 304:596-600. [DOI] [PubMed] [Google Scholar]
- 10.Carrondo, M. A. 2003. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 22:1959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conrad, R., C. Erkel, and W. Liesack. 2006. Rice cluster I methanogens, an important group of Archaea producing greenhouse gas in soil. Curr. Opin. Biotechnol. 17:262-267. [DOI] [PubMed] [Google Scholar]
- 12.Coulter, E. D., and D. M. Kurtz, Jr. 2001. A role for rubredoxin in oxidative stress protection in Desulfovibrio vulgaris: catalytic electron transfer to rubrerythrin and two-iron superoxide reductase. Arch. Biochem. Biophys. 394:76-86. [DOI] [PubMed] [Google Scholar]
- 13.Cruz, F., and J. G. Ferry. 2006. Interaction of iron-sulfur flavoprotein with oxygen and hydrogen peroxide. Biochim. Biophys. Acta 1760:858-864. [DOI] [PubMed] [Google Scholar]
- 14.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 15.Ellis, H. R., and L. B. Poole. 1997. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry 36:13349-13356. [DOI] [PubMed] [Google Scholar]
- 16.Erkel, C., D. Kemnitz, M. Kube, P. Ricke, K. J. Chin, S. Dedysh, R. Reinhardt, R. Conrad, and W. Liesack. 2005. Retrieval of first genome data for rice cluster I methanogens by a combination of cultivation and molecular techniques. FEMS Microbiol. Ecol. 53:187-204. [DOI] [PubMed] [Google Scholar]
- 17.Erkel, C., M. Kube, R. Reinhardt, and W. Liesack. 2006. Genome of rice cluster I Archaea—the key methane producers in the rice rhizosphere. Science 313:370-372. [DOI] [PubMed] [Google Scholar]
- 18.Eulberg, D., S. Lakner, L. A. Golovleva, and M. Schlomann. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180:1072-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, Y., N. Zhong, N. Rouhier, T. Hase, M. Kusunoki, J. P. Jacquot, C. Jin, and B. Xia. 2006. Structural insight into poplar glutaredoxin C1 with a bridging iron-sulfur cluster at the active site. Biochemistry 45:7998-8008. [DOI] [PubMed] [Google Scholar]
- 20.Fetzer, S., F. Bak, and R. Conrad. 1993. Sensitivity of methanogenic bacteria from paddy soil to oxygen and desiccation. FEMS Microbiol. Ecol. 12:107-115. [Google Scholar]
- 21.Fournier, M., Y. Zhang, J. D. Wildschut, A. Dolla, J. K. Voordouw, D. C. Schriemer, and G. Voordouw. 2003. Function of oxygen resistance proteins in the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 185:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiman, D. E., T. R. Raghunand, N. Agarwal, and W. R. Bishai. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerischer, U., A. Segura, and L. N. Ornston. 1998. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J. Bacteriol. 180:1512-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grunden, A. M., F. E. Jenney, Jr., K. Ma, M. Ji, M. V. Weinberg, and M. W. Adams. 2005. In vitro reconstitution of an NADPH-dependent superoxide reduction pathway from Pyrococcus furiosus. Appl. Environ. Microbiol. 71:1522-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guimaraes, B. G., H. Souchon, N. Honore, B. Saint-Joanis, R. Brosch, W. Shepard, S. T. Cole, and P. M. Alzari. 2005. Structure and mechanism of the alkyl hydroperoxidase AhpC, a key element of the Mycobacterium tuberculosis defense system against oxidative stress. J. Biol. Chem. 280:25735-25742. [DOI] [PubMed] [Google Scholar]
- 27.Hahn, J. S., S. Y. Oh, and J. H. Roe. 2002. Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 184:5214-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillas, P. J., F. S. del Alba, J. Oyarzabal, A. Wilks, and P. R. Ortiz De Montellano. 2000. The AhpC and AhpD antioxidant defense system of Mycobacterium tuberculosis. J. Biol. Chem. 275:18801-18809. [DOI] [PubMed] [Google Scholar]
- 29.Holmgren, A. 1989. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 264:13963-13966. [PubMed] [Google Scholar]
- 30.Holmgren, A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254:9627-9632. [PubMed] [Google Scholar]
- 31.Jenney, F. E., Jr., M. F. Verhagen, X. Cui, and M. W. Adams. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306-309. [DOI] [PubMed] [Google Scholar]
- 32.Johansson, C., K. L. Kavanagh, O. Gileadi, and U. Oppermann. 2007. Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J. Biol. Chem. 282:3077-3082. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, D. C., D. R. Dean, A. D. Smith, and M. K. Johnson. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247-281. [DOI] [PubMed] [Google Scholar]
- 34.Kiener, A., and T. Leisinger. 1983. Oxygen sensitivity of methanogenic bacteria. Syst. Appl. Microbiol. 4:305-312. [DOI] [PubMed] [Google Scholar]
- 35.Kim, T. H., J. S. Park, H. J. Kim, Y. Kim, P. Kim, and H. S. Lee. 2005. The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem. Biophys. Res. Commun. 337:757-764. [DOI] [PubMed] [Google Scholar]
- 36.Koshkin, A., C. M. Nunn, S. Djordjevic, and P. R. Ortiz de Montellano. 2003. The mechanism of Mycobacterium tuberculosis alkylhydroperoxidase AhpD as defined by mutagenesis, crystallography, and kinetics. J. Biol. Chem. 278:29502-29508. [DOI] [PubMed] [Google Scholar]
- 37.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefs Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 38.Leadbetter, J. R., and J. A. Breznak. 1996. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl. Environ. Microbiol. 62:3620-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeBlanc, J. J., R. J. Davidson, and P. S. Hoffman. 2006. Compensatory functions of two alkyl hydroperoxide reductases in the oxidative defense system of Legionella pneumophila. J. Bacteriol. 188:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leon, S., B. Touraine, C. Ribot, J. F. Briat, and S. Lobreaux. 2003. Iron-sulphur cluster assembly in plants: distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana. Biochem. J. 371:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lessner, D. J., L. Li, Q. Li, T. Rejtar, V. P. Andreev, M. Reichlen, K. Hill, J. J. Moran, B. L. Karger, and J. G. Ferry. 2006. An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. USA 103:17921-17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, Q., L. Li, T. Rejtar, B. L. Karger, and J. G. Ferry. 2005. Proteome of Methanosarcina acetivorans. Part I: an expanded view of the biology of the cell. J. Proteome Res. 4:112-128. [DOI] [PubMed] [Google Scholar]
- 43.Li, Q., L. Li, T. Rejtar, D. J. Lessner, B. L. Karger, and J. G. Ferry. 2006. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188:702-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lillig, C. H., C. Berndt, O. Vergnolle, M. E. Lonn, C. Hudemann, E. Bill, and A. Holmgren. 2005. Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc. Natl. Acad. Sci. USA 102:8168-8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorite, M. J., J. Sanjuan, L. Velasco, J. Olivares, and E. J. Bedmar. 1998. Characterization of Bradyrhizobium japonicum pcaBDC genes involved in 4-hydroxybenzoate degradation. Biochim. Biophys. Acta 1397:257-261. [DOI] [PubMed] [Google Scholar]
- 46.Lumppio, H. L., N. V. Shenvi, A. O. Summers, G. Voordouw, and D. M. Kurtz, Jr. 2001. Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J. Bacteriol. 183:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Major, T. A., H. Burd, and W. B. Whitman. 2004. Abundance of 4Fe-4S motifs in the genomes of methanogens and other prokaryotes. FEMS Microbiol. Lett. 239:117-123. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Galisteo, E., C. A. Padilla, C. Garcia-Alfonso, J. Lopez-Barea, and J. A. Barcena. 1993. Purification and properties of bovine thioredoxin system. Biochimie 75:803-809. [DOI] [PubMed] [Google Scholar]
- 49.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishio, K., and M. Nakai. 2000. Transfer of iron-sulfur cluster from NifU to apoferredoxin. J. Biol. Chem. 275:22615-22618. [DOI] [PubMed] [Google Scholar]
- 51.Nordlund, P., and H. Eklund. 1995. Di-iron-carboxylate proteins. Curr. Opin. Struct. Biol. 5:758-766. [DOI] [PubMed] [Google Scholar]
- 52.Parke, D. 1995. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J. Bacteriol. 177:3808-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poole, L. B. 2005. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433:240-254. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez-Manzaneque, M. T., J. Tamarit, G. Belli, J. Ros, and E. Herrero. 2002. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 13:1109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rouhier, N., H. Unno, S. Bandyopadhyay, L. Masip, S. K. Kim, M. Hirasawa, J. M. Gualberto, V. Lattard, M. Kusunoki, D. B. Knaff, G. Georgiou, T. Hase, M. K. Johnson, and J. P. Jacquot. 2007. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc. Natl. Acad. Sci. USA 104:7379-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seedorf, H., A. Dreisbach, R. Hedderich, S. Shima, and R. K. Thauer. 2004. F420H2 oxidase (FprA) from Methanobrevibacter arboriphilus, a coenzyme F420-dependent enzyme involved in O2 detoxification. Arch. Microbiol. 182:126-137. [DOI] [PubMed] [Google Scholar]
- 57.Shi, Y. Y., W. Tang, S. F. Hao, and C. C. Wang. 2005. Contributions of cysteine residues in Zn2 to zinc fingers and thiol-disulfide oxidoreductase activities of chaperone DnaJ. Biochemistry 44:1683-1689. [DOI] [PubMed] [Google Scholar]
- 58.Shima, S., A. Netrusov, M. Sordel, M. Wicke, G. C. Hartmann, and R. K. Thauer. 1999. Purification, characterization, and primary structure of a monofunctional catalase from Methanosarcina barkeri. Arch. Microbiol. 171:317-323. [DOI] [PubMed] [Google Scholar]
- 59.Soliveri, J. A., J. Gomez, W. R. Bishai, and K. F. Chater. 2000. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology 146:333-343. [DOI] [PubMed] [Google Scholar]
- 60.Sowers, K. R., S. F. Baron, and J. G. Ferry. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 62.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 63.White, R. H. 2006. The difficult road from sequence to function. J. Bacteriol. 188:3431-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeh, A. P., Y. Hu, F. E. Jenney, Jr., M. W. Adams, and D. C. Rees. 2000. Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states. Biochemistry 39:2499-2508. [DOI] [PubMed] [Google Scholar]
- 65.Zabinski, R., E. Munck, P. M. Champion, and J. M. Wood. 1972. Kinetic and Mossbauer studies on the mechanism of protocatechuic acid 4,5-oxygenase. Biochemistry 11:3212-3219. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, J. K., A. K. White, H. C. Kuettner, P. Boccazzi, and W. W. Metcalf. 2002. Directed mutagenesis and plasmid-based complementation in the methanogenic archaeon Methanosarcina acetivorans C2A demonstrated by genetic analysis of proline biosynthesis. J. Bacteriol. 184:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhilina, T. N. 1972. Death of Methanosarcina in the air. Mikrobiologiya 41:1105-1106. [PubMed] [Google Scholar]
- 68.Zinder, S. 1993. Physiological ecology of methanogens, p. 128-206. In J. G. Ferry (ed.), Methanogenesis. Chapman and Hall, New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.