Abstract
The KdpD sensor kinase and the KdpE response regulator control expression of the kdpFABC operon coding for the KdpFABC high-affinity K+ transport system of Escherichia coli. In search of a distinct part of the input domain of KdpD which is solely responsible for K+ sensing, sequences of kdpD encoding the transmembrane region and adjacent N-terminal and C-terminal extensions were subjected to random mutagenesis. Nine KdpD derivatives were identified that had lost tight regulation of kdpFABC expression. They all carried single amino acid replacements located in a region encompassing the fourth transmembrane helix and the adjacent arginine cluster of KdpD. All mutants exhibited high levels of kdpFABC expression regardless of the external K+ concentration. However, 3- to 14-fold induction was observed under extreme K+-limiting conditions and in response to an osmotic upshift when sucrose was used as an osmolyte. These KdpD derivatives were characterized by a reduced phosphatase activity in comparison to the autokinase activity in vitro, which explains constitutive expression. Whereas for wild-type KdpD the autokinase activity and also, in turn, the phosphotransfer activity to KdpE were inhibited by increasing concentrations of K+, both activities were unaffected in the KdpD derivatives. These data clearly show that the extension of the fourth transmembrane helix encompassing the arginine cluster is mainly involved in sensing both K+ limitation and osmotic upshift, which may not be separated mechanistically.
In bacteria K+ plays an important role in the maintenance of intracellular pH, cell turgor and control of cellular enzyme activities (for a review, see reference 5), and, together with glutamate, it is also known to positively regulate the expression of certain osmoresponsive genes (23, 32). The intracellular K+ level is maintained by a variety of K+ uptake and efflux systems. The KdpFABC complex (K+-dependent ATPase), encoded by the kdpFABC operon (1), is an inducible, high-affinity K+ transporter synthesized by Escherichia coli as an emergency system to scavenge K+ when other transporters cannot keep up with the cell's requirement for K+ (4). Expression of the kdpFABC operon is regulated by the products of the adjacent kdpDE genes (31). The membrane-bound sensor kinase KdpD and the cytosolic response regulator KdpE comprise a typical prokaryotic two-component signal transduction system (43). KdpD (894 amino acids; 99 kDa) consists of a cytoplasmic N-terminal domain and a cytoplasmic C-terminal domain interconnected by four transmembrane segments (46). Upon stimulus perception, KdpD undergoes autophosphorylation at His-673, and subsequently the phosphoryl group is transferred to Asp-52 of the response regulator KdpE (42). KdpE binds in its phosphorylated and dimerized form with high affinity at a 23-bp sequence immediately upstream of the canonical −35 and −10 regions of the kdpFABC promoter, thereby triggering kdpFABC transcription (41). The large input domain of the sensor kinase KdpD (about 660 amino acids), which is responsible for stimulus perception, is composed of an extended cytoplasmic N-terminal domain, the four transmembrane helices followed by a cluster of arginine residues, and about 140 amino acids of the cytoplasmic C-terminal domain. The N-terminal cytoplasmic domain consisting of about 400 amino acids, which is unique among sensor kinases, is highly conserved among KdpD proteins from different bacteria. It has a regulatory ATP-binding domain similar to the Walker A and B motifs (14) and a 120-amino-acid region which was shown to have high structural homology to Usp domains of the ATP-binding type (37). The other 230 amino acids of the cytoplasmic C-terminal part constitute a typical transmitter domain of histidine sensor kinases carrying all the conserved sequence motifs (29). In vitro KdpD exhibits not only kinase activity but also an activity that dephosphorylates phospho-KdpE (17). Brandon et al. (3) have suggested that the initiation of signal transduction by KdpD is mediated by the inhibition of the phospho-KdpE-specific phosphatase activity, leading to accumulation of phospho-KdpE, which in turn induces kdpFABC expression in vivo. In addition, a switch between kinase and phosphatase activity is proposed to be of electrostatic nature (15). Furthermore, the binding of ATP to the regulatory ATP-binding site in the N-terminal domain influences the enzymatic activity of KdpD (11). Experiments with KdpD in right-side-out membrane vesicles revealed an inhibition of KdpD autokinase activity by an increase of K+, whereas an increase of ionic strength had a stimulatory effect (19).
KdpD is activated under K+-limiting growth conditions and by an osmotic upshift imposed by ionic solutes. The stimulus that is sensed by KdpD still needs further clarification. Based on a large body of evidence, KdpD probably integrates various parameters: changes in turgor, in phospholipid composition of the membrane, in extra- and/or intracellular K+ concentration, in medium osmolality, in ionic strength of the cytoplasm, and in internal ATP concentration (for more details, see reference 16). These different stimuli could be sensed and integrated by the unusually large input domain.
It was postulated that the four transmembrane helices play a major role in stimulus perception and signal transduction. A KdpD protein lacking the four transmembrane helices and the adjacent arginine cluster is completely inactive in vitro and in vivo (33). However, deleting the four transmembrane helices while retaining the arginine cluster leads to a KdpD derivative which exhibits a semiconstitutive phenotype (10). This observation led to the hypothesis that the arginine cluster is involved in stimulus perception. Furthermore, analyses of kdpD mutants, mostly single amino acid replacements in or near the transmembrane region, led to the suggestion that the sensing mechanisms for K+ limitation and osmotic upshift may be mechanistically different (15, 40). To test the hypothesis that a specific domain of KdpD might be solely responsible for K+ sensing, the transmembrane region and adjacent N-terminal and C-terminal extensions of KdpD were subjected to random mutagenesis. Mutants with an altered response to the external K+ concentration were further characterized in vivo and in vitro.
MATERIALS AND METHODS
Media and growth conditions.
Complex KML medium (1% KCl, 1% tryptone, 0.5% yeast extract) and minimal phosphate-buffered medium identified by K+ content in millimolars (K115 medium has 115 mM K+, e.g.) have been described previously (6). In general, glucose (1%) served as a carbon and energy source in minimal medium. Complex TY medium consists of 1% tryptone, 0.5% yeast extract, 6.6 mM potassium (determined by flame photometry) and, if required, different concentrations of NaCl and sucrose as indicated. Other compounds, e.g., ampicillin (100 μg/ml) for plasmid selection and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside at 50 mg/liter) for β-galactosidase activity on agar plates, were added as needed.
Bacterial strains and plasmids.
E. coli strain JM109 (endA1 recA1 gyrA96 thi-1 hsdR17(rK− mK−) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15]) (44) was used as a carrier for the plasmids described. E. coli HAK006 [ΔkdpFABCD kdpFABCP::lacZ Δ(lac-pro) ara thi] (40) was used for determination of kdpFABC expression in vivo. TKR2000 (ΔkdpFABCDE trkA405 trkD1 atp706) (20) was used for preparation of everted membrane vesicles. BL21(DE3)/pLysS (Novagen Inc., Madison, WI) harboring plasmid pEE (12) was used for expression of kdpE from the T7 promoter. Plasmid pPV5-8 is a derivative of plasmid pPV5 (9, 18, 43), in which kdpD is under the control of the tac promoter and contains a total of eight additional unique restriction sites introduced by silent mutation (e.g., SpeI [1110], NotI [1213], KpnI [1402], MluI [1534], and Bsu36I [1582]; numbers in brackets correspond to the nucleotide position in the kdpD sequence). In plasmid pBD the kdpD gene was cloned into pBAD18 under the control of the arabinose promoter (18).
Oligonucleotide-directed site-specific mutagenesis.
To facilitate mutant construction an additional unique AatII restriction site (bp 1420) in the kdpD sequence was introduced into plasmid pPV5-8 by silent mutation, resulting in plasmid pPV5-9. The PCR-based site-specific mutation was directed by a synthetic oligonucleotide forward primer comprised of the newly created AatII site in addition to the unique KpnI recognition site (mentioned above). The PCR product was cloned into pPV5-8 using KpnI and MluI restriction enzymes, and the existence of the newly created AatII site in pPV5-9 was confirmed by restriction analysis and DNA sequencing.
Even under noninducing conditions, kdpD expression under the control of the strong tac promoter, e.g., in the overexpression vector pPV5-9, could not complement a chromosomal kdpD deletion. Therefore, we transferred the kdpD gene of plasmid pPV5-9 into pBAD18, under control of the inducible and repressible arabinose promoter (8), using XmaI and HindIII restriction sites, resulting in plasmid pBD5-9.
PCR-based random mutagenesis of kdpD gene fragments.
The three DNA fragments coding for region I (D375 to V403), region II (L408 to S473), and region III (L479 to R526) (Fig. 1) of KdpD were subjected to random mutagenesis. A modified PCR technique according to Leung et al. (24) was used to support a random mismatch of base pairs, typically at a frequency of one or a few mutations per DNA fragment. The standard PCR buffer for the used Taq DNA polymerase varied in such a way that 1.5 mM MgCl2 was replaced by 0.25 to 0.5 mM MnCl2, and in addition 50 to 100 mM mercaptoethanol and 0 to 5% dimethyl sulfoxide were added. In each case plasmid pBD5-9 was used as a template. Unique restriction enzyme recognition sites of pBD5-9 were used for cloning of the mutated PCR fragment as indicated in Table 1. In detail, with primer pair I the mutagenic PCR leads to an SpeI-NotI DNA fragment (135 bp), primer pair II resulted in an NotI-AatII fragment (195 bp), and primer pair III produced the AatII-Bsu36I fragment (207 bp). PCR products I to III were separated by agarose gel electrophoresis, purified from the gel, digested with appropriate restriction endonucleases, and then ligated separately to similarly treated vector pBD5-9, resulting in plasmids pBD5-9* (the asterisk stands for potential amino acid substitutions in KdpD). Each of the plasmids contained a mutated DNA fragment coding for one of the regions I to III mentioned above.
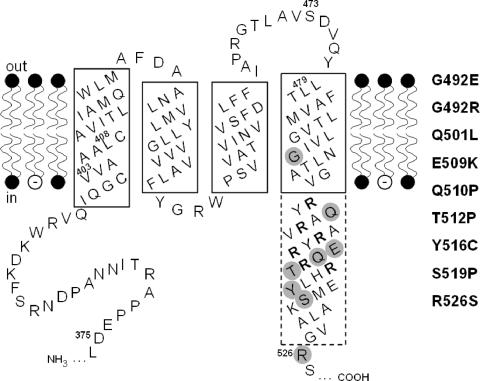
FIG. 1.
Secondary structure model of the transmembrane region of KdpD. The hydrophobic membrane-spanning domains in the phospholipid bilayer as well as parts of the adjacent hydrophilic domains are depicted. Amino acid numbers refer to positions in the KdpD protein. α-Helical regions believed to be embedded in the membrane are framed by a solid line, while the part of the fourth helix that extends into the cytoplasm is drawn with a dashed-line boundary. The arginine residues of the arginine cluster are shown in bold letters, and the three regions (I, II, and III) corresponding to DNA sequences that were subjected to random mutagenesis have been indicated by their first and last amino acid (for details, see Materials and Methods). The altered amino acids leading to a K+-insensitive phenotype are shaded gray, and the corresponding amino acid replacements are presented on the right hand.
TABLE 1.
PCR strategy used to amplify DNA sequences coding for regions I, II, and III of KdpD
| Amplified KdpD region (residues)a | Primer name | Sequence (5′ to 3′)b | Size of PCR fragment (bp) | Enzymes used for cloning |
|---|---|---|---|---|
| I (D375 to V403) | SpeIsense | 5′-CTCGATCAGGTACTAGTCGCGCTT-3′ | 135 | SpeI+NotI |
| NotIanti | 5′-AACGGCGCATAACGCGGCCGCCGCAAC-3′ | |||
| II (L408 to S473) | NotIsense | 5′-ATGCGTGGTTGCGGCCGC-3′ | 195 | NotI+AatII |
| AatIIanti | 5′-GTCAGCAGATATTGGACGTC-3′ | |||
| III (L479 to R526) | KpnIAatIIsense | 5′-CACGCGGTACCCTCGCCGTCTCTGACGTCCAATATCTGC-3′ | 207 | AatII+Bsu36I |
| Bsu36Ianti | 5′-TGGTGGCAGCAATATCCTGAGGACTGC-3′ |
Amino acid numbers refer to positions in the KdpD wild-type protein (Fig. 1). For details of the PCR method, see Materials and Methods.
Sites for restriction enzymes are shown in bold letters. The corresponding nucleotide positions in the kdpD gene are given in Materials and Methods.
Screening for K+-insensitive KdpD derivatives.
E. coli strain HAK006 carrying a kdpFABC-promoter-lacZ fusion was transformed with the mutated plasmid DNA of pBD5-9*, and transformants were spread on KML agar medium containing X-Gal. Cells of HAK006/pBD5-9 wild type form white colonies on this selection medium because wild-type kdpD cannot activate the kdpFABC-promoter-lacZ reporter gene at concentrations above 2 mM K+. A blue colony indicated that the kdpFABC operon in these cells was induced although K+ was not limited in the medium. The appropriate pBD5-9* plasmids, giving rise to blue colonies, were selected, and the mutated sequences of region I, II, or III of KdpD were subjected to DNA sequencing in order to identify the amino acid changes in KdpD. It is worth mentioning that the blue:white ratio of transformants was 1:50 for region I and II and 1:10 for region III.
Determination of kdpFABC expression in vivo.
In vivo signal transduction was probed with E. coli HAK006 transformed with the plasmids described. Cells were grown in TY medium supplemented with NaCl or sucrose or in minimal medium containing the indicated amounts of K+ (see Fig. 2). Cells were grown to mid-exponential growth phase and harvested by centrifugation. β-Galactosidase activity was determined as described previously (28) and is given in Miller units.
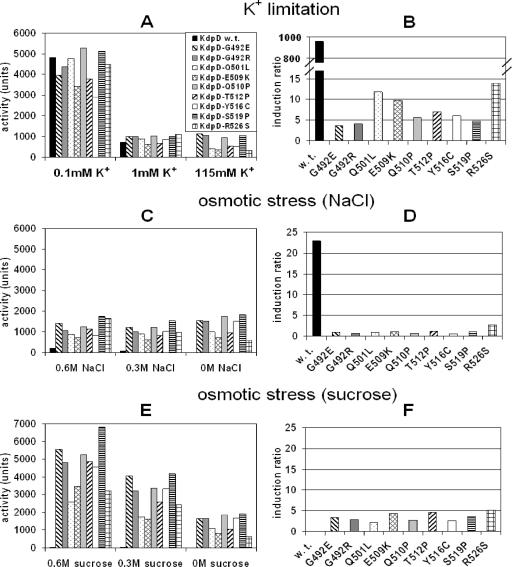
FIG. 2.
Influence of the nine single amino acid substitutions in KdpD on the regulation of kdpFABC expression under K+ limitation and osmotic stress. E. coli strain HAK006 was transformed with plasmid pBD5-9 or pBD5-9*, coding for KdpD or its derivatives carrying the indicated replacements. Cells were grown under K+-limiting conditions (A and B) or osmotic stress conditions induced by NaCl (C and D) or sucrose (E and F) (for details, see Materials and Methods). The corresponding K+, NaCl, and sucrose concentrations in the media are indicated. It should be noted that under osmotic stress conditions the medium still contains 6.6 mM K+. β-Galactosidase activities were determined as described in Materials and Methods and are given in Miller units. The data presented in graphs A, C, and E represent average values obtained in at least three independent experiments. In graphs B, D, and F, the corresponding induction ratios (inducing/noninducing conditions) are shown: 0.1 mM K+/115 mM K+ (B), 0.6 M NaCl/0 M NaCl (D), and 0.6 M sucrose/0 M sucrose (F).
Preparation of KdpD-containing everted membrane vesicles.
E. coli strain TKR2000 transformed with pBD5-9 wild-type plasmid or one of the mutated derivatives of pBD5-9* was grown aerobically at 37°C to an optical density at 600 nm of 0.5 in KML medium containing ampicillin. Expression of kdpD or kdpD derivatives (with one of the single amino acid substitutions listed in Fig. 1) was induced with 0.2% arabinose, and growth was continued for an additional 2 h. Cells were harvested and disrupted by passage through a Ribi cell fractionator. Membranes were prepared according to the method of Siebers and Altendorf (36) with the following modification: Tris-HCl was used in the same concentration instead of HEPES-Tris. Membrane vesicles in TG buffer (50 mM Tris-HCl, pH 7.5, 10% [vol/vol] glycerol) were frozen in liquid nitrogen and stored at −80°C until use.
Purification of KdpE.
Purification of the cytoplasmic His10-KdpE protein was carried out using E. coli strain BL21/pLysS/pEE as described previously (12).
Autophosphorylation, dephosphorylation, and phosphotransfer assays.
To test autophosphorylation activity, everted membrane vesicles (2 mg of protein ml−1) containing KdpD wild-type or KdpD mutant proteins were incubated at room temperature in standard phosphorylation buffer (50 mM Tris-HCl, pH 7.5, 10% glycerol, 0.5 M NaCl, 0.02 mM MgCl2 and 2 mM dithiothreitol). Phosphorylation was initiated by the addition of 20 μM [γ-32P]ATP (2.38 Ci mmol−1), and the reaction was stopped after 5 min by mixing aliquots with sodium dodecyl sulfate (SDS) sample buffer.
For dephosphorylation, assays were carried out as described previously (14, 33). Briefly, purified KdpE was first phosphorylated with wild-type KdpD by using [γ-32P]ATP in standard phosphorylation buffer, except that MgCl2 was replaced with CaCl2. Subsequently, KdpD-containing membrane vesicles were removed by centrifugation, and ATP was removed by gel filtration. Dephosphorylation of 32P-phosphorylated KdpE (KdpE∼32P) was initiated by the addition of MgCl2, ATPγS, and everted membrane vesicles containing KdpD derivatives. At different time points, aliquots were taken, and the reaction was stopped by the addition of SDS sample buffer.
For phosphotransfer measurements, autophosphorylation of membrane-bound KdpD (wild type or derivatives) was performed as indicated above. After incubation for 5 min, purified KdpE was added to a final concentration of 0.5 mg ml−1. The phosphotransfer capacity was observed by further incubation, and aliquots were removed at different time points. In addition, iso-osmolar phosphorylation buffers with different ionic compositions were used. Therefore, 0.5 M NaCl included in the phosphorylation buffer was partially replaced with different concentrations of KCl, K+-glutamate, or NH4Cl. Also the KdpE-containing sample used was adjusted to the appropriate buffer composition by dialyzing against the corresponding phosphorylation buffer.
All samples were immediately subjected to 9% or 11% SDS gels (21). Shortly before stopping SDS-polyacrylamide gel electrophoresis, a [γ-32P]ATP standard was loaded on the gel. Gels were dried and exposed to a storage phosphor screen. Phosphorylated proteins were quantified by image analysis using a PhosphorImager Storm 820 instrument (Molecular Dynamics).
Analytical procedures.
Protein was assayed by a modified Lowry method (30) using bovine serum albumin as a standard. Immunodetection of KdpD was performed with polyclonal antibodies against KdpD as described previously (42).
RESULTS
Secondary structure model of the transmembrane region of KdpD.
Theoretical and experimental analyses revealed that KdpD has four membrane-spanning segments in the middle of the polypeptide chain, whereas the N and C termini are both cytoplasmic (46). Furthermore, a cluster of arginine residues following the fourth transmembrane α-helix is found in all known KdpD sequences, and in the case of E. coli a substitution of amino acids in this cluster affects KdpD activities (15). A closer inspection of the secondary structure of the transmembrane region with the PSIPRED protein structure prediction method (26) revealed that the lengths of helices 1 to 3 are just sufficient for spanning the cytoplasmic membrane (Fig. 1). In contrast, the fourth membrane-spanning α-helix extends into the cytoplasm and consists of nearly 50 amino acids including the positive arginine cluster.
Distribution of amino acid replacements in KdpD which alter the response to K+.
Analyses of KdpD mutants led to the suggestion that the sensing mechanism for K+ limitation and osmotic upshift may be mechanistically different. Furthermore, various KdpD mutants which had lost sensitivity to the K+ signal turned out to be single amino acid replacements in close proximity to the cluster of positively charged residues (3, 40). To specifically address this issue, we screened the transmembrane region and the adjacent N-terminal and C-terminal cytoplasmic extensions of KdpD systematically for alterations that caused expression of kdpFABC in medium containing a high concentration of K+. Three DNA fragments encoding certain regions of this part of the protein (Fig. 1) were subjected to PCR-based random mutagenesis. These fragments were introduced into a plasmid containing wild-type kdpD by replacement of the corresponding wild-type sequence. A reporter strain transformed with these plasmids was used to screen for mutants that caused expression of kdpFABC at a high concentration of K+ in the medium, indicated by dark-blue colonies on KML-X-Gal agar plates. The distribution of the nine identified amino acid substitutions is presented in Fig. 1. As can be seen, no KdpD mutant with an altered response to K+ based on just a single amino acid replacement was found in regions I and II. It should be mentioned that we found some false-positive clones which were not as dark blue as expected. Indeed, they exhibited a wild-type-like K+-inducible phenotype as indicated by the quantitative β-galactosidase assay. In region II we isolated only one mutant with two amino acid substitutions, namely V438M and W446Q. In contrast, in region III, encompassing the fourth transmembrane helix and the adjacent arginine cluster (L479 to R526), we isolated nine K+-insensitive KdpD derivatives with a single amino acid replacement. In addition, six mutants contained two amino acid substitutions, and three mutants had three to four amino acid substitutions (data not shown). Those mutants were not further characterized. By taking a look at the nature of the substitutions, we found in four cases a change from a polar to a hydrophobic (Q501L, Q510P, T512P, and S519P) residue. In addition, it should be noted that in three cases proline was found as a replacement known as a helix breaker. Among the nine exchanged amino acids, there is only one (G492) located within the fourth transmembrane helix, whereas the others are presumably located within the cytoplasmic extension of that helix.
Influence of the amino acid replacements in KdpD on the regulation of kdpFABC expression under K+-limiting conditions and osmotic stress.
kdpFABC is expressed in a trk+ background (HAK006) growing in medium of standard osmolality (about 0.2 osM) when the K+ concentration of the medium falls below 2 mM or when the osmolality of the medium is increased by the addition of NaCl (22). To test the influence of the nine single amino acid replacements in KdpD on the regulation of kdpFABC expression, E. coli strain HAK006, which carries a transcriptional fusion of the upstream region of the kdpFABC operon and a promotorless lacZ gene, was used as a reporter strain. Since the amount of regulatory proteins is very critical in signal transduction, E. coli HAK006 was transformed with plasmids pBD5-9 and its derivative pBD5-9*, in which the kdpD gene is under the control of the arabinose promoter. When cells are grown in the absence of an inducer (arabinose) but in the presence of the repressor glucose, the small amount of KdpD synthesized is sufficient for complementation of the kdpD deletion strain, although the KdpD protein is not detectable in a Western blot (10). Under these conditions steady-state kdpFABC expression was determined indirectly by measuring β-galactosidase activities. As a control, however, cells were grown in the presence of arabinose. Under these conditions all KdpD derivatives were synthesized in similar amounts in comparison to wild-type KdpD (data not shown).
In a first set of experiments we tested the mutants under K+-limiting conditions (Fig. 2A). Apparently, maximal kdpFABC expression was reached only under these conditions. Both the KdpD wild-type and mutant cells responded with very high β-galactosidase activities up to 5,000 units. With sufficient K+ in the medium (K115) the mutants still exhibited appreciable activities, whereas the wild-type showed only a basal level of about 5 units. These studies revealed that the mutants exhibit a semiconstitutive phenotype with respect to K+. This can be seen clearly when the data are converted into kdpFABC induction ratios (ratio of β-galactosidase activities of inducing to noninducing conditions) (Fig. 2B). Whereas the wild type showed an induction ratio of about 1,000, the KdpD derivatives exhibited induction ratios ranging from only 4 to 14, reflecting the semiconstitutive phenotype. It is worthwhile mentioning that the magnitude of the β-galactosidase values of the mutant strains varied, but the induction ratios of the different mutants were quite constant.
In a second set of experiments we tested the response of the KdpD wild type and derivatives to an increase of osmolality. If the osmolality of the medium was increased with NaCl (Fig. 2C), cells synthesizing wild-type KdpD showed an inducible phenotype, but the induction ratio was significantly lower compared to K+ limitation (Fig. 2D). In contrast, cells synthesizing KdpD derivatives exhibited a constitutive phenotype with β-galactosidase activities of about 1,000 units. Furthermore, the very low induction ratio confirmed the constitutive expression of kdpFABC in the mutants. In the case of sucrose, an interesting kdpFABC expression pattern was found. For wild-type KdpD, kdpFABC expression is not observed under these conditions. In contrast, all of the nine KdpD mutants exhibited β-galactosidase activities that reached the same levels as under K+-limiting conditions (up to 6,000 units) (Fig. 2E). The induction ratios (Fig. 2F) revealed a low but significant induction in response to higher sucrose concentrations (semiconstitutive). It is worth mentioning that similar results were obtained in minimal medium (data not shown).
Autokinase and phosphatase activities of the KdpD derivatives with single amino acid replacements.
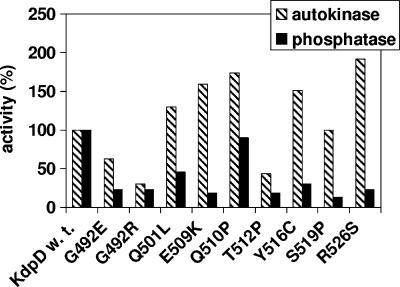
In order to investigate the autokinase and phosphatase activities of the KdpD derivatives in vitro, everted membrane vesicles of strain TKR2000 carrying plasmids with kdpD wild-type or mutated genes were used (see Materials and Methods). Membrane vesicles contained equal amounts of wild-type and mutant KdpD protein, as judged by Western blot analysis (data not shown). The results are presented as a percentage of wild-type activities (Fig. 3). Interestingly, five KdpD derivatives showed up to 90% higher autokinase activity [KdpD(Q501L), KdpD(E509K), KdpD(Q510P), KdpD(Y516C), and KdpD(R526S)], whereas three mutants [KdpD(G492E), KdpD(G492R), and KdpD(T512P)] exhibited up to 70% reduced autokinase activity in comparison to wild-type KdpD. Furthermore, in most cases a severely reduced phosphatase activity in relation to wild-type activity was observed. All KdpD derivatives were characterized by an altered ratio of the enzymatic activities, which was shifted toward the autokinase activity, implying high levels of KdpE∼P in vivo.
FIG. 3.
Autokinase and phosphatase activities of KdpD derivatives with single amino acid replacements. Kinase (gray bars) and phosphatase (black bars) activities were determined as described in Materials and Methods. Data are presented as percentages of the initial rates relative to wild-type KdpD. For wild-type KdpD (100% value) the autokinase activity was 0.1 pmol of KdpD∼P formed min−1, and the phosphatase activity was 0.02 pmol of Pi released from KdpE∼P min−1. The data presented represent average values obtained in at least three independent experiments.
Phosphotransfer activities of KdpD wild type and derivatives under different iso-osmotic conditions.
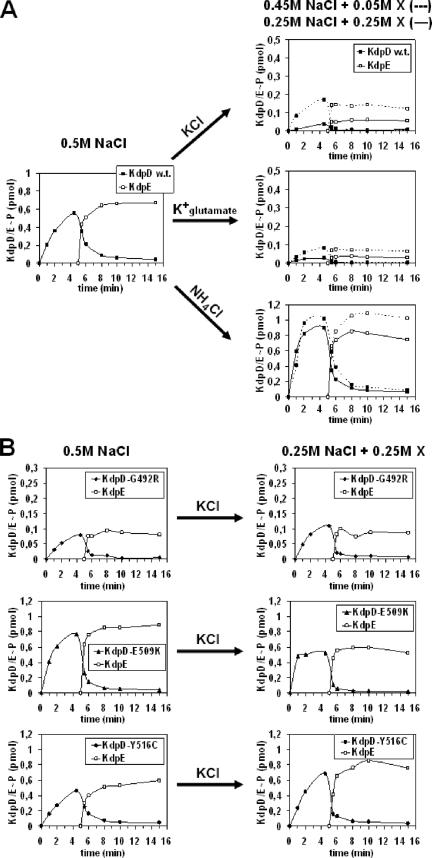
It has been shown that the autokinase activity and, in turn, the rapid phosphotransfer from KdpD∼P to KdpE depend on the ionic composition of the assay medium. High activity levels of KdpD are observed when a buffer with a high NaCl concentration (0.5 M NaCl) is used. Activity levels decrease with a decrease in the NaCl concentration. Furthermore, in the presence of KCl, KdpD autokinase activity is comparatively lower (42). Therefore, in this study we varied the ionic composition of the phosphorylation buffer without changing the osmolality. We found that partial substitution of 0.5 M NaCl with KCl, NH4Cl, or K+-glutamate had different effects on the kinase and, in turn, on the phosphotransfer activity of wild-type KdpD (Fig. 4A). In the presence of increasing KCl concentrations (50, 100, or 250 mM) (data for 100 mM are not shown), KdpD showed severely reduced autokinase and, in turn, reduced phosphotransfer activity. A continuous decrease of KdpD autokinase activity is already observed when the KCl concentration is increased from 0.1 to 10 mM (data not shown). Replacement of KCl with NH4Cl or K+-glutamate demonstrated that the effect is K+ specific (Fig. 4A). The partial replacement of NaCl with NH4Cl resulted in a slight increase in the amount of KdpD∼P, confirming previous results (45).
FIG. 4.
Autophosphorylation and phosphotransfer activities of KdpD wild type (A) and derivatives (B) under different iso-osmotic conditions. Time-dependent autophosphorylation of membrane vesicles containing KdpD, KdpD(G492R), KdpD(E509K), or KdpD(Y516C) was carried out as described in Materials and Methods. The 0.5 M NaCl of the phosphorylation buffer was partially replaced with KCl, K+-glutamate, or NH4Cl as indicated (represented by X). After 5 min of autophosphorylation, purified KdpE was added, and the phosphotransfer capacity was followed. At different time points aliquots were removed and subjected to SDS-polyacrylamide gel electrophoresis. The amounts of KdpD∼P and KdpE∼P were quantified with a phosphorimager, using [γ-32P]ATP as a standard. Please note the different scale for the enzymatic activities.
In contrast, the derivatives KdpD(G492R), KdpD(E509K), and KdpD(Y516C), which were selected on the basis of their location within the extended membrane-spanning α-helix and the nature of the amino acid substitution, exhibited modified autokinase and therefore also modified phosphotransfer capacities compared to the wild-type protein (Fig. 4B). Increasing the KCl concentration of the phosphorylation buffer did not obviously reduce the autokinase activity of the KdpD mutants (Fig. 4B). Similarly, increasing concentrations of K+-glutamate had no effect on the activities (data not shown). Furthermore, partial replacement of NaCl with NH4Cl also had no significant effect (data not shown). It should be noted that the level of KdpD(G492R∼P) is lower than the phosphorylation level of other KdpD proteins. This phenomenon could be explained by the overall reduced autokinase activity of this derivative (Fig. 3). It is worth mentioning that in all cases the phosphotransfer capacity from KdpD or its derivatives to KdpE is strongly correlated with its autokinase activity.
DISCUSSION
The processes of stimulus perception and transmembrane signaling are fundamental to bacterial sensory systems. However, little is known about how the membrane-bound histidine kinases sense their extra- or intracellular stimuli and propagate information across the cytoplasmic membrane. This is also true for KdpD. Analyses of KdpD mutants led to the suggestion that KdpD responds to two distinct stimuli, K+ limitation and high osmolality (15, 40). According to the secondary structure model (Fig. 1), most of the formerly isolated single amino acid replacements in KdpD leading to apparently constitutive kdpFABC expression (3, 15, 40) are found in the extended fourth membrane-spanning α-helix. Only a few mutants that have single amino acid replacements resulting in the same phenotype have been found in the third transmembrane helix (40) or in the conserved C-terminal domain (3). This raises the question of whether KdpD possesses a specific K+ binding site. With the method of PCR-based random mutagenesis, we systematically screened the transmembrane region and the adjacent cytoplasmic N-terminal and C-terminal extensions of KdpD. Nine KdpD mutants that had lost tight regulation of kdpFABC expression with respect to the external K+ concentration were isolated. Two of them are identical to previously isolated mutants: KdpD(G492E) (40) and KdpD(Q510P) (3). In three mutants [KdpD(G492R), KdpD(E509K), and KdpD(R526S)] the same amino acid is affected but found to be replaced by another amino acid [KdpD(G492E), KdpD(E509G), and KdpD(R526H)] (40). Among the nine alterations found, only one (G492) is located within the fourth membrane-spanning helix, whereas the others are presumably located within the cytoplasmic extension of that helix corresponding to a total of about 30 amino acids (encompassing the arginine-rich region R499 to R513). Among the six double mutants that were not further characterized (see Results), arginine substitutions were found in three derivatives (R506C, R508C, and R511H). This led to the hypothesis that this region is of structural importance for stimulus perception, perhaps by interaction of the positively charged arginine residues with the negatively charged phospholipids, as already indicated by Jung and Altendorf (15). In accord with this is the finding that KdpD autokinase activity is dependent on negatively charged phospholipids (38). It should be noted that the glutamate residue E509, replaced in previous experiments and also in our experiments, is the only acidic amino acid within the region containing the six arginine residues, which is relatively conserved among KdpD sequences. This is reminiscent of the BetP protein, which functions as a glycine betaine transport system of Corynebacterium glutamicum and which is activated by high osmolality. The C-terminal 25 amino acids of this transporter, which are involved in sensing osmolality via the internal K+ concentration, contain eight arginine residues and three glutamate residues, one of which (E572) is crucial for K+ sensing (35). The authors hypothesize that the negatively charged phospholipid headgroups interact with the terminal 25 amino acids, thus stabilizing a particular conformation of this domain important for K+ sensing. This observation lends support to the notion that different membrane proteins use the same general stimulus-sensing and/or signal-transmitting mechanism including a cluster of basic amino acids together with one or more acidic residues close to the cytoplasmic membrane. One must remember that the sensing of a stimulus might involve regions of a protein that are different from those that transmit the perceived signal (in the case of KdpD, to the kinase/phosphatase-controlling region of the conserved C-terminal domain). However, additional inter- and/or intramolecular interactions of the dimeric KdpD protein itself or with additional proteins are likely to contribute to the specificity of the KdpD sensing and/or signaling mechanism, as already shown for the cytoplasmic N-terminal and C-terminal domain of KdpD (11) and for the N-terminal domain of Mycobacterium tuberculosis KdpD with the lipoproteins LprJ and LprF (39).
Detailed studies of kdpFABC expression in vivo revealed that the sensing behavior of the KdpD derivatives isolated in this study was different for not only K+ (semiconstitutive phenotype in comparison to the inducible wild-type phenotype) but also osmotic stress. In contrast to wild-type KdpD, where inducible expression of kdpFABC is, under the applied conditions, observed only, when the osmolality of the medium is increased by salt and not by sucrose, all of the mutants exhibit a constitutive (NaCl) or semiconstitutive (sucrose) phenotype with respect to osmotic stress. These findings suggest that the cytoplasmic extension of the fourth transmembrane helix of KdpD is responsible for the sensing or transmitting properties per se and not necessarily for a specific stimulus. Moreover, it is possible that KdpD senses not two but only one overall stimulus, which exerts its effect under both growth conditions, K+ limitation and osmotic stress. Laimins et al. (22) already hypothesized that kdpFABC expression begins near the point where the growth rate starts to become limited for K+. Possibly, the cells' need for potassium or the K+ availability is changed under osmotic stress, resulting in a stimulus similar to that occurring under K+-limiting conditions. Furthermore, despite iso-osmotic stress conditions, the cells' need for potassium or the K+ availability might change depending on the osmolyte used (ionic or nonionic, respectively). Balaji et al. (2) proposed that only cations at elevated concentrations, e.g., Na+ but not nonpolar solutes such as sucrose, may compete with K+ for binding to the KdpD wild-type protein and therefore mimic K+ limitation. Malli and Epstein (25) showed that for trk+ and trk-deficient strains, both NaCl and glucose (instead of sucrose) induce kdpFABC expression, but in the case of NaCl, induction occurs at a higher K+ concentration in the medium. When wild-type KdpD was used under our experimental conditions with only NaCl as an osmolyte, kdpFABC expression was induced. This phenomenon has been shown also by others, e.g., Gowrishankar (7), Sugiura et al. (40), and Jung and Altendorf (15). In the case of our KdpD derivatives, the sensor kinase could be in a “locked-on state,” and independent of the medium conditions, expression of kdpFABC could take place, resulting in the observed constitutive phenotypes.
Since the induction ratios used with mutants in previous studies (3, 15, 40) gave rise to almost the same (semi-)constitutive phenotypes with respect to K+-limiting conditions and osmotic stress (ionic and nonionic osmolytes) as the mutants isolated in this study (data not shown), the hypothesis that KdpD responds in a fundamentally different way to a K+ signal and to an osmotic signal has to be modified. To our knowledge, no one thus far has succeeded in isolating KdpD derivatives in which kdpFABC induction ratios are modulated by only K+-limiting growth conditions and not osmotic stress. In a recent study, Rothenbücher et al. (34) reported that the C-terminal hydrophilic domain of KdpD is a K+ sensor that controls the transcription of kdpFABC. However, these studies are very difficult to interpret since they use kdpFABC induction ratios of the truncated KdpD derivative that are below 5, indicating an almost constitutive phenotype in contrast to the inducible KdpD wild-type phenotype.
In vitro phosphorylation assays using everted E. coli membrane vesicles revealed that all KdpD derivatives exhibited a severely reduced phosphatase activity in relation to their autokinase activity. Based on these results, the idea of a switch mechanism between autophosphorylation and phosphatase activity upon stimulus perception is further supported. Brandon et al. (3) already suggested that initiation of signal transduction by KdpD is mediated by the inhibition of the KdpE∼P-specific phosphatase activity. The reduced phosphatase activity of our KdpD derivatives probably results in the expression of the kdpFABC operon even under noninducing conditions, leading to the observed (semi-)constitutive phenotypes.
To exclude an osmotic effect on the phosphotransfer activity of KdpD, we developed an in vitro assay under iso-osmotic conditions. The studies of phosphotransfer from KdpD to KdpE under conditions with various ionic compositions but without changes in osmolality revealed severely reduced autokinase activity and also, in turn, reduced phosphotransfer activity in the presence of increasing K+ concentrations (from 0.1 mM up to 250 mM K+) for only wild-type KdpD. The intracellular K+ concentration in whole cells was close to 200 mM. This inhibitory effect of K+ on the phosphorylation capacity of KdpD in vitro (everted membrane vesicles) correlates with the inhibition of kdpFABC expression shown in vivo (β-galactosidase activity in growing cells). In contrast, the tested KdpD derivatives with different single amino acid alterations lost K+ specificity in vitro, which is in accord with the presented in vivo results (semiconstitutive phenotype with respect to K+).
In the oligomeric methyl-accepting chemotaxis proteins of the chemotaxis machinery of E. coli, the HAMP domain following the transmembrane signaling helix is involved in transmembrane signaling. The piston-triggered model supports the idea that attractant binding drives the signaling helix toward the cytoplasm by 1 to 2 Å (27). Recent structural studies of the HAMP domain (13) favored a model which implies helix rotation in transmembrane signaling. Upon stimulus perception from the environment, similar movement of the fourth transmembrane helix of kdpD could be envisaged, and/or the possible interaction between the N- and C-terminal domains could be affected, leading to a destabilization of the interaction between the arginine-rich region and the membrane, which, in turn, triggers changes in the kinase or phosphatase activity. Electron paramagnetic resonance studies to look for such structural changes are under way in our laboratory.
Acknowledgments
We thank Monika Nietschke for expert technical assistance and W. Epstein for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 431) and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print on 17 August 2007.
REFERENCES
- 1.Altendorf, K., and W. Epstein. 1996. The Kdp-ATPase of Escherichia coli, p. 403-420. In A. G. Lee (ed.), Biomembranes, vol. 5. JAI Press, London, United Kingdom. [Google Scholar]
- 2.Balaji, B., K. O'Connor, J. R. Lucas, J. M. Anderson, and L. N. Czonka. 2005. Timing of induction of osmotically controlled genes in Salmonella enterica serovar Typhimurium, determined with quantitative real-time reverse transcription-PCR. Appl. Environ. Microbiol. 71:8273-8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandon, L., S. Dorus, W. Epstein, K. Altendorf, and K. Jung. 2000. Modulation of KdpD phosphatase implicated in the physiological expression of the Kdp ATPase of Escherichia coli. Mol. Microbiol. 38:1086-1092. [DOI] [PubMed] [Google Scholar]
- 4.Epstein, W. 1992. Kdp, a bacterial P-type ATPase whose expression and activity are regulated by turgor pressure. Acta Physiol. Scand. 146:193-199. [PubMed] [Google Scholar]
- 5.Epstein, W. 2003. The roles and regulation of potassium in bacteria. Prog. Nucleic Acids Res. Mol. Biol. 75:293-320. [DOI] [PubMed] [Google Scholar]
- 6.Epstein, W., and B. S. Kim. 1971. Potassium transport loci in Escherichia coli K-12. J. Bacteriol. 108:639-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gowrishankar, J. 1985. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J. Bacteriol. 164:434-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heermann, R. 2001. Ph.D. thesis. University of Osnabrück, Osnabrück, Germany.
- 10.Heermann, R., A. Fohrmann, K. Altendorf, and K. Jung. 2003. The transmembrane domains of the sensor kinase KdpD of Escherichia coli are not essential for sensing K+ limitation. Mol. Microbiol. 47:839-848. [DOI] [PubMed] [Google Scholar]
- 11.Heermann, R., K. Altendorf, and K. Jung. 2000. The hydrophilic N-terminal domain complements the membrane-anchored C-terminal domain of the sensor kinase KdpD of Escherichia coli. J. Biol. Chem. 275:17080-17085. [DOI] [PubMed] [Google Scholar]
- 12.Heermann, R., K. Altendorf, and K. Jung. 2003. The N-terminal input domain of the sensor kinase KdpD of Escherichia coli stabilizes the interaction between the cognate response regulator KdpE and the corresponding DNA-binding site. J. Biol. Chem. 278:51277-51284. [DOI] [PubMed] [Google Scholar]
- 13.Hulko, M., F. Berndt, M. Gruber, J. U. Linder, V. Truffault, A. Schulz, J. Martin, J. E. Schulz, A. N. Lupas, and M. Coles. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929-940. [DOI] [PubMed] [Google Scholar]
- 14.Jung, K., and K. Altendorf. 1998. Truncation of amino acids 12-228 causes deregulation of the phosphatase activity of the sensor kinase KdpD of Escherichia coli. J. Biol. Chem. 273:17406-17410. [DOI] [PubMed] [Google Scholar]
- 15.Jung, K., and K. Altendorf. 1998. Individual substitutions of clustered arginine residues of the sensor kinase KdpD of Escherichia coli modulate the ratio of kinase to phosphatase activity. J. Biol. Chem. 273:26415-26420. [DOI] [PubMed] [Google Scholar]
- 16.Jung, K., and K. Altendorf. 2003. Stimulus perception and signal transduction by the KdpD/KdpE system of Escherichia coli, p. 53-58. In P. Dürre and B. Friedrich (ed.), Regulatory networks in prokaryotes. Horizon Scientific Press, Wymondham, United Kingdom.
- 17.Jung, K., B. Tjaden, and K. Altendorf. 1997. Purification, reconstitution and characterization of KdpD, the turgor sensor of Escherichia coli. J. Biol. Chem. 272:10847-10852. [DOI] [PubMed] [Google Scholar]
- 18.Jung, K., R. Heermann, M. Meyer, and K. Altendorf. 1998. Effect of cysteine replacements on the properties of the turgor sensor KdpD of Escherichia coli. Biochim. Biophys. Acta 1372:311-322. [DOI] [PubMed] [Google Scholar]
- 19.Jung, K., M. Veen, and K. Altendorf. 2000. K+ and ionic strength directly influence the autophosphorylation activity of the putative turgor sensor KdpD of Escherichia coli. J. Biol. Chem. 275:40142-40147. [DOI] [PubMed] [Google Scholar]
- 20.Kollmann, R., and K. Altendorf. 1993. ATP-driven potassium transport in right-side-out membrane vesicles via the Kdp-system of Escherichia coli. Proc. Natl. Acad. Sci. USA 75:3216-3219. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Laimins, L. A., D. B. Rhoads, and W. Epstein. 1981. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 78:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S. J., and J. D. Gralla. 2004. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell 14:153-162. [DOI] [PubMed] [Google Scholar]
- 24.Leung, D. W., E. Chen, and D. V. Goeddel. 1989. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1:11-15. [Google Scholar]
- 25.Malli, R., and W. Epstein. 1998. Expression of the Kdp ATPase is consistent with regulation by turgor pressure. J. Bacteriol. 180:5102-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 27.Miller, A. S., and J. F. Falke. 2004. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry 43:1763-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 30.Peterson, G. L. 1977. A simplification of the protein assay method of Lowry et al., which is more generally applicable. Anal. Biochem. 83:346-356. [DOI] [PubMed] [Google Scholar]
- 31.Polarek, J. W., G. Williams, and W. Epstein. 1992. The products of kdpDE operon are required for expression of the Kdp-ATPase of Escherichia coli. J. Bacteriol. 174:2145-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prince, W. S., and M. R. Villarejo. 1990. Osmotic control of proU transcription is mediated through direct action of K+ glutamate on the transcription complex. J. Biol. Chem. 265:17673-17679. [PubMed] [Google Scholar]
- 33.Puppe, W., P. Zimmann, K. Jung, M. Lucassen, and K. Altendorf. 1996. Characterization of truncated forms of the KdpD protein, the sensor kinase of the K+-translocating Kdp system of Escherichia coli. J. Biol. Chem. 271:25027-25034. [DOI] [PubMed] [Google Scholar]
- 34.Rothenbücher, M. C., S. J. Facey, D. Kiefer, M. Kossmann, and A. Kuhn. 2006. The cytoplasmic C-terminal domain of the Escherichia coli KdpD protein functions as a K+ sensor. J. Bacteriol. 188:1950-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiller, D., V. Ott, R. Krämer, and S. Morbach. 2006. Influence of membrane composition on osmosensing by the betaine carrier BetB from Corynebacterium glutamicum. J. Biol. Chem. 281:7737-7746. [DOI] [PubMed] [Google Scholar]
- 36.Siebers, A., and K. Altendorf. 1988. The K+-translocating Kdp-ATPase from Escherichia coli. Purification, enzymatic properties and production of complex- and subunit-specific antisera. Eur. J. Biochem. 178:131-140. [DOI] [PubMed] [Google Scholar]
- 37.Siegele, D. A. 2005. Universal stress proteins in Escherichia coli. J. Bacteriol. 187:6253-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stallkamp, I., W. Dowhan, K. Altendorf, and K. Jung. 1999. Negatively charged phospholipids influence the activity of the sensor kinase KdpD of Escherichia coli. Arch. Microbiol. 172:295-302. [DOI] [PubMed] [Google Scholar]
- 39.Steyn, A. J., J. Joseph, and B. R. Bloom. 2003. Interaction of the sensor module of Mycobacterium tuberculosis H37Rv KdpD with members of the Lpr family. Mol. Microbiol. 47:1075-1089. [DOI] [PubMed] [Google Scholar]
- 40.Sugiura, A., K. Hirokawa, K. Nakashima, and T. Mizuno. 1994. Signal-sensing mechanisms of the putative osmosensor KdpD in Escherichia coli. Mol. Microbiol. 14:929-938. [DOI] [PubMed] [Google Scholar]
- 41.Sugiura, A., K. Nakashima, K. Tanaka, and T. Mizuno. 1992. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol. Microbiol. 6:1769-1776. [DOI] [PubMed] [Google Scholar]
- 42.Voelkner, P., W. Puppe, and K. Altendorf. 1993. Characterisation of the KdpD protein, the sensor kinase of the K+-translocating system of Escherichia coli. Eur. J. Biochem. 217:1019-1026. [DOI] [PubMed] [Google Scholar]
- 43.Walderhaug, M. O., J. W. Polarek, P. Voelkner, J. M. Daniel, J. E. Hesse, K. Altendorf, and W. Epstein. 1992. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J. Bacteriol. 174:2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanish-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 45.Zimmann, P. 1995. Ph.D. thesis. University of Osnabrück, Osnabrück, Germany.
- 46.Zimmann, P., W. Puppe, and K. Altendorf. 1995. Membrane topology analysis of the sensor kinase KdpD of Escherichia coli. J. Biol. Chem. 270:28282-28288. [DOI] [PubMed] [Google Scholar]