Abstract
Yersinia enterocolitica serotype O:9 is a gram-negative enteropathogen that infects animals and humans. The role of lipopolysaccharide (LPS) in Y. enterocolitica O:9 pathogenesis, however, remains unclear. The O:9 LPS consists of lipid A to which is linked the inner core oligosaccharide, serving as an attachment site for both the outer core (OC) hexasaccharide and the O-polysaccharide (OPS; a homopolymer of N-formylperosamine). In this work, we cloned the OPS gene cluster of O:9 and identified 12 genes organized into four operons upstream of the gnd gene. Ten genes were predicted to encode glycosyltransferases, the ATP-binding cassette polysaccharide translocators, or enzymes required for the biosynthesis of GDP-N-formylperosamine. The two remaining genes within the OPS gene cluster, galF and galU, were not ascribed a clear function in OPS biosynthesis; however, the latter gene appeared to be essential for O:9. The biological functions of O:9 OPS and OC were studied using isogenic mutants lacking one or both of these LPS parts. We showed that OPS and OC confer resistance to human complement and polymyxin B; the OPS effect on polymyxin B resistance could be observed only in the absence of OC.
Yersinia enterocolitica serotypes O:3, O:5,27, O:8, and O:9 include strains that infect both humans and animals (16). Pathogenesis of these strains is associated with the presence of common virulence factors encoded on the chromosome and on pYV, the Yersinia virulence plasmid (73). Despite the genetic and phenotypic similarity, however, the serotypes display epidemiological and host range differences (17, 22). Biotyping and serotyping based on the antigenic O-polysaccharide (OPS or O-antigen) are important in studying Y. enterocolitica infections. OPS is a distal part of lipopolysaccharide (LPS), which is the major component of the outer membrane of gram-negative bacteria (55, 63). In addition to OPS, LPS comprises lipid A, a hydrophobic membrane anchor, and a core oligosaccharide, divided into a lipid A-proximal inner core and an outer core (OC). The latter typically provides the attachment site for the polymeric OPS (55). Heteropolymeric OPSs, such as that of Y. enterocolitica serotype O:8 (63), are the most frequent among gram-negative bacteria; they are made up of identical oligosaccharide repeat units comprising three to eight different monosaccharides (55). Y. enterocolitica serotypes O:3 and O:9, however, display a homopolymeric OPS composed of single-sugar repeating units of 6-deoxy-l-altrose or N-formylperosamine, respectively (24, 41). Furthermore, the structures of the O:3 and O:9 core oligosaccharides are identical (51, 63). They both have a branching hexasaccharide termed OC which in genetic and biosynthetic terms resembles more closely a nonpolymerized O unit than the classical hexose-containing OC of Escherichia coli or Salmonella species (67, 68). The linkage of both the OC and the homopolymeric OPS to the inner core is a unique feature shared between serotypes O:3 and O:9. The O:9 OPS is identical to that of Brucella species and thus gives false-positive reactions in serological assays for brucellosis in humans and animals (23, 28, 38, 53).
Homopolymeric OPSs are synthesized at the cytoplasmic face of the inner membrane. The OPS synthesis from nucleotide diphosphate-activated sugar precursors on the membrane-bound carrier undecaprenylphosphate is carried out by glycosyltransferases. Full-length polymer is then translocated to the periplasm by the ATP-binding cassette (ABC) transporter formed by Wzt and Wzm. Subsequently, the polymer is ligated to preformed lipid A-core compounds and further translocated to the outer membrane (55).
The genes encoding enzymes necessary for the OPS biosynthesis are usually clustered in the bacterial chromosome (60). In the genus Yersinia the locus between hemH and gsk contains the genes required for the biosynthesis of the heteropolymeric OPSs (57, 63, 64, 66). In serotypes O:9 and O:3, however, the hemH-gsk locus is occupied by the OC gene cluster, thereby indicating that the OPS gene cluster is located elsewhere in the genome (63, 65, 66).
In this work, we present the cloning and characterization of the Y. enterocolitica serotype O:9 OPS gene cluster. Due to the branched nature of the OC in LPS, mutants lacking OC but keeping the OPS and vice versa were constructed. We demonstrated that these mutants have both polymyxin B and serum resistance defects.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacteriophages, and culture conditions.
Bacterial strains, plasmids, and bacteriophages used in this work are listed in Table 1. All the Y. enterocolitica O:9 derivatives used in this work originate from the wild-type strain Ruokola/71 (62).
TABLE 1.
Bacterial strains, plasmids, bacteriophages, and bacteriocins used in this work
| Bacterial strain, plasmid, or bacteriophage or bacteriocin | Comments | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| SY327λpir | λ(lac pro) argE(Am) rif nalA recA56 (λpir) | 50 |
| Sm10λpir | thi thr leuB tonA lacY supE recA::RP4-2-Tc::Mu-Km (λpir) | 61 |
| S17-1 | thi pro hsdR hsdM+recA::RP4-2-Tc::Mu-Km::Tn7 Strr | 61 |
| S17-1λpir | thi pro hsdR hsdM+recA::RP4-2-Tc::Mu-Km::Tn7 Strr (λpir) | 29 |
| HB101/pRK2013 | Triparental conjugation helper strain; Kmr | 31 |
| C600 | thi thr leuB tonA lacY supE | 6 |
| Y. enterocolitica | ||
| Ruokola/71 | Patient isolate | 62 |
| Ruokola/71-c | Spontaneous virulence plasmid-cured derivative of Ruokola/71 | 62 |
| YeO9-OC-R | Phage φR1-37-resistant spontaneous OC-negative derivative of YeO9-R1 | This work |
| YeO9-R1 | Δper::KmGB; rough (OPS negative); Kmr derivative of Ruokola/71 | This work |
| YeO9-c-R3 | Δper::KmGB; rough (OPS negative); Kmr derivative of Ruokola/71-c | This work |
| YeO9-OC | Δ(wzx-wbcL)::KmGB; OC negative; derivative of Ruokola/71 | This work |
| YeO9-c-OC | Δ(wzx-wbcL)::KmGB; OC negative; derivative of Ruokola/71-c | This work |
| YeO9-OCP | Phage φR1-37-resistant spontaneous low-OC-expressing derivative of Ruokola/71 | This work |
| YeO9-galF | ΔgalF::KmGB; derivative of Ruokola/71; Kmr | This work |
| YeO9-c-galF | ΔgalF::KmGB; derivative of Ruokola/71; Kmr | This work |
| Plasmids | ||
| pRV1 | Suicide vector; Clmr | 68 |
| pRV1-galF | PvuII fragment of O:9 OPS gene cluster containing the galF gene cloned into pRV1 | This work |
| pRV1-galF-Nsi | pRV1-galF with the galF internal NsiI fragment deleted | This work |
| pRV1-galF-Nsi::GB | pRV1-galF with the galF internal NsiI fragment replaced by KmGB | This work |
| pRV1-galU | MscI fragment of O:9 OPS gene cluster containing the galU gene cloned into pRV1 | This work |
| pRV1-galU-del | pRV1-galU with a deletion in the galU gene | This work |
| p87/I | pHC79 carrying a genomic fragment of Ruokola/71-c containing the OPS gene cluster | This work |
| p46/III | pHC79 carrying a genomic fragment of Ruokola/71-c containing the OPS gene cluster | This work |
| p73/II | pHC79 carrying a genomic fragment of Ruokola/71-c containing the OPS gene cluster | This work |
| p77/II | pHC79 carrying a genomic fragment of Ruokola/71-c containing the OPS gene cluster | This work |
| pKK232-8 | Promoter trapping vector; Ampr | GE Healthcare |
| pPSL | 10-kb PstI fragment of Ruokola/71 genomic DNA cloned into pUC18 | This work |
| pWBO:9Sac | SacI fragment of pPSL | This work |
| pWBO:9SacGB | KmGB cloned into the EcoRV site within the per gene in pWBO:9Sac | This work |
| pRV1-WBO:9SacGB | 4-kb PvuII fragment of pWBO:9SacGB cloned into the EcoRV site of suicide vector pRV1 | This work |
| pHC79 | Cosmid cloning vector; Ampr | |
| pUC18 | Plasmid cloning vector; Ampr | |
| pUC4K | Source of the KmGB cassette; Ampr Kmr | Pharmacia |
| pRV19-GB | Suicide vector to inactivate OC gene cluster; Δ(wzx-wbcL)::KmGB; Clmr Kmr | 68 |
| Bacteriophages and bacteriocins | ||
| φR1-37 | OC-specific bacteriophage | 46, 67 |
| Enterocoliticin | OC-specific bacteriocin | 69, 70 |
Bacteria were routinely cultured in Luria-Bertani broth (LB) or on agar plates based on LB (LA plates). For bacteriophage cultures tryptic soy broth or agar plates were used. Yersinia selective agar plates (CIN agar; Oxoid, Basingstoke, England) supplemented with appropriate antibiotics were used for the selections of the transconjugants. Antibiotics were added to the growth media at following concentrations: ampicillin, 5 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 100 μg/ml in agar plates and 20 μg/ml in broth. E. coli strains were grown at 37°C and Y. enterocolitica strains at room temperature (RT; 22°C) unless otherwise indicated.
Recombinant DNA methods.
DNA isolations, restriction enzyme digestions, and DNA ligations were performed as described previously (7, 58). For PCR screening reactions, bacteria resuspended in 100 μl of water were incubated for 5 min at 95°C. Subsequently, 100 μl of phenol-chloroform-isoamyl alcohol (25:24:1) was added, and samples were vortexed and centrifuged for 5 min at 13,000 × g. A 5-μl aliquot of the upper DNA-containing phase was used as a PCR template. E. coli was transformed by electroporation or by the Hanahan method (36). Single-step screening of recombinant clones was performed as described earlier (13).
Construction of pUC and cosmid genomic libraries.
Genomic libraries of strain Ruokola/71-c were constructed in E. coli strain C600 using pUC18 as a cloning vector. Both the genomic and the vector DNA were digested with BamHI, ClaI, EcoRI, HindIII, PstI, HindIII/BamHI, or HindIII/EcoRI or with HindIII/PstI prior to ligation. A cosmid library was constructed by cloning partially Sau3AI-digested genomic DNA of Ruokola/71-c into cosmid pHC79 using the lambda packaging system (GE Healthcare).
PCR strategies.
Short-range PCR was performed using the thermostable DNA polymerase DynaZyme II (Finnzymes, Espoo, Finland) according to the manufacturer's instructions. Reaction conditions for PCR cycles were adjusted according to the oligonucleotide primers and the length of the amplified fragment.
The previously sequenced perosamine synthetase (per) genes of Vibrio cholerae (accession no. X59554), Vibrio anguillarum (accession no. AF025396), E. coli O157:H7 (accession no. AF061251), and Brucella melitensis biovar 1 (accession no. AF047478) (50 to 60% identical to each other) served to design degenerate primers (Table 2). These were used to amplify a 335-bp fragment of the per gene of Y. enterocolitica O:9. This PCR fragment was used as a probe to screen the libraries by colony hybridization and also for Southern hybridization as described below.
TABLE 2.
Primers used in this work
| Primer name | Sequence | 5′ position | Direction, accession no., use |
|---|---|---|---|
| Per-2 | GGMGATGAAGTBATTGTWCCRAC | 7148 | Forward, AJ605741, degenerate primer for amplifying per gene fragment of Ruokola/71-c for a probe |
| Per-4 | GTYTTRTTYCCAAARAARCTAAA | 7482 | Reverse, AJ605741, degenerate primer for amplifying per gene fragment of Ruokola/71-c for a probe |
| galU-f | GTTGTCATCGACGGGATATG | 4700 | Forward, AJ605741, plasmid PCR to delete part of galU gene |
| galU-r | ATTGAATCAGGGGCTTATCG | 3919 | Reverse, AJ605741, plasmid PCR to delete part of galU gene, cloning of the galU upstream promoter |
| PSL-F7a | TATCAACTGCAACTGCAAAG | 3454 | Forward, AJ605741, cloning of the galU upstream promoter |
| F7 | CTGAATCATTACTTCGAGTG | 3550 | Forward, AJ605741, cloning of the galU upstream promoter |
| galF-F | ACCGGGCAATCCTATAACTG | 5532 | Forward, AJ605741, cloning of the gmd upstream promoter |
| pSL-7B | GTTGTAACATCATCCATTGC | 6527 | Reverse, AJ605741, cloning of the gmd upstream promoter |
| pSL-7600Ra | TGTTGACAATGGCCATATGA | 8552 | Forward, AJ605741, cloning of the wzm upstream promoter |
| pSL-6A | AATTTATGAAAAGGCATGCC | 7528 | Reverse, AJ605741, cloning of the wzm upstream promoter |
Colony and Southern blotting.
The 335-bp per fragment was digoxigenin labeled using the High Prime DNA Labeling Kit (Roche Molecular Biochemicals, Boehringer-Mannheim, Germany) according to the instructions of the manufacturer. Colonies replica plated onto nylon membranes were lysed and processed for hybridization as described earlier (58). Slot blotting was performed using the Minifold II Slot Blot Manifold (Schleicher & Schuell, Dassel, Germany). Denatured genomic DNA was applied to the slots as instructed by the manufacturer. Southern blotting was performed as described previously (58). Hybridization and probe detection were performed using the DIG Luminescence Detection Kit for Nucleic Acids (Roche Molecular Biochemicals).
Nucleotide sequencing and sequence analysis.
Nucleotide sequencing reactions were performed on ABI373A and ABI377 automatic sequencers using the AmpliTaq FS dye terminator kit or the Prism BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Nucleotide sequence analysis was performed using the computer programs of the Wisconsin Package Version 10.0 (Genetics Computer Group, Madison, WI) and the EMBOSS programs and the HIBIO DNASIS program for Windows, Higgins and Sharp algorithm (CLUSTAL 4) (37). NCBI and EBI databases were searched using the BLAST programs (4). Similar amino acid sequences were aligned using the PILEUP program of the Genetics Computer Group package.
Construction of per, galF, and galU suicide vectors for allelic exchange.
pRV1, a Clmr derivative of the suicide vector pJM703.1, was used to construct genomic insertion derivatives by allelic exchange (68).
The suicide vector pRV1-WBO:9SacGB (Table 1) for the per gene inactivation was constructed by cloning the 2.6-kb SacI fragment containing the per gene from pPSL into pUC18. The resulting plasmid was named pWBO:9Sac and maintained in E. coli C600. The 1.2-kb kanamycin resistance gene block (KmGB) of pUC4K was cloned into the EcoRV site within the per gene of pWBO:9Sac to get plasmid pWBO:9SacGB. The 4-kb PvuII fragment of pWBO:9SacGB was cloned into the EcoRV site of the suicide vector pRV1 to get plasmid pRV1-WBO:9SacGB, which was maintained in E. coli SY327λpir.
The suicide vector pRV1-galF-Nsi:GB (Table 1) for the galF gene inactivation was constructed by cloning the galF-containing PvuII fragment of pPSL into pRV1. The resulting plasmid was named pRV1-galF. Deletion of the galF-internal 0.4-kb NsiI fragment resulted in plasmid pRV1-galF-Nsi. The KmGB was cloned into the NsiI site of the pRV1-galF-Nsi to get pRV1-galF-Nsi:GB, which was maintained in E. coli SY327λpir.
The suicide vector pRV1-galU-del (Table 1) for the galU gene inactivation was constructed by cloning the galU-containing MscI fragment of pPSL into pRV1. The resulting plasmid was named pRV1-galU. An 800-bp internal deletion in the galU gene of pRV1-galU was engineered by plasmid-PCR (20) using primers galU-f and galU-r (Table 2) to get plasmid pRV1-galU-del, which was maintained in E. coli SY327λpir and from there transformed into Sm10λpir.
The suicide vector pRV19-GB (Table 1) for the OC gene cluster inactivation has been described previously (68).
Inactivation of genes by allelic exchange.
Direct or triparental conjugation to Y. enterocolitica O:9 strains Ruokola/71 and Ruokola/71-c was performed to mobilize the suicide vectors pRV1-galF-NsiGB, pRV1-galU-del, pRV19-GB, and pRV1-WBO:9SacGB from E. coli SY327λpir. or Sm10λpir, as described earlier (68). Triparental conjugation was performed using the helper strain HB101/pRK2013 (Table 1). Transconjugants having the suicide vectors integrated by homologous recombination into the bacterial chromosome were subjected to cycloserine enrichment to select clones in which the second homologous recombination event had eliminated the suicide vector and the wild-type allele (34, 54). The constructed mutants were verified by PCR, Southern blotting, and/or sequencing. The LPS phenotype was analyzed by deoxycholate-polyacrylamide gel electrophoresis (DOC-PAGE; see below).
Isolation of spontaneous Y. enterocolitica O:9 OC mutants using φR1-37.
Bacteriophage φR1-37 was used to isolate spontaneous OC mutants. An overnight LB culture of the Y. enterocolitica O:9 strain was spread as a lawn on LA plates. A few drops of φR1-37 were pipetted on the dried bacterial lawn. After 2 days of incubation, individual phage-resistant colonies were picked from the lysis zone and subjected to the same treatment several times. To verify the loss of OC, LPSs from φR1-37-resistant bacteria were analyzed by DOC-PAGE.
PCR-based promoter identification.
PCR fragments containing selected intergenic regions, amplified using primers listed in Table 2, were cloned into the promoter trapping vector pKK232-8 (Table 1). The vector contains a promoterless cat gene encoding chloramphenicol acetyltransferase; thus, Clmr clones can be obtained only when a fragment containing promoter activity is cloned upstream of the cat gene. Transformants displaying promoter activity were selected on chloramphenicol plates. The inserted PCR fragments in the recovered plasmids were analyzed by sequencing.
Isolation and analysis of LPS.
LPSs from OC and OPS mutant candidates were checked by DOC-PAGE. For small-scale LPS isolation, a modified version of the protocol devised by Hitchcock and Brown was used (11, 39). Overnight 5-ml bacterial cultures were diluted to obtain an optical density at 540 nm (OD540) of <1. A 1.5-ml aliquot of the suspension was centrifuged in a microcentrifuge (13,000 × g, 3 min), and bacteria were resuspended in DOC lysis buffer (2% DOC, 4% 2-mercaptoethanol, 10% glycerol, and 0.002% bromophenol blue in 1 M Tris-HCl buffer, pH 6.8) in a volume adjusted according to the OD540 of the culture (100 μl of DOC lysis buffer/OD540 = 1.0). Samples were heated at 100°C for 10 min, and then 2 μl of proteinase K (20-mg/ml stock solution) was added. Samples were incubated at 55°C for ≥1 h. Samples were stored at −20°C until analyzed by DOC-PAGE as described previously (68).
Anti-Y. enterocolitica O:9 antiserum and slide agglutination.
Rabbit antiserum S-3 against formalin-killed Ruokola/71 bacteria was raised as described earlier (2). For the slide agglutination test the antiserum was diluted 1:100 in 0.9% NaCl.
Human serum and serum killing assay.
Blood was obtained from healthy human donors who were devoid of anti-Yersinia antibodies. Sera were pooled and stored in aliquots at −70°C as described earlier (14). The killing assay was performed as described previously (14). Briefly, ∼500 to 1,000 bacteria were incubated at 37°C for 30 and 120 min in 30 μl of 66.7% normal human serum (NHS), heat-inactivated serum, or EGTA-Mg serum. The latter contained 10 mM EGTA and 5 mM MgCl2. Surviving bacteria were cultured and counted after growth on LA plates. The serum bactericidal effect was calculated as the survival percentage using the bacterial counts obtained with bacteria incubated in heat-inactivated serum as 100%. The killing experiment was repeated for each strain at least three times starting from independent cultures.
Polymyxin B resistance.
Yersinia strains grown in 5 ml of LB either at 37°C or at 21°C were harvested (5,000 × g, 15 min, 5°C) in the exponential phase of growth. Bacteria were suspended in 1% (wt/vol) tryptone in phosphate-buffered saline (pH 7.4) to approximately 2.1 × 105 CFU/ml. Ten microliters of the suspension was mixed with various concentrations of polymyxin B in a volume of 200 μl and incubated at the original bacterial growth temperature for 30 min. Subsequently, 100 μl of the suspensions was directly plated on LB agar plates. The plates were incubated at 26°C, and colony counts were determined. The results were expressed as survival percentages, taking the colony counts of bacteria not exposed to antibacterial agents as 100%. The 50% inhibitory concentration of antimicrobial peptides (IC50) was defined as the concentrations showing a 50% reduction in the colony count compared with bacteria not exposed to the antibacterial agent (21). All experiments were done in duplicate and on four independent occasions.
Statistical methods.
Statistical analyses were performed using the analysis of variance or the two-sample t test or, when the requirements were not met, by the Mann-Whitney U test. A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession number.
The sequence data were annotated and submitted to the European Bioinformatics Institute (accession number AJ605741).
RESULTS
Identification, cloning, and characterization of the OPS gene cluster.
To clone the OPS gene cluster from Y. enterocolitica O:9, several genomic libraries of strain Ruokola/71-c were constructed in pUC18. Southern blotting analysis was performed to select appropriate restriction enzymes for the cloning. Degenerate primers Per-2 and Per-4 (Table 2) based on the perosamine synthetase (per) genes of other bacteria were used to amplify a per gene fragment of Y. enterocolitica O:9. This PCR product served as a probe in Southern hybridization and in screening the libraries by colony hybridization (42, 48). From the pUC18 libraries a colony containing a 10-kb PstI fragment was identified and characterized. Sequencing data revealed that the PstI fragment did not carry all the genes required for the O:9 OPS biosynthesis. However, from the cosmid library of Y. enterocolitica O:9 we isolated four hybridization-positive clones (Table 1). These were also positive in the slide agglutination assay using the O:9 OPS-specific antiserum S-3. Thus, the cosmids carried the genes required for the O:9 OPS expression in E. coli. These four clones were subsequently used to complete the sequencing of the OPS gene cluster.
Assignment of putative functions to open reading frames.
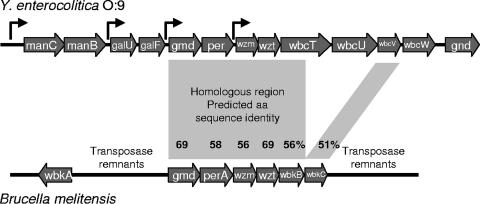
We identified 12 genes located upstream of the gnd gene. The genes were annotated based on similarity to sequences in the databases (Fig. 1; Table 3). The manB, manC, gmd, per, wbcV, and wbcT genes are predicted to encode enzymes involved in the biosynthesis of GDP-N-formylperosamine. The wbcU and wbcW genes are predicted to encode the N-formylperosaminyltransferases assembling the homopolymeric OPS onto undecaprenylphosphate carrier lipid while the wzm and wzt genes encode proteins for the transport of the OPS into the periplasm. The gnd gene encodes gluconate-6-phosphate dehydrogenase, which does not participate in OPS synthesis. The genes galU and galF, identified between the manB and gmd genes, however, were not assigned a clear function in the OPS biosynthesis.
FIG. 1.
OPS gene cluster of Y. enterocolitica O:9. The gene cluster figures were generated from the sequence files under the accession numbers AJ605741 for Y. enterocolitica O:9 and AF047478 for B. melitensis using the ggnVIEW sequence file viewer (http://colibase.bham.ac.uk/cgi-bin/fileprepare.cgi). The identified and predicted promoter locations of the O:9 gene cluster are indicated with bent arrows. The orf1 gene (Table 3), present in the reverse direction upstream of the manC gene, is not shown. The regions of homology between the gene clusters are indicated with shadowed quadrangles, and the percentages of identity between the predicted amino acid sequences are given.
TABLE 3.
OPS gene cluster of Y. enterocolitica O:9: genes, gene products, and similarities
| Gene | Protein | Size (no. of amino acids) | Similarity | Putative function(s) |
|---|---|---|---|---|
| orf1 | Orf1 | >117 | 80-98% identity | N terminus: similar to TerC-family proteins, possibly involved in tellurite resistance |
| manC | ManC | 472 | >70% identity to several ManC proteins | Mannose-1-phosphate guanylytransferase; involved in biosynthesis of GDP-mannose and GDP-N-formylperosamine |
| manB | ManB | 480 | >70% identity to several ManB proteins | Phosphomannomutase; involved in biosynthesis of GDP-mannose and GDP-N-formylperosamine |
| galU | GalU | 296 | 80-98% identity to several GalU proteins | UTP-glucose-1-phosphate uridylyltransferase |
| galF | GalF | 297 | 80-98% identity to several GalU proteins | UTP-glucose-1-phosphate uridylyltransferase; 66% identical to GalU of same strain. |
| gmd | Gmd | 373 | >80% identity to several Gmd proteins | GDP-mannose-4,6-dehydratase; involved in biosynthesis of GDP-N-formylperosamine |
| per | Per | 361 | 55-67% identity to several Per proteins | GDP-d-perosamine synthase; involved in biosynthesis of GDP-N-formylperosamine |
| wzm | Wzm | 260 | 55-71% identity to several Wzm proteins | Putative ABC transporter system integral membrane protein; translocation of OPS |
| wzt | Wzt | 251 | 45-78% identity to several Wzt proteins | Putative ATP binding protein involved in OPS export |
| wbcT | WbcT | 581 | 37% identity to glycosyltransferase of Aeromonas hydrophila | Involved in biosynthesis of O:9 OPS, N-terminal half 55% identical to WbkB, a putative glycosyltransferase of Brucella |
| wbcU | WbcU | 527 | 30-40% identity to mannosyltransferases | Putative N-formylperosaminyltransferase; amino acids 1 to 75 and 192 to 527 similar to Brucella mannosyltransferase |
| wbcV | WbcV | 260 | 40-50% identity to several N-formyltransferase proteins | Putative N-formyltransferase; involved in biosynthesis of GDP-N-formylperosamine |
| wbcW | WbcW | 370 | 45-55% identity to glycosyltransferases | Putative N-formylperosaminyltransferase |
| gnd | Gnd | 390 | 85-98% identity to several Gnd proteins | Gluconate-6-phosphate dehydrogenase |
Identification of promoters.
Sequence analysis revealed several putative promoters in the O:9 OPS gene cluster, one upstream from the entire cluster, in front of manC, and three intergenic promoters within the cluster in front of the galU, gmd, or wzm gene (Fig. 1). The intergenic regions were PCR amplified using primers given in Table 2; the manB-galU intergenic region was in 369- and 465-bp fragments, the galF-gmd region in a 995-bp fragment, and the per-wzm region in a 1,024-bp fragment. The fragments were cloned into the promoter trapping vector pKK232-8. Sequence analysis of the plasmids isolated from the transformants showed that Clmr colonies were obtained only when the fragments had been inserted into the vector in the correct orientation. This provided experimental evidence for the promoter activity upstream of the galU, gmd, and wzm genes.
Characterization of Y. enterocolitica O:9 galF and galU mutants.
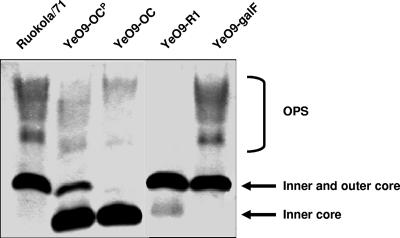
Two genes showing sequence similarity to the galU and galF genes were identified within the O:9 OPS gene cluster. The presence of these genes in the cluster was unexpected, as they both code for UTP-glucose-1-phosphate uridylyltransferase (EC 2.7.7.9; also called UDP-glucose pyrophosphorylase), which catalyzes the conversion of glucose-1-phosphate to UDP-glucose. UDP-glucose is a central nucleotide sugar in carbohydrate metabolism but redundant for the perosamine synthesis. We aimed to study the role of the galU and galF genes in the OPS gene cluster of Y. enterocolitica O:9. We thus inactivated the galU and galF genes and analyzed the LPS profiles and growth rates of the mutants. To inactivate the genes, suicide plasmids carrying the inactivated galU and galF genes were constructed and introduced into the Y. enterocolitica O:9 strains. No viable O:9 galU mutants were obtained. It suggested that there is only one copy of this gene in the chromosome and that it is essential for Y. enterocolitica O:9. On the other hand, we successfully constructed a galF mutant. The LPS profile of the galF mutant, analyzed by DOC-PAGE (Fig. 2), did not differ from that of the wild type. The growth of the mutant was not affected (data not shown).
FIG. 2.
DOC-PAGE analysis of the LPS of wild-type and mutant strains grown overnight at RT. Strains are indicated at the top, and the different parts of LPS are indicated at the right. The peculiar situation of the LPS of Y. enterocolitica O:9 is demonstrated here; the OC likely represents a nonpolymerized O unit attached to typical (inner) core oligosaccharide.
Characterization of Y. enterocolitica O:9 per mutant.
The per gene of the Y. enterocolitica serotype O:9 strain was inactivated by insertion of KmGB. The mutation was confirmed by Southern blotting, PCR, and sequencing. The LPS phenotype of the per mutant (YeO9-R1) was analyzed by DOC-PAGE, and as expected, the mutant did not express OPS (Fig. 2).
Polymyxin B resistance of Y. enterocolitica O:9 OPS and OC mutants.
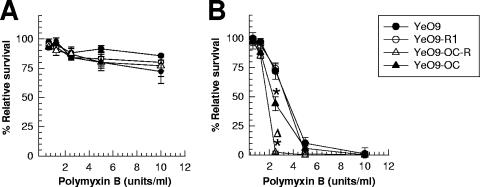
In several pathogens the LPS confers resistance to cationic antimicrobial peptides including polymyxin B (47, 67). We thus aimed to elucidate whether Y. enterocolitica O:9 LPS mediates resistance to polymyxin B. To this end we analyzed the polymyxin resistance phenotypes of the wild-type strain Ruokola/71 and OPS-negative (YeO9-R1), OC-negative (YeO9-OC), and OPS-negative and OC-negative (YeO9-OCR) strains in a survival assay. We also determined the IC50 values of the strains. In general, the resistance of the strains to polymyxin B was growth temperature dependent.
In the survival assay all strains grown at RT displayed high, wild-type-level resistance to polymyxin B (Fig. 3A). The IC50 values revealed, however, slight (less-than-twofold) differences between the RT-grown strains: the most sensitive was YeO9-OCR (IC50, 50 ± 6 units/ml), followed by YeO9-OC (74 ± 7 units/ml), YeO9-R1 (84 ± 5 units/ml), and the wild-type strain (90 ± 9 units/ml). The value of YeO9-OCR was significantly different (P < 0.05) from that of YeO9-R1 and the wild-type strains. The other differences were not significant.
FIG. 3.
Role of LPS phenotype in resistance of Y. enterocolitica O:9 to polymyxin B. Bacteria were grown either at RT (A) or at 37°C (B). *, P < 0.05, significant difference from wild-type strain; Δ, P < 0.05, significant difference from OC mutant.
In contrast, the survival assay revealed that the strains grown at 37°C were more sensitive to polymyxin B (Fig. 3B) and that there were significant differences between the strains. The wild-type strain and YeO9-R1 displayed equal and highest resistance against polymyxin B. Significantly more sensitive were YeO9-OC and YeO9-OCR, the latter being the most sensitive (Fig. 3B). Accordingly, the IC50 values also demonstrated the same differences between the 37°C-grown bacteria: the most sensitive was YeO9-OCR (IC50, 0.9 ± 0.6 units/ml), followed by YeO9-OC (2.1 ± 0.2 units/ml), YeO9-R1 (5.2 ± 0.4 units/ml), and the wild-type strain (5.6 ± 0.3 units/ml). The YeO9-OCR and YeO9-OC values differed significantly (P < 0.05) from those of YeO9-R1 and the wild type. On the other hand, the IC50 values of YeO9-R1 were not significantly different (P > 0.05) from those of the wild-type strain.
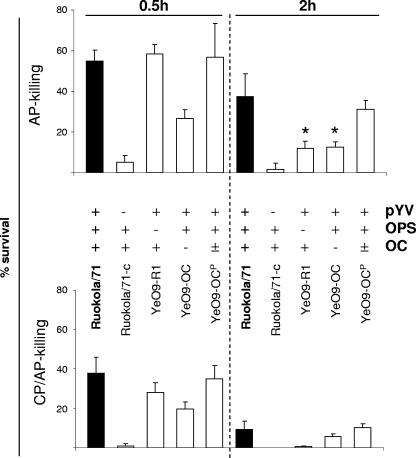
The role of Y. enterocolitica O:9 OPS and OC in serum resistance.
To examine the role of Y. enterocolitica O:9 LPS in serum resistance, the wild-type strain Ruokola/71, the pYV-negative strain Ruokola/71-c, and the LPS mutants, missing the OPS (YeO9-R1) or the OC (YeO9-OC) or expressing a reduced amount of OC (YeO9-OCP), were tested for the ability to resist alternative and classical complement pathway (AP and CP, respectively)-mediated killing (Table 1; Fig. 4).
FIG. 4.
Role of LPS phenotype in serum resistance of Y. enterocolitica O:9. Survival of bacteria in 66.7% NHS (CP/AP-killing bottom panel) and Mg-EGTA-treated serum (AP-killing, top panel) at 0.5- and 2-h time points. The columns indicate the mean survival percentage of the strain, and the bars indicate the ranges of standard errors. The filled columns show the results for the wild-type strain. In between the panels, the strains and their properties with respect to the presence of pYV (Yersinia virulence plasmid) and expression of OPS and OC are indicated. *, the AP killing results of YeO9-OC and YeO9-R1 at 2 h differed significantly (P < 0.001, Student's t test) from those of Ruokola/71.
In the bactericidal assay Ruokola/71-c was the most sensitive to both AP- and CP/AP-mediated killing, confirming the importance of pYV-encoded YadA (26) in protection of Y. enterocolitica O:9 against complement-mediated killing (Fig. 4). Both YeO9-OC and YeO9-R1 were less resistant to serum than the wild-type strain. They survived well; however, at the 0.5-h time point, YeO9-R1 was clearly more resistant than YeO9-OC. This suggested the role of OC in early resistance to complement (Fig. 4). At the 2-h time point the relationship was opposite and the YeO9-R1 mutant, especially in NHS, was more efficiently killed. Finally, YeO9-OCP showed resistance intermediate between that of the wild-type strain and that of YeO9-OC at 2 h of incubation in both types of serum (Fig. 4). Thus, even partial reduction in OC expression (YeO9-OCP, Fig. 2) resulted in reduced resistance to complement.
DISCUSSION
Cloning and sequencing of the Y. enterocolitica O:9 OPS gene cluster.
In this work, we describe the cloning and characterization of the OPS gene cluster of Y. enterocolitica serotype O:9. The cluster directs the biosynthesis of the homopolymeric O:9 OPS composed of α1,2-linked N-formylperosamine (4,6-dideoxy-4-formamido-d-mannopyranose) (24). In Yersinia spp. expressing heteropolymeric OPSs the OPS gene cluster is located between the hemH and gsk genes (63, 65, 66). In Y. enterocolitica serotypes O:3 and O:9, both encoding homopolymeric OPSs, the genomic locus between hemH and gsk genes is occupied by the OC gene cluster (63). We identified the O:9 OPS gene cluster located upstream from the gnd gene, similar to the OPS gene clusters of Salmonella and E. coli (12, 59) (Fig. 1). The gnd gene encodes gluconate-6-phosphate dehydrogenase, an enzyme not involved in the OPS biosynthesis.
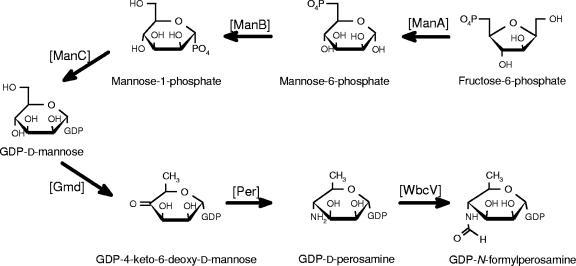
The O:9 gene cluster contains 12 genes. Based on sequence similarity and the predicted GDP-N-formylperosamine biosynthetic pathway (27, 33), manB, manC, gmd, per, and wbcV genes code for enzymes involved in the biosynthesis of GDP-N-formylperosamine, wbcU and wbcW code for N-formylperosaminyltransferases assembling the homopolymer onto undecaprenylphosphate carrier lipid, and wzm and wzt code for proteins for transporting the OPS homopolymer into periplasm. The putative pathway for the biosynthesis of the GDP-N-formylperosamine is given in Fig. 5. The WbcT protein is left without a function; therefore, it is likely that the functional assignments still need adjustments after biochemical evidence of the last steps of the pathway is found. The last step in the biosynthesis is shown to be the transfer of the formyl group by WbcV. However, it is not certain that the transfer of the formyl group takes place at the nucleotide sugar level; it could also take place after polymerization of OPS. Related to this, there is evidence for only one of the sugar nucleotides containing a formamido group, i.e., UDP-4-deoxy-4-formamido-l-arabinose (18, 72). In Y. enterocolitica O:9 the pathway could be verified either by identification of GDP-N-formylperosamine directly from bacterial cells or by characterization of the formyltransferase activity of WbcV in vitro.
FIG. 5.
Proposed biosynthesis pathway to GDP-N-formylperosamine in Y. enterocolitica O:9. The pathway up to GDP-4-keto-6-deoxy-d-mannose is identical to that of GDP-l-fucose biosynthesis; both Fcl (GDP-l-fucose synthetase) and Per (GDP-d-perosamine synthase) use this intermediate as a substrate.
The organization of the Y. enterocolitica O:9 OPS gene cluster is identical to that of B. melitensis over the region of the gmd, per, wzm, and wzt genes and the 5′ end of wbcT genes (Fig. 1); the gene products are 50 to 70% identical. Apparently the region of homology has originally extended further to the putative N-formyltransferase-encoding genes wbcV and wbkC. In Y. enterocolitica O:9, however, a DNA fragment containing the 3′ end of the wbcT gene and the wbcU gene was inserted upstream from the wbcV gene (Fig. 1). Otherwise there is no similarity between the clusters of these two organisms. Nucleotide sequence differences in the perosamine synthase (per) genes of Brucella and Y. enterocolitica O:9 were used to develop real-time PCR methods for identification of these bacteria (15, 42).
The OPS gene cluster also contains genes similar to galU and galF. These genes are involved in the synthesis of UDP-glucose and have no clear role in the OPS biosynthesis. Their presence in the OPS gene cluster was thus surprising. On the other hand, in Salmonella and E. coli the OPS gene clusters are usually located between the galF and gnd genes (12, 59). Our results showed that the galF mutation does not affect either the OPS synthesis or the viability of the bacterium. The galU gene, however, is essential for Y. enterocolitica O:9. Interestingly, in some bacteria galU mutants have been successfully constructed and displayed reduced virulence and resistance to antimicrobial agents (25, 32, 52).
Localization of the promoters of the OPS gene cluster.
Sequence analysis revealed several putative promoters in the OPS gene cluster, one upstream of the entire cluster, in front of manC, and three intergenic promoters inside the cluster in front of the galU, gmd, and wzm genes. Experimental data identified promoter activity upstream of the galU, gmd and wzm genes. The presence of an active promoter upstream of the manC gene is also very likely. The upstream region of the manC gene in Y. enterocolitica O:9 (Fig. 1) contains a 346-bp fragment more than 92% identical to the fragment containing the experimentally verified tandem promoters of the OPS gene cluster in Y. enterocolitica O:3 (74) (nucleotides 440 to 786, accession no AJ605741, versus nucleotides 1497 to 1844, accession no. Z18920, respectively). In fact, this is the only region of significant sequence similarity between the O:9 and O:3 OPS gene clusters. This region contains the JUMP start sequence (40) typical for all surface polysaccharide biosynthesis gene clusters and thus also present in all OPS gene clusters of Yersinia studied to date (63). Although more-detailed characterization of the promoters and regulation of transcription is warranted, our data indicate that the O:9 OPS gene cluster is organized into four transcriptional units.
Serum resistance.
The serum killing experiments confirmed the importance of pYV-encoded YadA in serum resistance of Y. enterocolitica O:9 (8). In contrast to serotype O:3 (14), however, O:9 LPS appears to play a clear role in serum resistance. Both OPS and OC protected Y. enterocolitica O:9 against complement killing (Fig. 4). The OPS seemed to provide long-term resistance to complement-mediated killing while OC was clearly more important at the early time point. The importance of long OPS chains in serum resistance has been shown in several studies (19, 35, 43). Long OPSs protect Salmonella and E. coli strains against insertion of the membrane attack complex into the membrane (44, 45). In Y. enterocolitica O:9, however, both OPS and OC seem to assist and strengthen the mainly YadA-dependent resistance against complement-mediated killing. Further work is warranted to elucidate the molecular mechanisms of this resistance.
Polymyxin B resistance.
The OPSs provide a steric hindrance for the access of antimicrobial peptides to inner LPS targets (9, 30, 47, 67). The serotype O:9 OPS, however, similarly to that of serotype O:3 (67), plays a minor role in the polymyxin B resistance. Moreover, the resistance is apparent only in the absence of the OC. On the other hand, as rough mutants of other bacterial species containing perosamine in their OPSs are more sensitive to antimicrobial peptides than the wild-type strains (49, 52, 71), the lack of contribution of serotype O:9 OPS to polymyxin B resistance is not likely due to its chemical composition. The available evidence, in fact, suggests that OPSs from yersiniae do not play a significant role in the resistance to antimicrobial peptides (11, 67; also unpublished data).
Temperature-dependent resistance to antimicrobial peptides, however, seems to be a common feature of yersiniae (5, 10, 56; also unpublished data). Even though both OPS expression and resistance to antimicrobial peptides in yersiniae are highest at RT (1, 11), our data suggest that OPS cannot account for this phenotype. We are currently investigating whether temperature-induced lipid A modifications affect the resistance to antimicrobial peptides.
Concluding remarks.
Isogenic OPS and OC mutants displayed decreased resistance to human serum complement and polymyxin B compared to the wild-type O:9 strain. Interestingly, the serum resistance phenotype, but not the polymyxin B resistance phenotype, differs from that of analogous OPS and OC mutants of serotype O:3 (3, 67). This points to different biological functions of the OPS and OC in serotypes O:3 and O:9. It is very likely that the chemical nature of the OPS may dictate the biological role that it plays in virulence. Since serotype O:9 strains more frequently infect animals than do serotype O:3 strains, one could further speculate that the LPS structures also influence the host preferences of the serotypes. Further studies are needed to address these questions.
Acknowledgments
This work has been supported by funding from the Nordic Joint Committee for Agricultural Research (project NKJ-115 to M.S. and J.H.), the Academy of Finland (projects 104361 and 50441 to M.S.), and the Fondo de Investigación Sanitaria (PI03/0881 to J.A.B.). M. Biedzka-Sarek has been supported by the University of Helsinki Graduate School in Biotechnology and Molecular Biology.
We thank Tatiana Bogdanovich and Kirsten Vestergaard for excellent technical assistance.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Al-Hendy, A., P. Toivanen, and M. Skurnik. 1991. The effect of growth temperature on the biosynthesis of Yersinia enterocolitica O:3 lipopolysaccharide: temperature regulates the transcription of the rfb but not of the rfa region. Microb. Pathog. 10:81-86. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hendy, A., P. Toivanen, and M. Skurnik. 1991. Expression cloning of Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb. Pathog. 10:47-59. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hendy, A., P. Toivanen, and M. Skurnik. 1992. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect. Immun. 60:870-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Anisimov, A. P., S. V. Dentovskaya, G. M. Titareva, I. V. Bakhteeva, R. Z. Shaikhutdinova, S. V. Balakhonov, B. Lindner, N. A. Kocharova, S. N. Senchenkova, O. Holst, G. B. Pier, and Y. A. Knirel. 2005. Intraspecies and temperature-dependent variations in susceptibility of Yersinia pestis to the bactericidal action of serum and to polymyxin B. Infect. Immun. 73:7324-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Appleyard, R. K. 1954. Segregation of new lysogenic types during growth of doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausubel, F. M., R. Brent, R. E. Kingston, O. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 8.Balligand, G., Y. Laroche, and G. Cornelis. 1985. Genetic analysis of a virulence plasmid from a serogroup 9 Yersinia enterocolitica strain: role of outer membrane protein P1 in resistance to human serum and autoagglutination. Infect. Immun. 48:782-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banemann, A., H. Deppisch, and R. Gross. 1998. The lipopolysaccharide of Bordetella bronchiseptica acts as a protective shield against antimicrobial peptides. Infect. Immun. 66:5607-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bengoechea, J. A., R. Díaz, and I. Moriyón. 1996. Outer membrane differences between pathogenic and environmental Yersinia enterocolitica biogroups probed with hydrophobic permeants and polycationic peptides. Infect. Immun. 64:4891-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bengoechea, J. A., H. Najdenski, and M. Skurnik. 2004. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol. Microbiol. 52:451-469. [DOI] [PubMed] [Google Scholar]
- 12.Berlyn, M. K. B., K. B. Low, K. E. Rudd, and M. Singer. 1996. Linkage map of Escherichia coli K-12, edition 9, p. 1715-1902. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, DC. [Google Scholar]
- 13.Beuken, E., C. Vink, and C. A. Bruggeman. 1998. One-step procedure for screening recombinant plasmids by size. BioTechniques 24:748-750. [DOI] [PubMed] [Google Scholar]
- 14.Biedzka-Sarek, M., R. Venho, and M. Skurnik. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 73:2232-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogdanovich, T., M. Skurnik, P. S. Lübeck, P. Ahrens, and J. Hoorfar. 2004. Validated 5′ nuclease PCR assay for rapid identification of the genus Brucella. J. Clin. Microbiol. 42:2261-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bottone, E. J. 1977. Yersinia enterocolitica: a panoramic view of a charismatic microorganism. Crit. Rev. Microbiol. 5:211-241. [DOI] [PubMed] [Google Scholar]
- 17.Bottone, E. J. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1:323-333. [DOI] [PubMed] [Google Scholar]
- 18.Breazeale, S. D., A. A. Ribeiro, A. L. McClerren, and C. R. Raetz. 2005. A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-L-arabinose. Identification and function of UDP-4-deoxy-4-formamido-L-arabinose. J. Biol. Chem. 280:14154-14167. [DOI] [PubMed] [Google Scholar]
- 19.Burns, S. M., and S. I. Hull. 1998. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect.Immun. 66:4244-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrappa, S., D. K. Gavin, and K. C. Gupta. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5:404-407. [DOI] [PubMed] [Google Scholar]
- 21.Campos, M. A., P. Morey, and J. A. Bengoechea. 2006. Quinolones sensitize gram-negative bacteria to antimicrobial peptides. Antimicrob. Agents Chemother. 50:2361-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carniel, E., I. Autenrieth, G. Cornelis, F. Guinet, H. Fukushima, R. Isberg, J. Pham, M. B. Prentice, M. Simonet, M. Skurnik, and G. Wauters. 2002. Y. enterocolitica and Y. pseudotuberculosis. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.11. Springer-Verlag, New York, NY.
- 23.Caroff, M., D. R. Bundle, M. B. Perry, J. W. Cherwonogrodzky, and J. R. Duncan. 1984. Antigenic S-type lipopolysaccharide of Brucella abortus 1119-3. Infect. Immun. 46:384-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caroff, M., D. R. Bundle, and M. B. Perry. 1984. Structure of the O-chain of the phenol-phase soluble cellular lipopolysaccharide of Yersinia enterocolitica serotype O:9. Eur. J. Biochem. 139:195-200. [DOI] [PubMed] [Google Scholar]
- 25.Chang, H. Y., J. H. Lee, W. L. Deng, T. F. Fu, and H. L. Peng. 1996. Virulence and outer membrane properties of a galU mutant of Klebsiella pneumoniae CG43. Microb. Pathog. 20:255-261. [DOI] [PubMed] [Google Scholar]
- 26.China, B., M. P. Sory, B. T. Nguyen, M. Debruyere, and G. R. Cornelis. 1993. Role of the YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect. Immun. 61:3129-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cloeckaert, A., M. Grayon, J. M. Verger, J. J. Letesson, and F. Godfroid. 2000. Conservation of seven genes involved in the biosynthesis of the lipopolysaccharide O-side chain in Brucella spp. Res. Microbiol. 151:209-216. [DOI] [PubMed] [Google Scholar]
- 28.Corbel, M. J. 1985. Recent advances in the study of Brucella antigens and their serological cross-reactions. Vet. Bull. 55:927. [Google Scholar]
- 29.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 30.Farnaud, S., C. Spiller, L. C. Moriarty, A. Patel, V. Gant, E. W. Odell, and R. W. Evans. 2004. Interactions of lactoferricin-derived peptides with LPS and antimicrobial activity. FEMS Microbiol. Lett. 233:193-199. [DOI] [PubMed] [Google Scholar]
- 31.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172:1-8. [DOI] [PubMed] [Google Scholar]
- 33.Godfroid, F., A. Cloeckaert, B. Taminiau, I. Danese, A. Tibor, X. de Bolle, P. Mertens, and J. J. Letesson. 2000. Genetic organisation of the lipopolysaccharide O-antigen biosynthesis region of brucella melitensis 16M (wbk). Res. Microbiol. 151:655-668. [DOI] [PubMed] [Google Scholar]
- 34.Gripenberg-Lerche, C., M. Skurnik, L. J. Zhang, K. O. Söderström, and P. Toivanen. 1994. Role of YadA in arthritogenicity of Yersinia enterocolitica serotype O:8: experimental studies with rats. Infect. Immun. 62:5568-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grossman, N., M. A. Schmetz, J. Foulds, E. N. Klima, V. Jiminez, L. L. Leive, and K. A. Joiner. 1987. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J. Bacteriol. 169:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 37.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 38.Hilbink, F., S. G. Fenwick, E. J. Thompson, R. Kittelberger, M. Penrose, and G. P. Ross. 1995. Non-specific seroreactions against Brucella abortus in ruminants in New Zealand and the presence of Yersinia enterocolitica O:9. N. Z. Vet. J. 45:175-178. [DOI] [PubMed] [Google Scholar]
- 39.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobbs, M., and P. R. Reeves. 1994. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol. Microbiol. 12:855-856. [DOI] [PubMed] [Google Scholar]
- 41.Hoffman, J., B. Lindberg, and R. R. Brubaker. 1980. Structural studies of the O-specific side-chains of the lipopolysaccharide from Yersinia enterocolitica Ye 128. Carbohydr. Res. 78:212-214. [DOI] [PubMed] [Google Scholar]
- 42.Jacobsen, N. R., T. Bogdanovich, M. Skurnik, P. S. Lübeck, P. Ahrens, and J. Hoorfar. 2005. A real-time PCR assay for the specific identification of serotype O:9 of Yersinia enterocolitica. J. Microbiol. Methods 63:151-156. [DOI] [PubMed] [Google Scholar]
- 43.Joiner, K. A., N. Grossman, M. Schmetz, and L. Leive. 1986. C3 binds preferentially to long-chain lipopolysaccharide during alternative pathway activation by Salmonella montevideo. J. Immunol. 136:710-715. [PubMed] [Google Scholar]
- 44.Joiner, K. A., C. H. Hammer, E. J. Brown, R. J. Cole, and M. M. Frank. 1982. Studies on the mechanism of bacterial resistance to complement-mediated killing. I. Terminal complement components are deposited and released from Salmonella minnesota S218 without causing bacterial death. J. Exp. Med. 155:797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joiner, K. A., C. H. Hammer, E. J. Brown, and M. M. Frank. 1982. Studies on the mechanism of bacterial resistance to complement-mediated killing. II. C8 and C9 release C5b67 from the surface of Salmonella minnesota S218 because the terminal complex does not insert into the bacterial outer membrane. J. Exp. Med. 155:809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiljunen, S., K. Hakala, E. Pinta, S. Huttunen, P. Pluta, A. Gador, H. Lönnberg, and M. Skurnik. 2005. Yersiniophage φR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology 151:4093-4102. [DOI] [PubMed] [Google Scholar]
- 47.Loutet, S. A., R. S. Flannagan, C. Kooi, P. A. Sokol, and M. A. Valvano. 2006. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J. Bacteriol. 188:2073-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lübeck, P. S., J. Hoorfar, P. Ahrens, and M. Skurnik. 2003. Cloning and characterization of the Yersinia enterocolitica serotype O:9 lipopolysaccharide O-antigen gene cluster, p. 207-209. In M. Skurnik, K. Granfors, and J. A. Bengoechea (ed.), The genus Yersinia: entering the functional genomic era. Kluwer Academic/Plenum Publishers, New York, NY. [DOI] [PubMed]
- 49.Martínez de Tejada, G., and I. Moriyón. 1995. The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect. Immun. 63:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 56:1242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller-Loennies, S., S. Rund, E. Ervelä, M. Skurnik, and O. Holst. 1999. The structure of the carbohydrate backbone of the core-lipid A region of the lipopolysaccharide from a clinical isolate of Yersinia enterocolitica O:9. Eur. J. Biochem. 261:19-24. [DOI] [PubMed] [Google Scholar]
- 52.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen, K., J. W. Cherwonogrodzky, J. R. Duncan, and D. R. Bundle. 1989. Enzyme-linked immunosorbent assay for differentiation of the antibody response of cattle naturally infected with Brucella abortus or vaccinated with strain 19. Am. J. Vet. Res. 50:5-9. [PubMed] [Google Scholar]
- 54.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 55.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rebeil, R., R. K. Ernst, B. B. Gowen, S. I. Miller, and B. J. Hinnebusch. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 52:1363-1373. [DOI] [PubMed] [Google Scholar]
- 57.Reeves, P. R., E. Pacinelli, and L. Wang. 2003. O antigen gene clusters of Yersinia pseudotuberculosis, p. 199-206. In M. Skurnik, K. Granfors, and J. A. Bengoechea (ed.), The genus Yersinia: entering the functional genomic era. Kluwer Academic/Plenum Publishers, New York, NY.
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 59.Sanderson, K. E., A. Hessel, S.-L. Liu, and K. E. Rudd. 1996. The genetic map of Salmonella typhimurium, edition VIII, p. 1903-1999. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. American Society for Microbiology, Washington, DC. [Google Scholar]
- 60.Schnaitman, C. A., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 62.Skurnik, M. 1984. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Appl. Bacteriol. 56:355-363. [DOI] [PubMed] [Google Scholar]
- 63.Skurnik, M. 2004. Lipopolysaccharides of Yersinia, p. 215-241. In E. Carniel and B. J. Hinnebusch (ed.), Yersinia: molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 64.Skurnik, M. 1999. Molecular genetics of Yersinia lipopolysaccharide, p. 23-51. In J. Goldberg (ed.), Genetics of bacterial polysaccharides. CRC Press, Boca Raton, FL.
- 65.Skurnik, M. 2003. Molecular genetics, biochemistry and biological role of Yersinia lipopolysaccharide, p. 187-189. In M. Skurnik, K. Granfors, and J. A. Bengoechea (ed.), The genus Yersinia: entering the functional genomic era. Kluwer Academic/Plenum Publishers, New York, NY.
- 66.Skurnik, M., and J. A. Bengoechea. 2003. The biosynthesis and biological role of lipopolysaccharide O-antigens of pathogenic Yersiniae. Carbohydr. Res. 338:2521-2529. [DOI] [PubMed] [Google Scholar]
- 67.Skurnik, M., R. Venho, J.-A. Bengoechea, and I. Moriyón. 1999. The lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3 is required for virulence and plays a role in outer membrane integrity. Mol. Microbiol. 31:1443-1462. [DOI] [PubMed] [Google Scholar]
- 68.Skurnik, M., R. Venho, P. Toivanen, and A. Al-Hendy. 1995. A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol. Microbiol. 17:575-594. [DOI] [PubMed] [Google Scholar]
- 69.Strauch, E., H. Kaspar, C. Schaudinn, C. Damasko, A. Konietzny, P. Dersch, M. Skurnik, and B. Appel. 2003. Analysis of enterocoliticin, a phage tail-like bacteriocin, p. 249-251. In M. Skurnik, K. Granfors, and J. A. Bengoechea (ed.), The genus Yersinia: entering the functional genomic era. Kluwer Academic/Plenum Publishers, New York, NY.
- 70.Strauch, E., H. Kaspar, C. Schaudinn, P. Dersch, K. Madela, C. Gewinner, S. Hertwig, J. Wecke, and B. Appel. 2001. Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl. Environ. Microbiol. 67:5634-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ugalde, J. E., C. Czibener, M. F. Feldman, and R. A. Ugalde. 2000. Identification and characterization of the Brucella abortus phosphoglucomutase gene: role of lipopolysaccharide in virulence and intracellular multiplication. Infect. Immun. 68:5716-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams, G. J., S. D. Breazeale, C. R. Raetz, and J. H. Naismith. 2005. Structure and function of both domains of ArnA, a dual function decarboxylase and a formyltransferase, involved in 4-amino-4-deoxy-L-arabinose biosynthesis. J. Biol. Chem. 280:23000-23008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wren, B. W. 2003. The yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1:55-64. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, L., A. Al-Hendy, P. Toivanen, and M. Skurnik. 1993. Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: similarities to the dTDP-L-rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol. Microbiol. 9:309-321. [DOI] [PubMed] [Google Scholar]