Abstract
During sporulation of Bacillus subtilis, four regulatory proteins act in the order σE, SpoIIID, σK, and GerE to temporally control gene expression in the mother cell. σE and σK work sequentially with core RNA polymerase to transcribe different sets of genes. SpoIIID and GerE are small, sequence-specific DNA-binding proteins that activate or repress transcription of many genes. Previous studies showed that transcriptionally active σK RNA polymerase inhibits early mother cell gene expression, reducing accumulation of SpoIIID late in sporulation. Here, the effects of perturbing the mother cell gene regulatory network by maintaining the SpoIIID level late during sporulation are reported. Persistent expression was obtained by fusing spoIIID to the σK-controlled gerE promoter on a multicopy plasmid. Fewer heat- and lysozyme-resistant spores were produced by the strain with persistent spoIIID expression, but the number of spores resistant to organic solvents was unchanged, as was their germination ability. Transmission electron microscopy showed structural defects in the spore coat. Reporter fusions to σK-dependent promoters showed lower expression of gerE and cotC and higher expression of cotD. Altered expression of cot genes, which encode spore coat proteins, may account for the spore structural defects. These results suggest that one role of negative feedback by σK RNA polymerase on early mother cell gene expression is to lower the level of SpoIIID late during sporulation in order to allow normal expression of genes in the σK regulon.
Upon sensing nutrient limitation, the gram-positive bacterium Bacillus subtilis initiates an adaptive process called sporulation that culminates in the formation of a dormant spore (reviewed in references 12 and 28). During sporulation, B. subtilis undergoes an asymmetric division, forming a larger mother cell (MC) compartment and a smaller forespore (FS) compartment. Different genes are expressed in each compartment. Signaling between the MC and the FS ensures precise temporal and spatial control of gene expression. The products of genes expressed in the two cell types cause further morphological changes. The FS is engulfed by the MC membrane, surrounding the FS with two membranes and pinching it off as a protoplast within the MC. Then, cell wall-like material called cortex is deposited between the two membranes of the FS. Next, the spore coat is assembled on the surface of the FS and its interior or core is dehydrated. The mature spore, which is resistant to environmental insults such as heat, UV light, lytic enzymes, and chemicals, is released by lysis of the MC. Under favorable environmental conditions, the dormant spore germinates and forms a growing cell.
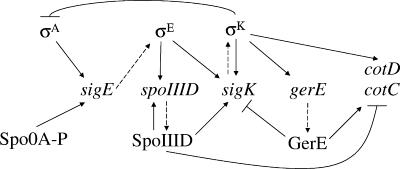
In the MC, a hierarchical cascade of four regulatory factors, σE, SpoIIID, σK, and lastly GerE, governs gene expression during sporulation (55) (Fig. 1). The sigE gene is transcribed by σA RNA polymerase (RNAP), the major form of RNAP in growing cells, prior to asymmetric division, but initiation of transcription at the promoter of the spoIIG operon, which includes sigE, requires phosphorylated Spo0A, a key transcription factor that governs entry into the sporulation process (reviewed in reference 20). After formation of the asymmetric septum, phosphorylated Spo0A persists in the MC and the product of sigE, pro-σE, accumulates primarily in the MC (15, 16). Pro-σE is cleaved in response to a signal from the FS (23, 27, 33), forming active σE RNAP in the MC, where it transcribes more than 260 genes, including those that encode SpoIIID and pro-σK (13, 14, 46) (Fig. 1). SpoIIID is a small, sequence-specific DNA-binding protein (18) that positively regulates as least eight transcription units in the σE regulon, including the sigK gene (Fig. 1), and negatively regulates at least 62 transcription units (13). SpoIIID also negatively regulates genes in the σK regulon, including cotC and cotD (18, 25, 29) (Fig. 1), which encode spore coat proteins (9). GerR (not shown in Fig. 1) is a second transcription factor under σE RNAP control that negatively regulates 10 transcription units but is not known to affect the expression of the four regulators in the cascade (13). The sigK gene encodes pro-σK (49). Like pro-σE, pro-σK is activated by proteolytic cleavage in response to an FS signal (6, 34), although the mechanisms of signaling and proteolysis are different (reviewed in reference 28). σK RNAP transcribes more than 100 genes, including many cot genes that encode spore coat proteins, and the gene encoding GerE (13, 46) (Fig. 1). Like SpoIIID, GerE is a small, sequence-specific DNA-binding protein (54). It positively regulates at least 27 transcription units, including the cotC and cotD genes, and negatively regulates at least 36 transcription units (13), including the sigK gene (24, 54) (Fig. 1).
FIG. 1.
Gene regulatory network in the MC during sporulation of B. subtilis. Dashed arrows indicate gene-to-protein relationships. Solid arrows or lines with barred ends represent positive or negative regulation, respectively. See the text for explanation and references.
All of the regulatory interactions depicted in Fig. 1 are direct effects based on in vitro studies, with one exception. The negative-feedback loop from σK to σA is inferred from in vivo studies (17, 52, 53). Transcriptionally active σK RNAP inhibits early MC gene expression, including transcription of sigE by σA RNAP (53). This results in reduced accumulation of σE and SpoIIID late during sporulation (17, 52). Neither the mechanism nor the significance of this negative-feedback loop is known. To access its significance, we engineered ectopic expression of spoIIID such that the SpoIIID level was maintained late during sporulation. This resulted in fewer heat- and lysozyme-resistant spores being produced, and most of the spores exhibited structural changes in their coats. Perturbing the MC gene regulatory network in this way also uncovered a novel connection that was unexpected from in vitro studies; persistent spoIIID expression lowered expression of a gerE-lacZ fusion. These results demonstrate the importance of lowering the SpoIIID level late during sporulation, which is one consequence of the σK-dependent negative-feedback loop.
MATERIALS AND METHODS
Construction of plasmids.
A DNA fragment containing the gerE promoter region was synthesized by PCR using primers 5′-GGGAATTCCTGGACGGATGAGGAGAAAG-3′ and 5′-GGGAAGCTTGCGAATCAGAAACGAATGG-3′, which contain EcoRI and HindIII restriction sites (underlined), respectively. This fragment spans from bp −96 to bp +33 relative to the transcriptional start site of gerE and does not include the ribosomal binding site (7). The PCR product was digested with EcoRI and HindIII and ligated into EcoRI-HindIII-digested pDG364 (26), resulting in plasmid pRB1 (R. Burri and L. Kroos, unpublished data). The sequence of this insert was determined by the Research Technology Support Facility at Michigan State University in order to ensure that no errors occurred during the PCR. A DNA fragment containing the spoIIID gene, from bp +145 to bp +463 relative to the transcriptional start site of spoIIID (50), including the ribosomal binding site and the entire coding region, was amplified by PCR using primers 5′-GGAAGCTTAGGGAGGTCGAGTGGTGTG-3′ and 5′-CGGATCCCAAGAAGGCAATGCCAGG-3′, which contain HindIII and BamHI restriction sites (underlined), respectively. The PCR product was digested with HindIII and BamHI and ligated with HindIII-BamHI-digested pRB1, resulting in pRB2 (R. Burri and L. Kroos, unpublished). The sequence of the entire PgerE-spoIIID fusion in pRB2 was determined as described above. Next, pRB2 was digested with EcoRI and BamHI, the PgerE-spoIIID fragment was purified, and it was ligated with a 6.6-kb fragment of EcoRI (partial digestion)-BamHI-digested pDG148 (48), resulting in pJP1. To construct a control plasmid containing only PgerE, pJP1 was digested with BamHI and HindIII to remove the spoIIID insert. The resulting 6.7-kb fragment was treated with the Klenow fragment of DNA polymerase I, and the blunt ends were ligated by T4 DNA ligase, resulting in pJP2.
Bacterial strains.
Escherichia coli strain AG115 [araD139 Δ(ara leu)7697 ΔlacX74 galU galK hsr hsm+ strA (F′ proAB lacIqZ::Tn5)] was obtained from A. Grossman (Massachusetts Institute of Technology). It was used during construction and maintenance of plasmids. Luria-Bertani (LB) medium (44) was used to grow E. coli and B. subtilis and was supplemented with appropriate antibiotics. PY79 (Spo+ prototroph) (51), BK556 (spoIVCB23) (31), and OR825 (PY79 SPβ::cotC-lacZ) (5) were provided by R. Losick (Harvard University). EUDC9901 (trpC2 pheA1 gerE::kan) (4) was provided by C. Moran (Emory University). pJP1 was transformed into PY79 and OR825 as described previously (21), with selection by addition of kanamycin sulfate (5 μg/ml) to the medium, generating strains BJP1 and BJP3, respectively. Similarly, pJP2 was transformed into PY79 and OR825 to generate strains BJP2 and BJP4, respectively. SPβ::cotD-lacZ and SPβ::gerE-lacZ have been described previously (6, 7). Specialized transduction was used to move lacZ fusions into BJP1, BJP2, and PY79 as described previously (21). Transductants were selected on LB agar containing chloramphenicol (5 μg/ml). In each case, at least 10 candidates were transferred onto a DSM agar (21) plate with 5-bromo-4-cholro-3-indolyl-β-d-galactopyranoside (40 μg/ml). Three or more isolates with average blue color were saved for further analysis, excluding occasional isolates with abnormally high or low β-galactosidase activity.
Cell growth and sporulation.
Sporulation was induced by resuspension of cells in SM medium as described previously (21). The time of resuspension is defined as the onset of sporulation (T0).
Western blot analysis.
Starting at 3 h into sporulation (T3) and at hourly intervals thereafter until T9, 0.5-ml samples were subjected to centrifugation (14,000 × g for 1 min), the supernatants were removed, and cell pellets were stored at −70°C. Preparation of whole-cell extracts, electrophoresis, and electroblotting were as described previously (17, 34). The blots were probed with anti-SpoIIID antiserum diluted 1:10,000 (17) or polyclonal anti-pro-σK antiserum diluted 1:10,000 (34). Immunodetection of primary antibodies was as described previously (30).
Analysis of β-galactosidase activity.
Samples were collected during sporulation as described above. Cell pellets were stored at −70°C prior to the assay. Cells were resuspended and then treated with lysozyme and permeabilized by toluene as described previously (38). The β-galactosidase specific activity was determined as described previously (38), using o-nitrophenol-β-d-galactopyranoside as the substrate. One unit of the enzyme hydrolyzed 1 μmol of substrate per minute per unit of initial-culture optical density at 595 nm.
Spore purification and germination and resistance assays.
Spores were harvested at T24 by centrifugation at 7,000 × g for 10 min, washed with 4°C water once, and stored at 4°C overnight. The next day, spores were purified on a step gradient of 20% to 50% RenoCal-76 (Bracco Diagnostics Inc.) as described previously (22). The purity of the spores was verified by microscopy. The germination assay was performed with purified spores, using l-alanine (10 mM) as the germinant as described previously (39). Assays for resistance to heat, lysozyme, and organic solvents were performed at T24 without spore purification, as described previously (21).
Transmission electron microscopy of spores.
Spores were harvested at T24, washed with water, and immediately fixed as described previously (37).
RESULTS
Engineering B. subtilis to maintain the SpoIIID level late during sporulation.
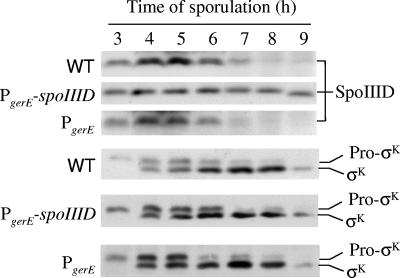
In wild-type PY79, the level of SpoIIID reaches a maximum at 5 h into sporulation and decreases thereafter (17) (Fig. 2). To test whether the SpoIIID decrease is important, we initially constructed a B. subtilis strain with a single copy of a PgerE-spoIIID fusion integrated at the amyE locus in the chromosome. Unlike the native spoIIID promoter, which is under σE RNAP control, the gerE promoter is under the control of the later-acting σK RNAP (Fig. 1). However, at single copy, the PgerE-spoIIID fusion had no apparent effect on the level of SpoIIID during sporulation (R. Burri and L. Kroos, unpublished). Therefore, we constructed pJP1, a multicopy plasmid bearing the PgerE-spoIIID fusion. pJP1 was transformed into PY79, resulting in B. subtilis BJP1. As a control, a plasmid with only PgerE, pJP2, was also transformed into PY79, resulting in strain BJP2. During sporulation, the SpoIIID level in BJP2 decreased after 5 h, as observed for PY79, but in BJP1 the level of SpoIIID remained about the same at least until 9 h into sporulation (Fig. 2). We conclude that BJP1 maintains the level of SpoIIID late during sporulation.
FIG. 2.
Levels of SpoIIID pro-σK, and σK during sporulation. B. subtilis wild-type (WT) PY79, BJP1 bearing pJP1 with the PgerE-spoIIID insert, and BJP2 bearing pJP2 with the PgerE insert were induced to sporulate by resuspension in SM medium, and samples were collected at hourly intervals beginning at 3 h after the onset of sporulation. Equal volumes (5 μl) of whole-cell lysates were fractionated on SDS-14% polyacrylamide gels and subjected to Western blot analysis with anti-SpoIIID or anti-pro-σK serum.
Pro-σK and σK levels are unchanged in B. subtilis strains that maintain the SpoIIID level late during sporulation.
SpoIIID activates or represses many genes in the MC (13, 18, 19). One key gene that it activates is sigK (Fig. 1), which encodes pro-σK (49). SpoIIID activates sigK transcription by RNAP containing either σE or σK (18). Hence, maintaining the SpoIIID level late during sporulation might elevate the pro-σK and/or σK level at late times if SpoIIID is limiting for transcription of sigK and/or for processing of pro-σK to σK.
The levels of pro-σK and σK were indistinguishable in PY79, BJP1, and BJP2 (Fig. 2). This result indicates that SpoIIID is not the limiting factor for pro-σK or σK production late during sporulation. Negative-feedback loops in the MC network (Fig. 1) may limit σE and σK production late during sporulation, counteracting potential activation by SpoIIID of sigK transcription.
Maintaining the SpoIIID level late during sporulation alters gene expression.
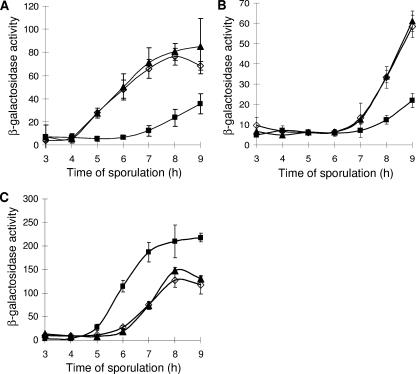
SpoIIID strongly represses transcription of cotD (18, 19) and cotC (25) by σK RNAP in vitro and weakly activates transcription of gerE by σK RNAP in vitro (19). Using lacZ reporter fusions to these three promoters, we examined the effect of persistent spoIIID expression late during sporulation. Surprisingly, gerE-lacZ expression was reduced in the strain that maintains the SpoIIID level late during sporulation, compared with the gerE-lacZ levels in the wild type and the control strain with multicopy PgerE, which were indistinguishable (Fig. 3A). This result was unanticipated from the in vitro study (19). It suggests that SpoIIID can directly or indirectly inhibit gerE expression.
FIG. 3.
Effects on gene expression. β-Galactosidase activity during sporulation after resuspension in SM medium was measured for B. subtilis containing lacZ fused to the gerE (A), cotC (B), or cotD (C) promoter in wild-type PY79 (⋄) or strains bearing multicopy PgerE-spoIIID (▪) or PgerE (▴). Each point is the average for three determinations, and error bars show 1 standard deviation of the data.
Since SpoIIID represses cotC transcription by σK RNAP in vitro (25), it was not surprising that cotC-lacZ expression was lower in the strain with multicopy PgerE-spoIIID than in the wild type or the control strain bearing multicopy PgerE (Fig. 3B). Moreover, GerE activates cotC transcription by σK RNAP in vitro (54), so diminished gerE expression in the PgerE-spoIIID strain (Fig. 3A) may also contribute to its diminished cotC-lacZ expression (Fig. 3B).
Expression of cotD-lacZ rose earlier and reached a higher level in the PgerE-spoIIID strain than in the wild type or the PgerE control strain (Fig. 3C). This may not be surprising in light of the diminished gerE expression in the PgerE-spoIIID strain (Fig. 3A), because a low level of GerE activates cotD transcription by σK RNAP in vitro, but at a higher level, GerE represses (24). Although SpoIIID strongly represses transcription of cotD by σK RNAP in vitro (18, 19), the SpoIIID level in the PgerE-spoIIID strain may not be high enough for repression to be the dominant effect.
We conclude that maintaining the SpoIIID level late during sporulation lowers gerE-lacZ expression and alters the expression levels of other genes in the σK regulon, increasing or decreasing their expression levels depending on the effects of SpoIIID and GerE at particular promoters.
Persistent spoIIID expression affects certain spore resistance properties.
The effects of maintaining the SpoIIID level late during sporulation on the numbers of heat-, lysozyme-, phenol-, ethanol-, and chloroform-resistant spores were measured. B. subtilis strain BK556 (spoIVCB23), which fails to make σK (34), served as a negative control. In each assay, BK556 produced at least 105 fewer resistant spores (data not shown) than the wild type (PY79 in Fig. 4).
FIG. 4.
Resistance properties of spores. Ratios of CFU counts after the indicated treatments to those before the treatments were determined for B. subtilis wild-type PY79 (white bars), strain BJP1 bearing multicopy PgerE-spoIIID (gray bars), and strain BJP2 bearing multicopy PgerE (black bars) at 24 h after resuspension in SM medium. Bars show the averages for three determinations, and error bars show 1 standard deviation of the data. Asterisks indicate P values of <0.05 in the comparisons between BJP1 and both PY79 and BJP2.
About 70% of the viable cells for wild-type PY79 and the multicopy PgerE control strain BJP2 at 24 h after the onset of sporulation were heat resistant (Fig. 4). However, for the multicopy PgerE-spoIIID strain BJP1, only about 30% of the viable cells were heat resistant. Statistical analysis of the data indicated no significant difference between PY79 and BJP2, but the differences between these two strains and BJP1 were significant (P values of less than 0.05 in Student t tests).
About 80% of the viable PY79 and BJP2 cells were lysozyme resistant at 24 h after the onset of sporulation, while the number was only about 50% for BJP1 (Fig. 4). Again, statistical analysis indicated a significant difference (P values of less than 0.05 in Student t tests) between BJP1 and either PY79 or BJP2 but no significant difference between the two control strains.
Resistance to three types of organic solvents was tested: the water-immiscible solvent chloroform, the water-miscible solvent ethanol, and the organic acid phenol. On average, BJP1 exhibited slightly less resistance than the two control strains for each treatment (Fig. 4), but these differences were not considered statistically significant, because no pairwise comparison between the data obtained for two strains treated with the same organic solvent yielded a P value of less than 0.05 in Student t tests.
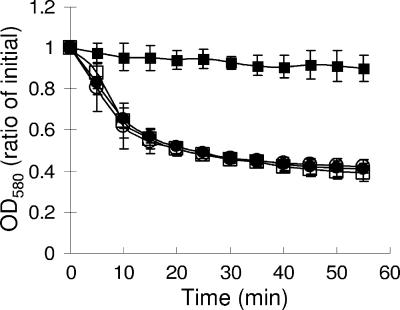
Maintaining the SpoIIID level late during sporulation does not impair spore germination.
Spores were harvested 24 h after the onset of sporulation and purified. Germination was assayed as the change in absorbance at 580 nm over time after exposure to the germinant l-alanine. A germination mutant, B. subtilis EUDC9901 (gerE), showed very little change in absorbance even after 55 min (Fig. 5). In contrast, the absorbance levels of wild-type PY79 spores decreased rapidly after exposure to l-alanine, and similar results were observed for strains bearing multicopy PgerE-spoIIID or PgerE. We conclude that spores produced by a strain with persistent spoIIID expression germinate normally.
FIG. 5.
Spore germination. B. subtilis wild-type PY79 (□), strain BJP1 bearing multicopy PgerE-spoIIID (•), strain BJP2 bearing multicopy PgerE (○), and gerE mutant EUDC9901 (▪) were induced to sporulate by resuspension in SM medium; samples were collected after 24 h; spores were purified; and quantitative germination assays were performed. Each point is the average for three determinations, and error bars show 1 standard deviation of the data. OD580, optical density at 580 nm.
Persistent spoIIID expression causes a defect in spore coat structure.
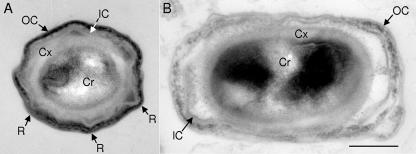
To assess the consequences of persistent spoIIID expression on the assembly of cellular structures, we carried out thin-section transmission electron microscopy of spores. The spores from the wild type (data not shown) and the control strain with multicopy PgerE (Fig. 6A) were indistinguishable. As is characteristic of mature spores, the core, cortex, and inner and outer coat layers were readily identified, and the coats had ridges. Approximately 10% of the spores from the strain with multicopy PgerE-spoIIID resembled wild-type spores (data not shown). The remaining 90% showed significant defects in coat assembly. While structures resembling the inner and outer coat layers were present, these were thinner and disorganized (Fig. 6B). Additionally, the coat was no longer in close apposition to the cortex and ridges were less evident.
FIG. 6.
Thin-section electron microscopic analysis of spores. Spores from B. subtilis BJP2 bearing multicopy PgerE (A) or BJP1 bearing multicopy PgerE-spoIIID (B) were harvested 24 h after resuspension in SM medium, washed, and immediately fixed before further processing. In panel A, a section through the short axis of the spore is shown. In panel B, the cross section is through the long axis. Therefore, the difference in the shapes of the two spores is insignificant (i.e., both strains, like wild-type PY79, showed mixtures of spherical and ovoid sections). The core (Cr), cortex (Cx), inner coat (IC), outer coat (OC), and ridges (R) are indicated. The bar applies to both panels and indicates 200 nm.
DISCUSSION
Our results demonstrate that maintaining the SpoIIID level late during sporulation alters the expression levels of genes in the σK regulon, lowers the numbers of heat- and lysozyme-resistant spores produced, and causes a defect in spore coat assembly. Clearly, it is important that the SpoIIID level decreases in order for spore formation to proceed normally during the late stages. One contributor to the SpoIIID decrease is a negative-feedback loop initiated by σK RNAP that inhibits early gene expression in the MC (17, 52, 53). Our finding that a single copy of the PgerE-spoIIID fusion was insufficient to detectably boost the SpoIIID level late during sporulation was surprising, as was the possibly related observation that SpoIIID can negatively regulate gerE expression. While multiple copies of PgerE-spoIIID did allow the SpoIIID level to be maintained late during sporulation, the levels of pro-σK and σK were unchanged, indicating that the MC regulatory network is somewhat resistant to perturbation. Such robustness is a common feature of regulatory networks (1, 47). Our results reveal elements of both robustness and susceptibility to perturbation in the MC regulatory network.
A single copy of PgerE-spoIIID did not result in perceptibly higher accumulation of SpoIIID late during sporulation. We chose the gerE promoter because it is strongly transcribed by σK RNAP in vitro (54) with little effect of SpoIIID (19). However, we discovered that in cells bearing multicopy PgerE-spoIIID, the level of SpoIIID is elevated late during sporulation (Fig. 2) and gerE-lacZ expression is reduced (Fig. 3A). Hence, SpoIIID might negatively autoregulate PgerE-spoIIID expression, necessitating a high copy number to maintain the SpoIIID level late during sporulation. Alternatively or in addition, one or more posttranscriptional mechanisms might inhibit SpoIIID accumulation late during sporulation. If so, such a mechanism(s) would presumably contribute to the decrease in SpoIIID level observed in wild-type cells late during sporulation (17).
Multicopy PgerE exhibited none of the effects of multicopy PgerE-spoIIID. Both plasmids were derived from pUB110, which is maintained at about 50 copies/cell (36). Most of these copies would presumably be in the MC (owing to its larger size) after polar septation. Considering that σK RNAP transcribes approximately 71 genes or operons in the MC during sporulation (13, 46), the PgerE-containing plasmid is expected to significantly increase the number of σK-dependent promoters in the MC. Yet, the strain containing this plasmid showed no differences from the wild type in any of our assays. Moreover, a multicopy PgerE-cotC fusion did not lower the numbers of heat- and lysozyme-resistant spores produced (L. Wang, J. Perpich, and L. Kroos, unpublished data), as did multicopy PgerE-spoIIID (Fig. 4). We conclude that neither PgerE nor PgerE-cotC titrates σK RNAP or other cellular resources sufficiently to inhibit sporulation. An earlier study showed that when σK is produced at a much lower level than normal, it is sufficient for considerable σK-dependent gene expression and sporulation (35). Taken together, our results suggest that σK is normally made in excess during sporulation.
Our results also suggest that the regulatory network resists elevating the levels of pro-σK and σK late during sporulation. Transcription of sigK depends absolutely on SpoIIID (32). However, maintaining the SpoIIID level late during sporulation did not elevate the pro-σK or σK level (Fig. 2). This suggests that some other factor limits sigK expression late during sporulation. By that time, the level of σE is decreased due to negative feedback by σK RNAP (52), so only σK RNAP can transcribe sigK. One factor known to limit sigK transcription is GerE (24, 54). In a gerE mutant, sigK expression is elevated about twofold (24). A mutation in the GerE binding site in the sigK promoter region elevates the pro-σK and σK levels approximately twofold during sporulation (L. Wang, J. Perpich, and L. Kroos, unpublished). However, no further enhancement of the pro-σK or σK level was observed when multicopy PgerE-spoIIID was introduced into the binding site mutant (L. Wang, J. Perpich, and L. Kroos, unpublished). Under these conditions, neither SpoIIID (acting positively) nor GerE (acting negatively) should limit sigK expression late during sporulation. Perhaps under these conditions, the ability to process pro-σK to active σK or degradation of σK limits sigK expression.
Given the unchanged level of σK in cells engineered to maintain the SpoIIID level late during sporulation, we would not have predicted the observed decrease in gerE-lacZ expression (Fig. 3A), because σK RNAP transcribes gerE and SpoIIID has little effect, based on in vitro studies (19). The same gerE promoter-containing fragment (bp −96 to +170) used in the in vitro study was used to create the gerE-lacZ translational fusion used here, so it is unlikely that the gerE-lacZ fusion has an additional SpoIIID binding site. Nevertheless, further experiments are warranted to test whether maintaining the SpoIIID level late during sporulation inhibits gerE expression (e.g., by measuring gerE mRNA and GerE protein levels), since this appears to be a previously unknown connection in the MC regulatory network, coordinating the disappearance of SpoIIID with the appearance of GerE.
Maintaining the SpoIIID level late during sporulation decreased expression of cotC-lacZ (Fig. 3B) and increased that of cotD-lacZ (Fig. 3C). These effects can be understood from the effects of SpoIIID and GerE on transcription of cotC (25, 54) and cotD (18, 19, 24) by σK RNAP in vitro, as noted in Results. We predict that the expression levels of other genes in the σK regulon whose transcription is influenced by SpoIIID and/or GerE are also altered in cells engineered to maintain the SpoIIID level late during sporulation. Only 4 genes in the σK regulon have been shown to be influenced by SpoIIID (18, 25), but at least 53 genes or operons have been shown to be positively or negatively regulated by GerE, based on genome-wide DNA microarray expression profiling experiments (13, 46).
Altered expression of genes in the σK regulon presumably causes the altered spore resistance properties (Fig. 4) and the spore coat structural defects (Fig. 6B) observed for spores derived from cells with persistent spoIIID expression. Interestingly, the coat protein compositions of these spores are not detectably altered, as determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis of SDS-extractable coat proteins (data not shown). A possible interpretation of this is that proper coat assembly depends on regulation of coat protein levels in the MC cytoplasm prior to deposition around the FS and not just on the final composition of the coat. In this view, the normal changes in SpoIIID and GerE levels regulate coat protein levels so that interactions interfering with coat assembly are suppressed. Persistent expression of spoIIID might lead to the abnormal simultaneous appearance of proteins that can interact nonproductively and, as a result, affect the usual course of coat formation without altering its final composition. Alternatively, it is possible that coat composition is altered in spores produced by the PgerE-spoIIID fusion-bearing strain. This would be the case if the changes were in protein species that were not extracted by SDS or did not enter the SDS-polyacrylamide gel. That such species do exist is highly likely (37, 40). Regardless of whether the coat composition is altered by persistent spoIIID expression, our data support the notion that proper regulation of coat protein levels during sporulation is required for proper coat assembly. Recently, it was shown that the timing of cotE expression is important for normal assembly of the spore outer coat (3).
The nature of the spore defect caused by maintaining the SpoIIID level late during sporulation is intriguing. The resistance of the spore to environmental insults is due both to the complex structure of its cortex and coat and to the unique physiological state of the core (reviewed in references 2 and 12). It is believed that the cortex confines the core of the spore by forming a woven fabric-like structure, maintaining a highly dehydrated state that is resistant to heat. On the other hand, lysozyme resistance is largely due to the coat, which shields the cortex from the enzyme. The defect in lysozyme resistance of spores derived from cells with persistent spoIIID expression (Fig. 4) might be explained by their defect in spore coat structure (Fig. 6B). Although gross cortex and core defects were not evident by electron microscopic analysis of these spores, subtle defects are implied since more than half lost their ability to resist the heat treatment (which does not depend on the coat) (Fig. 4).
The electron microscopic analysis of spores derived from cells with persistent spoIIID expression revealed that typically much of the coat did not contact the cortex and that ridges were less evident than for wild-type spores (Fig. 6). These observations suggest an inability of the coat to maintain the folded, contracted state that appears to be responsible for the ridges (2, 10, 11, 41-43). Previous work implicated CotE (possibly indirectly) in coat folding (2). While that data indicate that the absence of a single protein can prevent ridge formation, the present study suggests that the ability of the wild-type coat to fold requires proper global regulation of coat protein levels. This supports the view that the flexibility of the coat is a function of multiple coat proteins.
The impact of maintaining the SpoIIID level late during sporulation depends to some extent on the conditions used to initiate the sporulation process. In the experiments documented in Results, sporulation was initiated by centrifugal collection of growing cells followed by resuspension in SM medium lacking nutrients. When sporulation was instead initiated by growth in Difco sporulation (DS) medium followed by nutrient exhaustion (21), results similar to those shown in Fig. 2 and 3 were observed, but the effects of persistent spoIIID expression on spore resistance properties were diminished such that none of the differences were statistically significant by the criterion (P value of less than 0.05 in a Student t test) that we used (data not shown). These results indicate that the method of sporulation initiation influences susceptibility to perturbation of the SpoIIID level late during sporulation, at least in terms of spore resistance properties. A connection between the method of sporulation initiation and expression of cotC late during sporulation was observed previously (55). Recently, aconitase, a Krebs cycle enzyme, was shown to be required for efficient late-sporulation gene expression, apparently because it binds to the 3′ untranslated region of gerE mRNA, facilitating its accumulation (45). Aconitase provides a plausible link between early and late events during sporulation since the citB gene encoding aconitase is induced during late exponential phase in nutrient-exhausted medium (8). Our preliminary results suggest that aconitase accumulates to a considerably higher level late during sporulation in nutrient-exhausted DS medium than after resuspension in SM medium (L. Wang and L. Kroos, unpublished data). The higher level of aconitase under nutrient exhaustion conditions might lead to a higher level of GerE, counteracting some of the effects of maintaining the SpoIIID level late during sporulation. Obviously, the MC gene regulatory network is more complex than that depicted in Fig. 1 and much remains to be learned about how entry into sporulation influences network characteristics and output.
Acknowledgments
We thank R. Burri for constructing pRB1 and pRB2 and conducting preliminary studies with PgerE-spoIIID at single copy. We also thank R. Losick, A. Grossman, and C. Moran for providing bacterial strains.
This work was supported by NIH grant GM43585 and by the Michigan Agricultural Experiment Station.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Alon, U. 2003. Biological networks: the tinkerer as an engineer. Science 301:1866-1867. [DOI] [PubMed] [Google Scholar]
- 2.Chada, V. G., E. A. Sanstad, R. Wang, and A. Driks. 2003. Morphogenesis of Bacillus spore surfaces. J. Bacteriol. 185:6255-6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa, T., M. Serrano, L. Steil, U. Volker, C. P. Moran, Jr., and A. O. Henriques. 2007. The timing of cotE expression affects Bacillus subtilis spore coat morphology but not lysozyme resistance. J. Bacteriol. 189:2401-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crater, D. L., and C. P. Moran, Jr. 2002. Two regions of GerE required for promoter activation in Bacillus subtilis. J. Bacteriol. 184:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crater, D. L., K. H. Wade, O. Resnekov, H. T. Ichikawa, L. Kroos, J. A. Brannigan, and C. P. Moran, Jr. 2002. A mutation in GerE that affects cotC promoter activation in Bacillus subtilis. Biochim. Biophys. Acta 1576:30-38. [DOI] [PubMed] [Google Scholar]
- 6.Cutting, S., V. Oke, A. Driks, R. Losick, S. Lu, and L. Kroos. 1990. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis. Cell 62:239-250. [DOI] [PubMed] [Google Scholar]
- 7.Cutting, S., S. Panzer, and R. Losick. 1989. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J. Mol. Biol. 207:393-404. [DOI] [PubMed] [Google Scholar]
- 8.Dingman, D. W., M. S. Rosenkrantz, and A. L. Sonenshein. 1987. Relationship between aconitase gene expression and sporulation in Bacillus subtilis. J. Bacteriol. 169:3068-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donovan, W., L. Zheng, K. Sandman, and R. Losick. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J. Mol. Biol. 196:1-10. [DOI] [PubMed] [Google Scholar]
- 10.Driks, A. 2003. The dynamic spore. Proc. Natl. Acad. Sci. USA 100:3007-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driks, A. 2004. From rings to layers: surprising patterns of protein deposition during bacterial spore assembly. J. Bacteriol. 186:4423-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driks, A. 2002. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 10:251-254. [DOI] [PubMed] [Google Scholar]
- 13.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:1664-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feucht, A., L. Evans, and J. Errington. 2003. Identification of sporulation genes by genome-wide analysis of the σE regulon of Bacillus subtilis. Microbiology 149:3023-3034. [DOI] [PubMed] [Google Scholar]
- 15.Fujita, M., and R. Losick. 2002. An investigation into the compartmentalization of the sporulation transcription factor σE in Bacillus subtilis. Mol. Microbiol. 43:27-38. [DOI] [PubMed] [Google Scholar]
- 16.Fujita, M., and R. Losick. 2003. The master regulator for entry into sporulation in Bacillus subtilis becomes a cell-specific transcription factor after asymmetric division. Genes Dev. 17:1166-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halberg, R., and L. Kroos. 1992. Fate of the SpoIIID switch protein during Bacillus subtilis sporulation depends on the mother-cell sigma factor, σK. J. Mol. Biol. 228:840-849. [DOI] [PubMed] [Google Scholar]
- 18.Halberg, R., and L. Kroos. 1994. Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J. Mol. Biol. 243:425-436. [DOI] [PubMed] [Google Scholar]
- 19.Halberg, R., V. Oke, and L. Kroos. 1995. Effects of Bacillus subtilis sporulation regulatory protein SpoIIID on transctiption by σK RNA polymerase in vivo and in vitro. J. Bacteriol. 177:1888-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, England.
- 22.Henriques, A. O., B. W. Beall, K. Roland, and C. P. Moran, Jr. 1995. Characterization of cotJ, a σE-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 177:3394-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmeister, A. E. M., A. Londono-Vallejo, E. Harry, P. Stragier, and R. Losick. 1995. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell 83:219-226. [DOI] [PubMed] [Google Scholar]
- 24.Ichikawa, H., R. Halberg, and L. Kroos. 1999. Negative regulation by the Bacillus subtilis GerE protein. J. Biol. Chem. 274:8322-8327. [DOI] [PubMed] [Google Scholar]
- 25.Ichikawa, H., and L. Kroos. 2000. Combined action of two transcription factors regulates genes encoding spore coat proteins of Bacillus subtilis. J. Biol. Chem. 275:13849-13855. [DOI] [PubMed] [Google Scholar]
- 26.Karmazyn-Campelli, C., C. Bonamy, B. Savelli, and P. Stragier. 1989. Tandem genes encoding σ-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 3:150-157. [DOI] [PubMed] [Google Scholar]
- 27.Karow, M. L., P. Glaser, and P. J. Piggot. 1995. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 92:2012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroos, L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. [Epub ahead of print.] http://arjournals.annualreviews.org/doi/pdf/10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed]
- 29.Kroos, L., B. Kunkel, and R. Losick. 1989. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science 243:526-529. [DOI] [PubMed] [Google Scholar]
- 30.Kroos, L., Y.-T. Yu, D. Mills, and S. Ferguson-Miller. 2002. Forespore signaling is necessary for pro-σK processing during Bacillus subtilis sporulation despite the loss of SpoIVFA upon translational arrest. J. Bacteriol. 184:5393-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel, B., L. Kroos, H. Poth, P. Youngman, and R. Losick. 1989. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev. 3:1735-1744. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel, B., K. Sandman, S. Panzer, P. Youngman, and R. Losick. 1988. The promoter for a sporulation gene in the spoIVC locus of Bacillus subtilis and its use in studies of temporal and spatial control of gene expression. J. Bacteriol. 170:3513-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Londoño-Vallejo, J. A., and P. Stragier. 1995. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 9:503-508. [DOI] [PubMed] [Google Scholar]
- 34.Lu, S., R. Halberg, and L. Kroos. 1990. Processing of the mother-cell σ factor, σK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc. Natl. Acad. Sci. USA 87:9722-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, S., and L. Kroos. 1994. Overproducing the Bacillus subtilis mother-cell sigma factor precursor, pro-σK, uncouples σK-dependent gene expression from dependence on intercompartmental communication. J. Bacteriol. 176:3936-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maciag, I. E., J. F. Viret, and J. C. Alonso. 1988. Replication and incompatibility properties of plasmid pUB110 in Bacillus subtilis. Mol. Gen. Genet. 212:232-240. [DOI] [PubMed] [Google Scholar]
- 37.McPherson, D., H. Kim, M. Hahn, R. Wang, P. Grabowski, P. Eichenberger, and A. Driks. 2005. Characterization of the Bacillus subtilis spore morphogenetic coat protein CotO. J. Bacteriol. 187:8278-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Naclerio, G., L. Baccigalupi, R. Zilhao, M. De Felice, and E. Ricca. 1996. Bacillus subtilis spore coat assembly requires cotH gene expression. J. Bacteriol. 178:4375-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandey, N. K., and A. I. Aronson. 1979. Properties of the Bacillus subtilis spore coat. J. Bacteriol. 137:1208-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plomp, M., T. J. Leighton, K. E. Wheeler, and A. J. Malkin. 2005. Architecture and high-resolution structure of Bacillus thuringiensis and Bacillus cereus spore coat surfaces. Langmuir 21:7892-7898. [DOI] [PubMed] [Google Scholar]
- 42.Plomp, M., T. J. Leighton, K. E. Wheeler, and A. J. Malkin. 2005. The high-resolution architecture and structural dynamics of Bacillus spores. Biophys. J. 88:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plomp, M., T. J. Leighton, K. E. Wheeler, M. E. Pitesky, and A. J. Malkin. 2005. Bacillus atrophaeus outer spore coat assembly and ultrastructure. Langmuir 21:10710-10716. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Serio, A. W., K. B. Pechter, and A. L. Sonenshein. 2006. Bacillus subtilis aconitase is required for efficient late-sporulation gene expression. J. Bacteriol. 188:6396-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steil, L., M. Serrano, A. O. Henriques, and U. Volker. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151:399-420. [DOI] [PubMed] [Google Scholar]
- 47.Stelling, J., U. Sauer, Z. Szallasi, F. J. Doyle III, and J. Doyle. 2004. Robustness of cellular functions. Cell 118:675-685. [DOI] [PubMed] [Google Scholar]
- 48.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 49.Stragier, P., B. Kunkel, L. Kroos, and R. Losick. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243:507-512. [DOI] [PubMed] [Google Scholar]
- 50.Tatti, K. M., C. H. Jones, and C. P. Moran. 1991. Genetic evidence for interaction of σE with the spoIIID promoter in Bacillus subtilis. J. Bacteriol. 173:7828-7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, B., and L. Kroos. 1997. A feedback loop regulates the switch from one sigma factor to the next in the cascade controlling Bacillus subtilis mother cell gene expression. J. Bacteriol. 179:6138-6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, B., P. Struffi, and L. Kroos. 1999. σK can negatively regulate sigE expression by two different mechanisms during sporulation of Bacillus subtilis. J. Bacteriol. 181:4081-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, L., R. Halberg, S. Roels, H. Ichikawa, L. Kroos, and R. Losick. 1992. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J. Mol. Biol. 226:1037-1050. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, L., and R. Losick. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645-660. [DOI] [PubMed] [Google Scholar]