Abstract
Using genomic analysis, researchers previously identified genes coding for proteins homologous to the structural proteins of nitrogenase (J. Raymond, J. L. Siefert, C. R. Staples, and R. E. Blankenship, Mol. Biol. Evol. 21:541-554, 2004). The expression and association of NifD and NifH nitrogenase homologs (named NflD and NflH for “Nif-like” D and H, respectively) have been detected in a non-nitrogen-fixing hyperthermophilic methanogen, Methanocaldococcus jannaschii. These homologs are expressed constitutively and do not appear to be directly involved with nitrogen metabolism or detoxification of compounds such as cyanide or azide. The NflH and NflD proteins were found to interact with each other, as determined by bacterial two-hybrid studies. Upon immunoisolation, NflD and NflH copurified, along with three other proteins whose functions are as yet uncharacterized. The apparent presence of genes coding for NflH and NflD in all known methanogens, their constitutive expression, and their high sequence similarity to the NifH and NifD proteins or the BchL and BchN/BchB proteins suggest that NflH and NflD participate in an indispensable and fundamental function(s) in methanogens.
Nitrogenase is a two-component metalloenzyme complex that catalyzes the ATP-dependent reduction of dinitrogen to ammonia (nitrogen fixation) (2, 11, 21, 52). The fixation of atmospheric nitrogen is an essential process for the survival of life on earth and a critical part of the global nitrogen cycle (14, 22, 39). The components that make up the nitrogenase complex are dinitrogenase (called the FeMo protein or component I) and dinitrogenase reductase (called the Fe protein or component II). Dinitrogenase is an α2β2 heterotetramer of the proteins NifD and NifK (encoded by the genes nifD and nifK, respectively) (33). Dinitrogenase reductase is a homodimer of the protein NifH (encoded by the gene nifH) (18). Residing at the α/β subunit interface of dinitrogenase is an unusual [Fe8S7] cluster called the P cluster (7, 42, 51), which has been shown to transfer electrons and protons to the active site [Fe7S8MoN(homocitrate)] cluster (called FeMo-co), which is the active site of substrate reduction. FeMo-co lies within the α subunit (NifD) (8, 12, 26, 27, 29, 36, 54).
It has been proposed that a key event in the evolution of dinitrogenase was a paralogous gene duplication of a NifD/NifK precursor (13). Subsequent divergence resulted in the heterotetrameric form found in diazotrophs. It has also been suggested that the ancestral nitrogenase complex might have existed as a homotetramer of the NifD/NifK ancestor (13). Subsequent NifDK paralogous operon duplication and divergence is proposed to have given rise both to NifEN (13), a NifDK homolog that serves as a scaffold during the biosynthesis of FeMo-co (19, 28), and to dinitrogenase.
Another essential biochemical process on earth is photosynthesis, the ability of microorganisms and plants to convert solar energy to chemical energy. This process is the primary producer of energy for life on this planet and is also a part of the global carbon cycle, as photosynthetic organisms utilize the chemical energy generated by photosynthesis to fix carbon. There are two nitrogenase homolog systems involved in late steps in the biosynthetic pathway of chlorophyll and bacteriochlorophyll (5, 16, 17). The transformation of protochlorophyllide into chlorophyllide through the reduction of ring D is accomplished by an enzymatic complex of BchLNB, where BchL is a NifH homolog and BchN and BchB are NifD and NifK homologs, respectively (16). The BchLNB complex is also known as the light-independent protochlorophyllide oxidoreductase. The transformation of chlorophyllide into bacteriochlorophyllide through the reduction of ring B is accomplished by the system of BchXYZ, where BchX is a NifH homolog and BchY/BchZ are NifD/NifK homologs, respectively. Because the nitrogenase and bacteriochlorophyllide/chlorophyllide synthesis proteins described here are almost certainly homologous, it is likely that there was ancestral gene duplication and divergence in function to produce enzymes that utilize very different substrates (44). The ancient reductase enzyme from which all the extant Nif and Bch reductases are derived may well have had an entirely different function. It is of great interest, both with regard to the evolution of life on earth and to the evolution of substrate specificity, to determine when this diversification of function occurred and what the function of the ancestral system was.
Recently, the analysis of the published whole genomes of 101 organisms for the presence of NifH or NifD/NifK homologs uncovered numerous atypical sequences of both nifH-like and nifD/nifK-like genes scattered among all known methanogens and some phototrophs (44); these genes are distinct from bona fide nifH and nifD homologs that allow nitrogen fixation in several methanogens. The same general relationship holds true for both NifH homolog and NifD/NifK homolog trees. Phylogenetic analyses show that these sequences lie basal in the tree, between the Nif and Bch clades (44). Due to the similarity of these genes to nif genes and to the lack of knowledge of a specific function for the proteins, these genes were termed nflH and nflD, and the proteins they code for were termed NflH and NflD (nfl is for “nif-like”). Two of the organisms containing these atypical sequences do not perform either photosynthesis or nitrogen fixation. These organisms are Methanocaldococcus jannaschii and Methanopyrus kandleri, both hyperthermophilic methanogenic members of the Euryarchaeota. In this study, M. jannaschii was utilized because it has been much more widely studied and the methods for cultivating this organism with precision have been established (40). The goals of this study were to assess whether the nfl genes present in the M. jannaschii genome are expressed and, if so, under what conditions; to determine whether these genes are involved in nitrogen metabolism; and to determine whether the NflD protein is associated with the NflH protein in vivo. In the course of this study, we determined that NflH and NflD are constitutively expressed. NflH and some other proteins are associated with NflD, suggesting a possible functional role for these proteins in M. jannaschii.
MATERIALS AND METHODS
Strains and plasmids.
The plasmid pRSETA (Invitrogen) was used to construct the heterologous expression vectors. Escherichia coli strain BL21-CodonPlus(DE3)-RIL (Stratagene, La Jolla, CA) was the expression host. M. jannaschii nflH or MJ0879 coding sequence was PCR amplified by the use of Vent polymerase (New England Biolabs, Beverly, MA) and the primer pair (5′ to 3′) MJ0879 (NflH)/F (GAGCTCATGAGAAAATTTTGTGTCTATG; SacI; restriction site is underlined) and MJ0879 (NflH)/R (GAATTCTTATCCTTTAACACTCTCTTTTA). The amplified DNA was digested with SacI and EcoRI and was cloned into similarly digested pRSETA to obtain the plasmid pRSETA-MjNflH. The plasmid pRSETA-MjNflD was constructed similarly, and for this purpose, M. jannaschii nflD or MJ1423 coding sequence was amplified using the primer pairs MJ1423 (NflD)/F, GAGCTCATCATATTCCATCCAAGA (SacI), and MJ1423 (NflD)/R, GAATTCTTATTCCAATGCATAATCCAATATTTCAC (EcoRI). Each of these constructs allowed the expression of the recombinant protein with a polyhistidine (His6) tag on the N terminus. Standard techniques were used for DNA manipulations (48). The sequences of cloned DNA segments were confirmed by determining the nucleotide sequences of both strands.
For bacterial two-hybrid experiments using the BacterioMatch system (Stratagene, La Jolla, CA), the E. coli XL1-Blue host was used. Sequences for the specific primers used to PCR amplify nflH from Methanocaldococcus jannaschii genomic DNA for insertion into the pBT bait vector were 5′-AGATGGATCCATGAGAAAATTTTGTGTCTAT-3′ (BamHI) and 5′-GATAGGATCCTTATCCTTTAACACTCTCTTT-3′ (BamHI). The sequences for the primers designed to amplify nflD for cloning into the pTRG target vector were 5′-AGATGGATCCATGATATTCCAT CCAAGACCT-3′ (BamHI) and 5′-GATAGGATCCTTATTCCAATGCATAATCCAA-3′ (BamHI). The PCR amplified nflH and nflD fragments were digested with the BamHI enzyme and ligated with the similarly digested pBT and pTRG plasmids, respectively. The pBT-nflH and pTRG-nflD plasmids were then used for cotransformation of the E. coli XL1-Blue cells. These cotransformants were further used for the measurement of the β-galactosidase activity in order to detect the protein-protein interaction between NflH and NflD.
E. coli cell growth and harvest.
E. coli BL21-CodonPlus(DE3)-RIL, carrying either pRSETA-MjNflH or pRSETA-MjNflD, was grown aerobically in 1 liter LB medium (48), with vigorous agitation at 37°C until the culture reached an optical density at 600 nm (OD600) of 0.5 to 0.6. Protein expression was then induced with IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM, and the cultivation was continued for another 5 h. From this culture, the cells were pelleted by centrifugation at 7,140 × g for 20 min at 4°C. The resulting cell pellet was frozen in liquid nitrogen and stored at −80°C. The E. coli XL-1 Blue strain was grown at 37°C in 2YT medium (48). Ampicillin, chloramphenicol, and tetracycline were used to final concentrations of 50, 34, and 5 μg/ml, respectively, wherever selection was necessary.
Recombinant NflH and NflD purification.
Frozen cells were thawed in an anaerobic chamber (Coy Laboratory Products, Inc., Grass Lake, MI), maintained under 3% hydrogen with 97% ultra-high-purity nitrogen, and suspended in 2.5× the cell mass of anaerobic 20 mM sodium phosphate (pH 7.5), 500 mM NaCl, 1 mM imidazole that also included DNase, RNase, lysozyme, and phenylmethylsulfonyl fluoride (PMSF). Cells were homogenized by vortex homogenization (modified Cuisinart daiquiri maker; Cuisinart, East Windsor, NJ), transferred to 40-ml Nalgene tubes with sealed modified caps (for use on a Schlenk line), and then removed from the glove box and placed on ice. The cell suspension was then sonicated five times using a Branson sonifier (Branson, Danbury, CT) cell disruptor under streams of ultra-high-purity nitrogen. After cooling on ice, the disrupted cell lysate was centrifuged at 47,800 × g for 20 min. The cell extract supernatant was collected in the anaerobic chamber and passed through a 0.45-μm syringe filter to remove particulate matter.
Purification of recombinant NflH.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed that upon centrifugation of cell lysate, most of the overexpressed NflH was recovered in the supernatant. Therefore, this protein was expressed in the soluble fraction in high abundance. The cell supernatant was heat treated (anaerobically) within a sealed Beckman centrifuge tube (Beckman-Coulter, Fullerton, CA) for 2 h in a 65°C water bath to denature most E. coli proteins. The precipitated proteins were removed via centrifugation at 47,800 × g for 20 min, and the supernatant was collected within the anaerobic chamber. All subsequent work was performed inside the anaerobic chamber. The supernatant was filtered and loaded onto a Pharmacia HiTrap chelating HP column (Pfizer-Pharmacia, New York, NY) charged with nickel by using a Rainin Dynamax peristaltic pump (Rainin Instrument Co., Alameda, CA). Upon loading, the column exhibited a brown color indicative of bound protein containing iron-sulfur clusters. The column was washed with 20 mM phosphate, pH 7.5, 500 mM NaCl, and 100 mM imidazole. The protein was eluted with 500 mM imidazole (with the major portion eluting at 200 mM imidazole). If the column was run aerobically, some NflH began eluting at 50 mM imidazole, which is the imidazole concentration utilized to remove proteins with nonspecific binding affinity from the column. Protein was eluted directly into a 20× elution volume containing 25 mM Tris HCl, pH 7.5, 100 mM NaCl. The diluted protein solution was then concentrated anaerobically to 2 ml by an Amicon ultraconcentrator (Millipore, Billerica, MA). It retained the tan color when concentrated.
Purification of recombinant NflD.
Like NflH, NflD was also expressed in the soluble fraction in high abundance. NflD purification was similar to that of NflH, with some exceptions. Extracts of E. coli overexpressing NflD were not heat treated at 65°C, but after filtration through a 0.45-μm filter, they were loaded directly onto the HiTrap column, which was subsequently washed with 1 mM, 30 mM, and 50 mM imidazole in 20 mM phosphate, pH 7.5, 500 mM NaCl. The protein was eluted (using 200 mM imidazole in 20 mM phosphate, pH 7.5, 500 mM NaCl) directly into 20× the elution volume of 25 mM Tris HCl, pH 7.5 (no NaCl), with 1 mM EDTA added to avoid the protein precipitation that occurs soon after elution if the eluted protein is not diluted, and to prepare for the next column. This mixture was then loaded onto a 5-ml DE-52 column. The protein was eluted from the column with 500 mM NaCl in 25 mM Tris HCl, pH 7.5, in one step, after washing with 50 mM NaCl in Tris HCl, pH 7.5, for 10 column volumes. The eluted protein was tan in color.
Production and purification of antibodies.
NflD antigen was sent for antibody production in two forms, folded (0.2 mg) and denatured (0.4 mg), to make sure that the resultant antibodies would recognize protein by using both native PAGE and SDS-PAGE. The same rabbit was used so that the anti-natured NflD and anti-denatured NflD were in the same serum. Antibodies for NflD were obtained from Rockland Immunochemicals (Gainesville, PA). NflH antigen (1.5 mg) was sent only in the folded form to ProSci Incorporated (Poway, CA). NflD and NflH antibodies were cleaned and purified from rabbit serum using Bio-Rad Econo-Pac DG disposable chromatography columns and Bio-Rad Econo-Pac DEAE Blue cartridges according to the manufacturer's recommended protocols.
Construction of a cross-linked protein A-anti-NflD immunoglobulin G (IgG) agarose column.
NflD antibodies (in Tris HCl) were buffer exchanged by using an Amicon ultrafiltration device and dilution into 50 mM sodium borate, pH 8.2. A 3-ml suspension of ImmunoPure immobilized protein A-agarose (Pierce, Rockford, IL) was equilibrated with the borate buffer and then very gently stirred overnight at 4°C with 20 ml of NflD antibody solution. The column matrix was allowed to settle, and the remaining antibody solution was decanted. Sodium borate buffer was added to the matrix, and the matrix was poured into a 10-ml Bio-Rad disposable column. Twenty micrograms of dimethyl pimelimidate · 2 HCl (DMP) was dissolved in 2 ml of 0.2 M triethanolamine, pH 8.0 (cross-linking buffer), and the solution was immediately added to the column. Then the column was allowed to sit for 2 h at room temperature. The column was washed with 10 ml of cross-linking buffer. The column was loaded with 3 ml of 0.1 ethanolamine, pH 8.2 (blocking buffer), and allowed to sit for 30 min. Afterwards, 10 ml of 0.1 M glycine, pH 2.5 (elution buffer), was applied. The column was stored in the dark at 4°C until used.
Western blotting of denatured proteins.
All SDS-denaturing polyacrylamide (Bio-Rad) Laemmli gels contained 10% polyacrylamide in the resolving gel and 4.5% polyacrylamide in the stacking gel and were run on a Bio-Rad Mini Protean 3 cell. The transfer was performed in Tris-glycine-methanol according to recommended protocols (Bio-Rad, Hercules, CA) by using a Bio-Rad Mini Trans-Blot cell. Rabbit anti-NflD IgG was used at a 1:20,000 dilution, and a 1:1,000 dilution was used for rabbit anti-NflH IgG. A 1:10,000 dilution was used for the secondary antibody (goat anti-rabbit IgG-alkaline phosphatase [AP] conjugate).
Growth of M. jannaschii.
M. jannaschii cells were grown in a mineral salts medium under an atmosphere of H2 with CO2 (80:20, vol/vol) at a total pressure of 236 kPa as described previously (40), but with the following modifications. For generating cell mass for the purification of Nfl proteins, a 16-liter (12-liter working volume) stirred-tank reactor was used and the growth medium contained 22 mM ammonium chloride and 50 μM sodium selenate (40). For studying the effect of cyanide, isothiocyanate, or azide on the expression of NflD and NflH, the organism was cultivated in a sealed 530-ml serum bottle containing 200 ml growth medium (40) with 0.8 mM ammonium chloride and 1.5 μM VSO4. Before inoculation, the sterile medium was supplemented with cyanide, isothiocyanate, or azide to a final concentration of 1 mM from an anaerobic filter-sterilized aqueous stock solution. This organism is normally grown with 22 mM ammonium chloride (40), and under this condition, a 530-ml serum bottle-based culture attained a final OD600 of 1.0. Since the ammonium level in the test medium was suboptimal, growth ceased when the culture reached an OD600 of 0.2. An assay utilizing Nessler's reagent on samples withdrawn from the serum bottles showed that at this stage, the culture medium was devoid of free ammonium ions, a condition conducive for the derepression or activation of the nifD and nifH genes in organisms capable of diazotrophic growth (35) and cyanide and azide detoxification genes in certain bacteria (9, 30, 43, 46). For testing the possibility of diazotrophic growth upon the exhaustion of NH4+, nitrogen was supplied in the headspace at a partial pressure of 50 kPa. Diazotrophic growth was also tested using medium devoid of ammonium from the time of inoculation, but with 50 kPa nitrogen in the headspace.
Immunoisolation of NflD and associated proteins.
The immobilized, cross-linked, rabbit anti-NflD IgG-protein A column was brought to room temperature and equilibrated with anaerobic 10 mM Tris HCl, pH 7.5 (load buffer), in an anaerobic chamber (Coy Laboratory Products, Inc., Grass Lake, MI). Two to 3 g of M. jannaschii frozen cell paste was thawed inside the anaerobic chamber, where 30 ml of anaerobic 10 mM Tris HCl, pH 7.5, containing DNase, RNase, lysozyme, PMSF, and dithiothreitol (1 mM) was added. The cell suspension was then homogenized to break the M. jannaschii cell walls. The broken cell slurry was centrifuged at 47,800 × g for 20 min in a sealed centrifuge tube. The supernatant was collected inside the anaerobic chamber and passed through a 0.45-μm syringe filter to remove particulate matter. The clarified supernatant was applied to the antibody column under a gravity flow. After the sample passed through, the column was washed with 20 ml of load buffer. Subsequent washes were with 20 ml each of 100 mM MES (morpholineethanesulfonic acid), pH 3.5, 4.5, and 5.5, and finally with 10 ml of elution buffer.
Analysis of proteins eluted from anti-NflD IgG column.
The NflH and NflD proteins were identified using Western blotting and also by using matrix-assisted laser desorption-time of flight (MALDI-TOF) mass spectroscopy and trypsin fingerprinting as described below. Proteins other than NflH and NflD persisted in binding to the column through several washings and relatively harsh acidic conditions (buffers at pH 5.5, 4.5, and 3.5). Three prominent bands from the resolved elution buffer fraction were excised from a Coomassie-stained gel in portions of approximately 2 mm by 1 mm by 1 mm. Each denatured polypeptide band in a Coomassie blue-stained gel slice (2 mm by 1 mm by 1 mm) was digested with trypsin by using an in-gel digestion protocol as instructed by Promega (Madison, WI). Mass spectra were collected on an Applied Biosystems Voyager MALDI DE-STR mass spectrometer (Applied Biosystems, Foster City, CA). Data processing to obtain a list of monoisotopic peak masses was performed using Data Explorer software (version 4.0; Applied Biosystems, Foster City, CA). Monoisotopic masses obtained by this procedure are accurate to within 0.1 Da over the mass range of peptides examined in this work.
Tryptic fragment data obtained from mass spectral analysis were analyzed using the MASCOT peptide mass fingerprint tool available online from Matrix Science (Matrix Science, Inc., Boston, MA). Monoisotopic peak masses were searched against the MASCOT Archaea database.
β-Galactosidase assay.
The β-galactosidase activity assay was performed as described elsewhere previously (34, 48). β-Galactosidase units were defined as the activity that hydrolyzes 1 μmol of o-nitrophenyl-β-d-galactopyranoside (ONPG) to o-nitrophenol and d-galactose per minute. β-Galactosidase activity in Miller units was calculated as 1,000 × [OD420 − (1.75× OD550)/(t × V × OD600)], where t is the elapsed time (in minutes) of incubation, V is the volume of cells (including the concentration factor), and OD600 is the optical density of 1 ml of culture at 600 nm. The negative controls used for the β-galactosidase assay were E. coli XL1 Blue reporter cells cotransformed with the following plasmid combinations: (i) N-terminal α-RNAP of pTRG with NflD protein and pBT (full-length λCI only), (ii) full-length λCI with NflH protein and pTRG (N-terminal α-RNAP only), and (ii) λCI of pBT and α-RNAP of pTRG vectors without any translational fusions. The positive control was the E. coli XL1 Blue reporter cells cotransformed with the following plasmids: pBT-LGF2 containing the full-length λCI protein of pBT vector fused to the LGF2 (dimerization domain of the yeast transcriptional activator Gal4) and pTRG-Gal11P containing the N-terminal domain of the α-RNAP of the pTRG vector fused to the Gal11P (a mutant form of Gal11 protein) (BacterioMatch two-hybrid instruction manual; Stratagene, La Jolla, CA).
RESULTS AND DISCUSSION
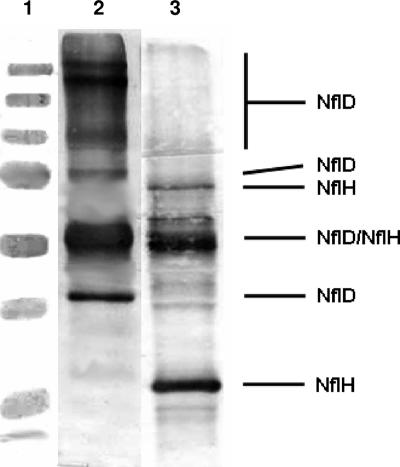
As described in Materials and Methods, an extract of M. jannaschii cell supernatant was passed through an anti-NflD IgG agarose column. The goal was to trap NflD and the proteins that associated strongly with NflD onto the column and then to wash the column with a low-salt buffer and elute the bound proteins with a high-ionic-strength solution or low-pH solution. The fractions were examined for NflD and NflH via Western blot analysis. Most of the loaded M. jannaschii proteins were found in the flowthrough and in the 10-mM Tris HCl wash. NflD and NflH bound to the column and did not elute under the high-salt wash. Therefore, we used a solution at pH 2.5 for elution. At a very low pH, iron-sulfur clusters are largely destroyed because the sulfur of iron-sulfur clusters is acid labile. Thus, in our work, the eluted proteins were not suitable for spectroscopic characterization. While a very small amount of NflH was eluted in the wash at pH 3.5, the majority of this protein associated with NflD and was eluted in 100 mM glycine, pH 2.5. An assessment of the presence of NflD and NflH was performed using Western blot analysis. The Western blot (Fig. 1) showed a prominent NflH band, a much less prominent NflD band, and an apparent NflD/NflH dimer combination band. The NflD/NflH dimer appeared above the 50-kDa molecular mass marker, although the predicted molecular mass for this NflD/NflH combination band is 69 kDa. Typically, if a protein has unreduced areas of incompletely disrupted secondary structure, it cannot unfold to full length and it tends to run faster than expected in a typical SDS gel. In addition to the apparent NflD/NflH dimer, there may be other combinations that represent an NflH dimer (slightly below the 75-kDa molecular mass marker), an NflD dimer (at the 75-kDa molecular mass marker), and higher order multimeric NflD forms (at the 100-kDa molecular mass marker and slightly below and at the 250-kDa molecular mass marker). A dimer, trimer, and tetramer of NflD would be expected to have molecular masses of 78.8, 118.2, and 157.6 kDa, respectively. Since the samples were treated with 20 percent β-mercaptoethanol and boiled for 20 min (conditions that would normally break protein complexes into their corresponding monomers), the appearance of multimeric forms was a surprise. However, it is often observed that multimeric protein complexes from hyperthermophilic organisms such as M. jannaschii persist under SDS-PAGE analysis in even the harshest of conditions (6).
FIG. 1.
Anti-NflD and anti-NflH Western blots of the elution buffer fraction of the cross-linked anti-NflD IgG-protein-A agarose column. Lane 1, molecular mass markers (250, 150, 100, 75, 50, 37, 25, and 20 kDa); lane 2, anti-NflD (1:20,000 dilution) primary/Novagen goat anti-rabbit IgG-AP conjugate; lane 3, anti-NflH (1:1,000 dilution) primary/Novagen goat anti-rabbit IgG-AP conjugate.
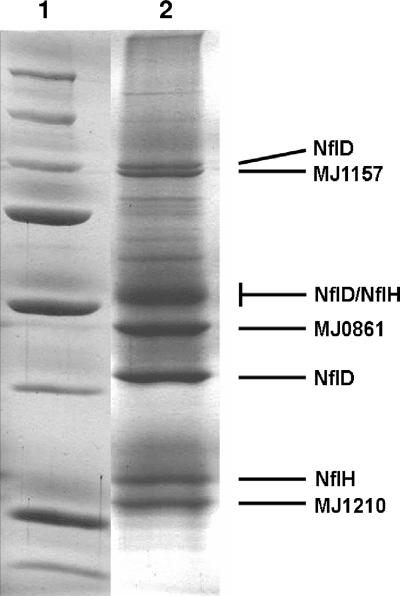
Because the Coomassie-stained SDS gel did not show a prominent band for NflD, even though this was an anti-NflD rabbit IgG polyclonal antibody column, it was speculated that much of NflD remained on the column even after harsh washing in high-salt (not shown) or low-pH conditions. This possibility was tested by treating the column matrix with acetonitrile-trichloracetic acid (TCA) solution and loading a suspension of the matrix and precipitated or released proteins on an SDS-polyacrylamide gel. As shown in Fig. 2, a significant increase in NflD was observed but also other proteins described below remained associated with NflD on the column matrix even after extensive washing. These other proteins may form a complex with NflD, although we cannot rule out the possibility that these proteins have a fortuitous affinity for the anti-NflD antibody. These proteins were identified by MALDI-TOF mass spectroscopy and trypsin fingerprinting as MJ1210 (NCBI accession no. A64451), MJ0861 (NCBI accession no. E64407), and MJ1157 (NCBI accession no. E64444) (see the supplemental material).
FIG. 2.
Coomassie-stained 10% acrylamide SDS-PAGE of M. jannaschii proteins remaining tightly associated to a cross-linked polyclonal anti-NflD IgG-protein-A agarose after extensive washing with elution buffer and 2 M NaCl. MJ1157, a 100.3-kDa protein (NCBI accession no. E64444); MJ0861, a 50.03-kDa protein (NCBI accession no. E64407); MJ1210, a 28.68-kDa protein (NCBI accession no. A64451). Lane 1, molecular mass markers (250, 150, 100, 75, 50, 37, 25, and 20 kDa); lane 2, proteins extracted from the slurry of column matrix using trichloracetic acid-acetonitrile-SDS-sample buffer as described in the text.
Detection of protein-protein interaction between NflH and NflD by bacterial two-hybrid analysis.
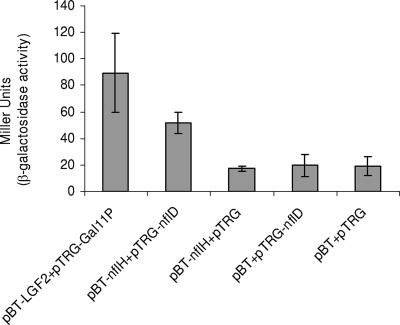
The BacterioMatch two-hybrid system (Stratagene, La Jolla, CA) is a molecular genetic approach for the detection of protein-protein interactions in vivo in Escherichia coli based on the principle of transcriptional activation (BacterioMatch two-hybrid system manual; Stratagene, La Jolla, CA). As shown in Fig. 3, we found that when the E. coli XL1-Blue reporter strain was cotransformed with the plasmids pBT-nflH and pTRG-nflD, the β-galactosidase activity of the cotransformants corresponded to 52 ± 8 Miller units. In comparison, the cotransformants containing (i) pBT and pTRG, (ii) pBT-nflH and pTRG, and (iii) pBT and pTRG-nflD were each found to have β-galactosidase activities at least 2.5-fold lower. As expected, strong interaction was demonstrated between the LGF2 and Gal11P proteins, as observed by high β-galactosidase activity units (89 ± 28). Therefore, the interaction between NflH and NflD proteins was further validated by this quantitative method.
FIG. 3.
Results of the liquid β-galactosidase assay with ONPG as a substrate to demonstrate protein-protein interaction. Miller units were calculated as 1,000 × [OD420 − (1.75 × OD550)/(t × V × OD600)], where t is the elapsed time (in minutes) of incubation, V is 0.1 ml times the concentration factor, and OD600 is the absorbance of 1 ml of culture at 600 nm. Assays were repeated a minimum of four times with various cotransformants, and the β-galactosidase activity units shown are an average of four independent observations. Values are means ± standard errors.
Expression of NflH and NflD in M. jannaschii under various nitrogen nutrition and toxicity conditions.
Western blots of SDS-resolving polyacrylamide gels of whole-cell extracts were used to examine the expression of NflD and NflH in M. jannaschii. Cells generated with the following nitrogen sources were analyzed: ammonium chloride at a concentration of 22 mM (sufficient) or 0.8 mM (limiting) as the sole nitrogen source; 1.0 mM of sodium azide, sodium isothiocyanate, or sodium cyanide, along with a sufficient or limiting amount of ammonium; and nitrogen in the headspace at 50 kPa, along with a sufficient or limiting amount of ammonium. When normalized for cell density (not shown), the results showed that NflD and NflH were expressed at equal levels under all conditions tested. Therefore, we concluded that in M. jannaschii, NflD and NflH were constitutively expressed. M. jannaschii, in a medium without any fixed nitrogen available, but with nitrogen gas in the headspace, did not exhibit any growth, nor did M. jannaschii, in a medium with nitrogen gas in the headspace, continue to grow upon the depletion of reduced nitrogen from the medium.
We have detected the expression of one of the uncharacterized group IV nitrogenase homologs in methanogens, and in particular in a non-nitrogen-fixing species, so these genes appear to be functional and do not function as nitrogenases. We have also established that these group IV NifH and NifD homologs associate with each other, as would be expected if they form a functional complex with analogy to the NifHDK complex. The evolution of the nitrogen cycle (including nitrogen fixation) and photosynthesis (including chlorophyll and bacteriochlorophyll biosynthesis) are two of the most important events in the history of life on earth. Methanogens typically lie basal in the phylogenetic tree of life and may have had an ancient origin (1, 15, 55), as evidence of methanogenesis dates back 2.8 billion years (24, 47). The observation of group IV nitrogenase homologs basal to the phylogenetic Nif homolog tree and intermediary between group V (bacteriochlorophyll synthesis) and groups I to III (nitrogen fixation) suggested that the genes giving rise to those involved in both of these important processes may have originated in methanogens (44). In this work, Nfl proteins from M. jannaschii were chosen to be studied because (i) M. jannaschii does not have the complicating factor of having additional Nif homologs involved in nitrogen fixation or photosynthesis; (ii) M. jannaschii lives in an environment that may mimic that of the early earth; (iii) genome trees based on shared gene pairs have shown a strong monophyletic clustering of M. jannaschii with M. kandleri, and both represent very deeply branching methanogenic lineages (4); (iv) the genome is completely sequenced (3); and (v) M. jannaschii is amenable to physiological and enzymological studies (32, 40, 41).
There are several possible reasons why nflH and nflD are present in the genome of M. jannaschii. One possibility that fits well with phylogenetic analysis is that nflD and nflH may represent relic prenitrogenase genes or another branch off a more ancient, possibly extinct, precursor. If this possibility were the case, nflD and nflH might still have the same function in M. jannaschii that they had before the evolution of nitrogenase or photosynthesis. One must allow for the possibility that they were laterally transferred from another nitrogen-fixing or photosynthetic organism and recruited to perform a completely different type of enzymatic activity. However, it has been proposed that nifDK evolved from a paralogous duplication of a single nifD/nifK precursor gene. The nifD-like gene, here named nflD, in M. jannaschii is a single copy, so apparently it has not been duplicated. Indeed, it has been shown (44) that NflD contains conserved cysteine residues (among group IV) that correspond to cysteines in both NifD and NifK, suggesting that NflD predates both NifD and NifK, rather than it being a laterally transferred NifD (or NifK). This possibility may have evolutionary ramifications if it can be proven that NflD predates both dinitrogenase and BchNB/BchYZ. A complicating fact is that homologs fitting into group IV, while present in all methanogens whose genomes have been sequenced, are also present scattered in some phototrophs. In phototrophs, these homologs may have a different as yet unknown function.
In 2000, Fani and colleagues proposed two models for the origin of nitrogenase from an ancestor protein (13). In the first case, assuming a neutral atmosphere, the ancestral protein might have been a rather nonspecific enzyme utilizing a variety of substrates. The first nitrogenase would then have been very slow, inefficient, and with low substrate specificity, able to react with a wide range of compounds with a triple bond (such as acetylene, hydrogen azide, hydrogen cyanide, nitrous oxide, and dinitrogen). In the second case, assuming a reducing atmosphere, the possible broad substrate specificity may have been useful in detoxifying cyanides and other chemicals present in the primitive reducing atmosphere. In this scenario, the primitive protein encoded by the ancestral gene would have been a detoxyase, perhaps detoxifying cyanide and other reduced chemicals. If NflD and NflK represented relics, preduplication versions of dinitrogenase and dinitrogenase reductase, they may still be performing the ancient functions predicted by Fani et al. (13). If these ancient functions were as a detoxyase or a primitive nitrogenase, one would expect the expression to be regulated dependent upon the presence of toxifying agents or on the availability of reduced nitrogen (ammonium), as the expression of nitrogenase in most organisms is highly regulated (20, 22, 23, 25, 31, 37, 38, 53), being completely repressed in the presence of ammonium and derepressed upon ammonium depletion. We found, however, that the expression of NflD and NflH is constitutive, independent of either the availability of nitrogen or the presence of a range of toxic agents.
The observations of the expression patterns of NflD and NflH from this work suggest that they (i) are not directly involved in nitrogen metabolism in M. jannaschii, and (ii) are important for cell function as they are constitutively expressed. The other system that contains proteins homologous to NifH and NifD consists of the group V (1) protochlorophyllide reductase (BchLNB) and chlorophyllide reductase (BchXYZ). These proteins both reduce heterocyclic macrocyclic chlorin-type rings whose synthesis pathways diverge off porphyrin synthesis. Thus, the functions of BchLNB and BchXYZ are broadly similar to that of nitrogenase (i.e., reducing a multiple bond), while the substrate is very different. However, M. jannaschii does not contain a photosynthetic apparatus. Therefore, NflD and NflH cannot be involved in photosynthesis as BchLNB and BchXYZ are. Nor are they involved in the reduction of dinitrogen to ammonium, as M. jannaschii is not diazotrophic (as was previously known and is confirmed in this work). However, NflD and NflH might be utilized in a similar chemistry, namely a bond reduction. Also, as validated by bacterial two-hybrid analysis, NflH and NflD interact with each other, although the functional implications of this interaction are not yet fully understood.
The sequence of NflH shows striking similarity to the sequence of NifH, particularly in what are known to be the metal-binding and ATP-binding regions of NifH. The sequence of NflH also shows a high degree of similarity to the NifH homologs BchL and BchX (the obligate electron donors in the bacteriochlorophyll synthesis pathway) (5, 16). Due to the high level of sequence identity in functionally important regions, we think that it is likely that this protein is individually performing a role in M. jannaschii similar to its role in the Nif and Bch pathways, namely, as an ATP-dependent obligate electron donor. That NflH associates with NflD suggests that it may be donating electrons to NflD in a manner similar to the nitrogenase system or donating the electrons to one or more of the other proteins described in this work.
It has been found that all known methanogenic archaea rely on methyl-coenzyme M reductase for the terminal step in methanogenesis (45). The active site of methyl-coenzyme M reductase contains F430, a nickel-containing heterocyclic macrocycle cofactor with external cyloketone and lactam rings (10, 49). However, very little is known about the biosynthetic pathway for F430 (50). Considering the role of NifH and NifD homologs in chlorin synthesis, it may be speculated that NflD and NflH are similarly involved in methanogens in the synthesis of F430, possibly through ring reductions. Perhaps the other proteins associated with NflD are also involved. For instance, the N-terminal lactamase domain predicted in the 50-kDa protein (see the supplemental material) may be involved in forming the external lactam ring of F430. Further investigations, including extensive genetic manipulations of and mechanistic studies with NflD and NflH, are required to assess these possibilities.
It is interesting to note that NflH and NflD are both expressed as soluble proteins in aerobically grown E. coli. We were able to overexpress the dinitrogenase reductase or a dinitrogenase homolog in E. coli as a soluble protein rather than an inclusion body. Even when grown aerobically, a significant portion of iron-sulfur clusters (as judged by the brown coloration) are assembled. Furthermore, it is likely that the anaerobic expression of NflH (or possibly NflD) in E. coli could yield fully active protein. If this possibility is the case, future structural, biochemical, and spectroscopic studies would be greatly facilitated.
Supplementary Material
Acknowledgments
We thank Wesley Swingley (ASU) and Eric Johnson for helpful discussions.
This work was supported by a National Astrobiology Institute/National Research Council Research Associateship to C.S. (NRC Associateship no. 0253240), a NASA Astrobiology: Exobiology and Evolutionary Biology grant (NNG05GP24G) to B.M., an NSF Small Grant for Exploratory Research grant to R.B. (0415283), and a grant from the Monsanto/Washington University Plant Science Program to R.B.
Footnotes
Published ahead of print on 27 July 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bapteste, E., C. Brochier, and Y. Boucher. 2005. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea 1:353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bulen, W. A., and J. R. LeComte. 1966. The nitrogenase system from Azotobacter: two-enzyme requirement for N2 reduction, ATP-dependent H2 evolution, and ATP hydrolysis. Proc. Natl. Acad. Sci. USA 56:979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J.-F. Tomb, M. D. Adams, C. L. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, J. F. Weidman, J. L. Fuhrmann, D. Nguyen, T. R. Utterback, J. M. Kelley, J. D. Peterson, P. W. Sadow, M. C. Hanna, M. D. Cotton, K. M. Roberts, M. A. Hurst, B. P. Kaine, M. Borodovsky, H.-P. Klenk, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 4.Burggraf, S., K. O. Stetter, P. Rouviere, and C. R. Woese. 1991. Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst. Appl. Microbiol. 14:346-351. [DOI] [PubMed] [Google Scholar]
- 5.Burke, D. H., J. E. Hearst, and A. Sidow. 1993. Early evolution of photosynthesis: clues from nitrogenase and chlorophyll iron proteins. Proc. Natl. Acad. Sci. USA 90:7134-7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cacciapuoti, G., S. Gorassini, M. F. Mazzeo, R. A. Siciliano, V. Carbone, V. Zappia, and M. Porcelli. 2007. Biochemical and structural characterization of mammalian-like purine nucleoside phosphorylase from the Archaeon Pyrococcus furiosus. FEBS J. 274:2482-2495. [DOI] [PubMed] [Google Scholar]
- 7.Chan, M. K., J. Kim, and D. C. Rees. 1993. The nitrogenase FeMo-cofactor and P-cluster pair: 2.2 A resolution structures. Science 260:792-794. [DOI] [PubMed] [Google Scholar]
- 8.Conradson, S. D., B. K. Burgess, W. E. Newton, A. Di Cicco, A. Filipponi, Z. Y. Wu, C. R. Natoli, B. Hedman, and K. O. Hodgson. 1994. Selenol binds to iron in nitrogenase iron-molybdenum cofactor: an extended X-ray absorption fine structure study. Proc. Natl. Acad. Sci. USA 91:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dilworth, M. J., and R. N. Thorneley. 1981. Nitrogenase of Klebsiella pneumoniae. Hydrazine is a product of azide reduction. Biochem. J. 193:971-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duin, E. C., L. Signor, R. Piskorski, F. Mahlert, M. D. Clay, M. Goenrich, R. K. Thauer, B. Jaun, and M. K. Johnson. 2004. Spectroscopic investigation of the nickel-containing porphinoid cofactor F(430). Comparison of the free cofactor in the +1, +2 and +3 oxidation states with the cofactor bound to methyl-coenzyme M reductase in the silent, red, and ox forms. J. Biol. Inorg. Chem. 9:563-576. [DOI] [PubMed] [Google Scholar]
- 11.Eady, R. R., B. E. Smith, K. A. Cook, and J. R. Postgate. 1972. Nitrogenase of Klebsiella pneumoniae. Purification and properties of the component proteins. Biochem. J. 128:665-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Einsle, O., F. A. Tezcan, S. L. Andrade, B. Schmid, M. Yoshida, J. B. Howard, and D. C. Rees. 2002. Nitrogenase MoFe-protein at 1.16 A resolution: a central ligand in the FeMo-cofactor. Science 297:1696-1700. [DOI] [PubMed] [Google Scholar]
- 13.Fani, R., R. Gallo, and P. Lio. 2000. Molecular evolution of nitrogen fixation: the evolutionary history of the nifD, nifK, nifE, and nifN genes. J. Mol. Evol. 51:1-11. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson, S. J. 1998. Nitrogen cycle enzymology. Curr. Opin. Chem. Biol. 2:182-193. [DOI] [PubMed] [Google Scholar]
- 15.Forterre, P., C. Brochier, and H. Philippe. 2002. Evolution of the Archaea. Theor. Popul. Biol. 61:409-422. [DOI] [PubMed] [Google Scholar]
- 16.Fujita, Y., and C. E. Bauer. 2000. Reconstitution of light-independent protochlorophyllide reductase from purified BchL and BchN-BchB subunits. In vitro confirmation of nitrogenase-like features of a bacteriochlorophyll biosynthesis enzyme. J. Biol. Chem. 275:23583-23588. [DOI] [PubMed] [Google Scholar]
- 17.Fujita, Y., H. Matsumoto, Y. Takahashi, and H. Matsubara. 1993. Identification of a nifDK-like gene (ORF467) involved in the biosynthesis of chlorophyll in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 34:305-314. [PubMed] [Google Scholar]
- 18.Georgiadis, M. M., H. Komiya, P. Chakrabarti, D. Woo, J. J. Kornuc, and D. C. Rees. 1992. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257:1653-1659. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin, P. J., J. N. Agar, J. T. Roll, G. P. Roberts, M. K. Johnson, and D. R. Dean. 1998. The Azotobacter vinelandii NifEN complex contains two identical [4Fe-4S] clusters. Biochemistry 37:10420-10428. [DOI] [PubMed] [Google Scholar]
- 20.Haaker, H., C. Laane, K. Hellingwerf, B. Houwer, W. N. Konings, and C. Veeger. 1982. Short-term regulation of the nitrogenase activity in Rhodopseudomonas sphaeroides. Eur. J. Biochem. 127:639-645. [DOI] [PubMed] [Google Scholar]
- 21.Hageman, R. V., and R. H. Burris. 1978. Nitrogenase and nitrogenase reductase associate and dissociate with each catalytic cycle. Proc. Natl. Acad. Sci. USA 75:2699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halbleib, C. M., and P. W. Ludden. 2000. Regulation of biological nitrogen fixation. J. Nutr. 130:1081-1084. [DOI] [PubMed] [Google Scholar]
- 23.Halbleib, C. M., Y. Zhang, and P. W. Ludden. 2000. Regulation of dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase-activating glycohydrolase by a redox-dependent conformational change of nitrogenase Fe protein. J. Biol. Chem. 275:3493-3500. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, J. 1992. Global methanotrophy at the archaean-proterozoic transition, p. 220-236. In H. Baltscheffsky, S. Bengtson, J. Bergström, G. Vidal, and A. H. Knoll (ed.), Early life on earth, Columbia University Press, New York, NY.
- 25.Helber, J. T., T. R. Johnson, L. R. Yarbrough, and R. Hirschberg. 1988. Regulation of nitrogenase gene expression in anaerobic cultures of Anabaena variabilis. J. Bacteriol. 170:552-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinnemann, B., and J. K. Norskov. 2003. Modeling a central ligand in the nitrogenase FeMo cofactor. J. Am. Chem. Soc. 125:1466-1467. [DOI] [PubMed] [Google Scholar]
- 27.Hinnemann, B., and J. K. Norskov. 2004. Chemical activity of the nitrogenase FeMo cofactor with a central nitrogen ligand: density functional study. J. Am. Chem. Soc. 126:3920-3927. [DOI] [PubMed] [Google Scholar]
- 28.Hu, Y., A. W. Fay, and M. W. Ribbe. 2005. Identification of a nitrogenase FeMo cofactor precursor on NifEN complex. Proc. Natl. Acad. Sci. USA 102:3236-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huniar, U., R. Ahlrichs, and D. Coucouvanis. 2004. Density functional theory calculations and exploration of a possible mechanism of N2 reduction by nitrogenase. J. Am. Chem. Soc. 126:2588-2601. [DOI] [PubMed] [Google Scholar]
- 30.Hwang, J. C., and R. H. Burris. 1972. Nitrogenase-catalyzed reactions. Biochim. Biophys. Acta 283:339-350. [DOI] [PubMed] [Google Scholar]
- 31.Jacobitz, S., and P. E. Bishop. 1992. Regulation of nitrogenase-2 in Azotobacter vinelandii by ammonium, molybdenum, and vanadium. J. Bacteriol. 174:3884-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, E. F., and B. Mukhopadhyay. 2005. A new type of sulfite reductase, a novel coenzyme F420-dependent enzyme, from the methanarchaeon Methanocaldococcus jannaschii. J. Biol. Chem. 280:38776-38786. [DOI] [PubMed] [Google Scholar]
- 33.Kim, J., D. Woo, and D. C. Rees. 1993. X-ray crystal structure of the nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 3.0-A resolution. Biochemistry 32:7104-7115. [DOI] [PubMed] [Google Scholar]
- 34.Lahiri, S., L. Pulakat, and N. Gavini. 2005. Functional NifD-K fusion protein in Azotobacter vinelandii is a homodimeric complex equivalent to the native heterotetrameric MoFe protein. Biochem. Biophys. Res. Commun. 337:677-684. [DOI] [PubMed] [Google Scholar]
- 35.Lee, J. S., and E. H. Sng. 1982. A rapid, simple and reliable test for the routine identification of urease producing gram-negative bacteria. Ann. Acad. Med. Singap. 11:94-97. [PubMed] [Google Scholar]
- 36.Lovell, T., T. Liu, D. A. Case, and L. Noodleman. 2003. Structural, spectroscopic, and redox consequences of a central ligand in the FeMoco of nitrogenase: a density functional theoretical study. J. Am. Chem. Soc. 125:8377-8383. [DOI] [PubMed] [Google Scholar]
- 37.Lowe, D. J., K. Fisher, R. N. Thorneley, S. A. Vaughn, and B. K. Burgess. 1989. Kinetics and mechanism of the reaction of cyanide with molybdenum nitrogenase from Azotobacter vinelandii. Biochemistry 28:8460-8466. [DOI] [PubMed] [Google Scholar]
- 38.Ludden, P. W., and G. P. Roberts. 1989. Regulation of nitrogenase activity by reversible ADP-ribosylation, p. 23-55. In B. Horecker, E. Stadtman, P. Boon Chock, and A. Levitzki, (ed.), Current topics in cellular regulation, vol. 30. Academic Press, Inc., Orlando, FL. [DOI] [PubMed] [Google Scholar]
- 39.Mancinelli, R. L., and C. P. McKay. 1988. The evolution of nitrogen cycling. Origins Life Evol. Biosph. 18:311-325. [DOI] [PubMed] [Google Scholar]
- 40.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 1999. Reactor-scale cultivation of the hyperthermophilic methanarchaeon Methanococcus jannaschii to high cell densities. Appl. Environ. Microbiol. 65:5059-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 2000. A novel pH2 control on the expression of flagella in the hyperthermophilic strictly hydrogenotrophic methanarchaeaon Methanococcus jannaschii. Proc. Natl. Acad. Sci. USA 97:11522-11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters, J. W., M. H. Stowell, S. M. Soltis, M. G. Finnegan, M. K. Johnson, and D. C. Rees. 1997. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry 36:1181-1187. [DOI] [PubMed] [Google Scholar]
- 43.Pratt, J. M. 1978. The chemistry and biochemistry of nitrogen. Horiz. Biochem. Biophys. 5:119-160. [PubMed] [Google Scholar]
- 44.Raymond, J., J. L. Siefert, C. R. Staples, and R. E. Blankenship. 2004. The natural history of nitrogen fixation. Mol. Biol. Evol. 21:541-554. [DOI] [PubMed] [Google Scholar]
- 45.Rouviere, P. E., and R. S. Wolfe. 1987. Use of subunits of the methylreductase protein for taxonomy of methanogenic bacteria. Arch. Microbiol. 148:253-259. [Google Scholar]
- 46.Rubinson, J. F., J. L. Corbin, and B. K. Burgess. 1983. Nitrogenase reactivity: methyl isocyanide as substrate and inhibitor. Biochemistry 22:6260-6268. [DOI] [PubMed] [Google Scholar]
- 47.Rye, R., and H. D. Holland. 2000. Geology and geochemistry of paleosols developed on the Hekpoort Basalt, Pretoria Group, South Africa. Geology 28:483-486. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Shima, S., M. Goubeaud, D. Vinzenz, R. K. Thauer, and U. Ermler. 1997. Crystallization and preliminary X-ray diffraction studies of methyl-coenzyme M reductase from Methanobacterium thermoautotrophicum. J. Biochem. (Tokyo) 121:829-830. [DOI] [PubMed] [Google Scholar]
- 50.Thauer, R. K., and L. G. Bonacker. 1994. Biosynthesis of coenzyme F430, a nickel porphinoid involved in methanogenesis. Ciba Found. Symp. 180:210-222. [DOI] [PubMed] [Google Scholar]
- 51.Tittsworth, R. C., and B. J. Hales. 1993. Detection of EPR signals assigned to the 1-equiv-oxidized P-clusters of the nitrogenase MoFe-protein from Azotobacter vinelandii. J. Am. Chem. Soc. 115:9763-9767. [Google Scholar]
- 52.Tso, M. Y. 1974. Some properties of the nitrogenase proteins from Clostridium pasteurianum. Arch. Microbiol. 99:71-80. [DOI] [PubMed] [Google Scholar]
- 53.Yakunin, A. F., and P. C. Hallenbeck. 2000. Regulation of nitrogenase activity in Rhodobacter capsulatus under dark microxic conditions. Arch. Microbiol. 173:366-372. [DOI] [PubMed] [Google Scholar]
- 54.Yang, S., W.-H. Pan, G. D. Friesen, B. K. Burgess, J. L. Corbin, E. I. Stiefel, and W. E. Newton. 1982. Iron-molybdenum cofactor from nitrogenase. Modified extraction methods as probes for composition. J. Biol. Chem. 257:8042-8048. [PubMed] [Google Scholar]
- 55.Zorzopulos, J. 2003. Birth of the domains Bacteria, Archaea and Eucarya and of major taxa within them: a hypothesis. Rev. Argent. Microbiol. 35:175-182. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.