Abstract
Phosphocholine (PCho) is an important substituent of surface structures expressed by a number of bacterial pathogens. Its role in virulence has been investigated in several species, in which it has been shown to play a role in bacterial adhesion to mucosal surfaces, in resistance to antimicrobial peptides, or in sensitivity to complement-mediated killing. The lipopolysaccharide (LPS) structure of Pasteurella multocida strain Pm70, whose genome sequence is known, has recently been determined and does not contain PCho. However, LPS structures from the closely related, virulent P. multocida strains VP161 and X-73 were shown to contain PCho on their terminal galactose sugar residues. To determine if PCho was involved in the virulence of P. multocida, we used subtractive hybridization of the VP161 genome against the Pm70 genome to identify a four-gene locus (designated pcgDABC) which we show is required for the addition of the PCho residues to LPS. The proteins predicted to be encoded by pcgABC showed identity to proteins involved in choline uptake, phosphorylation, and nucleotide sugar activation of PCho. We constructed a P. multocida VP161 pcgC mutant and demonstrated that this strain produces LPS that lacks PCho on the terminal galactose residues. This pcgC mutant displayed reduced in vivo growth in a chicken infection model and was more sensitive to the chicken antimicrobial peptide fowlicidin-1 than the wild-type P. multocida strain.
Pasteurella multocida is a gram-negative coccobacillus that is the causative agent of a wide range of diseases in animals, including fowl cholera, a disease of poultry with worldwide economic importance (6). P. multocida strains can be differentiated into 5 serogroups (A, B, D, E, and F), based on capsule antigens, and into 16 serotypes based on lipopolysaccharide (LPS) structures (7, 14, 26). Fowl cholera is generally caused by the A:1, A:3, or A:4 strains (37). The pathogenesis of fowl cholera is not well understood at the molecular level, but it is likely that susceptible birds are colonized via the trachea and/or lungs, and once bacteria penetrate to the bloodstream, they multiply rapidly in the liver and spleen (3). Toward the end stages of the disease, high levels of bacteremia often occur (4).
Survival of the bacteria in the blood is critical for pathogenesis, and the P. multocida capsule has been identified as the major virulence determinant that allows the bacteria to survive complement-mediated killing and to evade phagocytosis (2, 8). In addition to the role of the capsule, LPS also plays a critical role in virulence, as mutants expressing truncated LPS are highly attenuated (13).
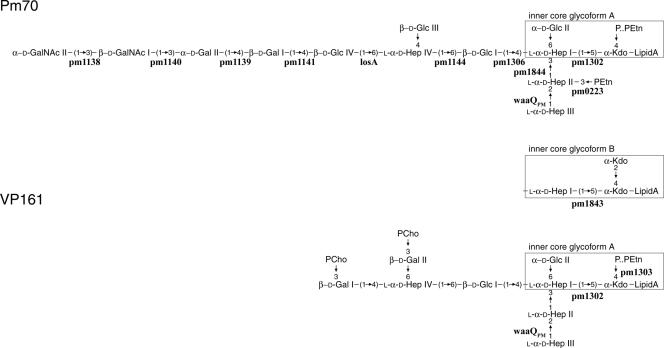
The LPS structure has been determined for three fowl cholera-causing isolates of P. multocida, namely VP161 (34), X-73 (33), and the genome-sequenced strain Pm70 (35). All three strains produce two conserved LPS core glycoforms (Fig. 1, inner core glycoforms A and B), but Pm70 and VP161 LPS structures differ significantly in their oligosaccharide extensions (Fig. 1). Unlike Pm70, strains VP161 and X-73 have phosphocholine (PCho) residues on each of their terminal galactose residues.
FIG. 1.
LPS structures of P. multocida strains Pm70 and VP161. Two LPS inner core forms are observed for both strains; one is as shown (inner core glycoform A), while the other has the boxed residues replaced by the boxed residues shown between the Pm70 and VP161 structures and labeled inner core glycoform B. The Pm70 and VP161 genes that are either known or predicted to encode transferases for each addition are shown below or beside the appropriate linkages. The LPS expressed by P. multocida strain X-73 is identical to the VP161 LPS molecule shown except that a phosphoethanolamine residue is linked to the 6 position of each of the terminal galactose residues. Residues are Glc, glucose; Hep, heptose; Gal, galactose, GlcNAc, N-acetylglucosamine; PEtn, phosphoethanolamine; KDO, 3-deoxy-d-mannooctulosonate; P, phosphate.
PCho-substituted surface components have been identified in a number of mucosal pathogens including Haemophilus influenzae, Neisseria spp., and Streptococcus pneumoniae (9, 21, 30). PCho substitutions have been observed with LPS, fimbriae, capsules, and teichoic and lipoteichoic acids (11, 17, 21, 30), and PCho can contribute to pathogenicity via a number of mechanisms. In H. influenzae infection, PCho mediates adhesion to, and uptake by, 16HBE14 human bronchial epithelial cells via the interaction of PCho with platelet-activating factor receptor (36). Similarly, PCho is critical for the invasion of S. pneumoniae into lung and brain via interactions with platelet-activating factor (9, 27). Furthermore, PCho has been shown to reduce the susceptibility of nontypeable H. influenzae strains to antimicrobial peptides expressed in the human lower respiratory tract (22). In contrast, PCho is a target for C-reactive protein, and the presence and/or position of PCho affects susceptibility to complement-mediated killing in both H. influenzae and commensal N. meningitidis strains (21, 29).
In this study, we identified and characterized a four-gene operon responsible for the addition of PCho to the LPS of P. multocida strains VP161 and X-73. We show that inactivation of one of the identified genes (pcgC) leads to an inability of the bacteria to add PCho to the LPS and the growth of these mutants is reduced in chickens but not in vitro. Furthermore, the lack of phosphocholine results in an increase in susceptibility to the chicken antimicrobial peptide fowlicidin-1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and routine culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown in 2YT broth. P. multocida liquid cultures were grown in brain heart infusion (BHI) broth (Oxoid, Hampshire, England). For P. multocida culture on solid medium, 1.5% agar was added to either BHI, nutrient broth with 0.3% yeast extract, or Columbia agar base (Oxoid) with 5% horse blood. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml (E. coli); tetracycline, 12.5 μg/ml (E. coli) or 8 μg/ml (P. multocida); kanamycin, 50 μg/ml; streptomycin, 100 μg/ml; and spectinomycin, 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Details and genotype | Reference or source |
|---|---|---|
| E. coli | ||
| SM10 λpir | Strain for propagation of pUA826 and its derivatives; thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km | 31 |
| AL562 | E. coli SM10 λpir containing pAL297 | This study |
| DH5α | General laboratory cloning strain; deoR endA1 gyrA96 hsdR17(rK− mK+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argF)U169 φ80lacZΔM15 F− | Bethesda Research Laboratories |
| P. multocida | ||
| VP161 | Serotype A:1 wild-type strain isolated from a chicken in Vietnam | 40 |
| AL435 | VP161 carrying a Tn916 insertion in the pml417 gene; still fully virulent | This study |
| AL571 | PCho−; pcgC mutant of AL435 | This study |
| AL829 | PCho+; AL571 pcgC mutant complemented with pAL293 | This study |
| AL831 | PCho−; AL571 pcgC mutant harboring pAL99 | This study |
| Plasmids | ||
| pAL99 | P. multocida expression vector; cloned genes are expressed from the strong constitutive P. multocida tpiA promoter | 13 |
| pUA826 | Mob+, R6K replicon, Apr Strepr Specr | 1 |
| pAL297 | Internal fragment of pcgC cloned into pUA826 | This study |
| pAL293 | Intact copy of pcgC cloned into the P. multocida expression vector pAL99 | This study |
| pPCR2.1 | E. coli cloning vector; Apr Kanr | Clontech |
DNA manipulations.
Restriction enzymes and Taq polymerase were purchased from either Roche Molecular Biochemicals (Hilden, Germany) or New England Biolabs (Ipswich, MA) and used with the buffers supplied, in accordance with the manufacturers' instructions. PCR-amplified DNA was purified using Real Biotech Corp. (Taipei, Taiwan) PCR DNA fragment extraction kit spin columns. DNA sequence was determined using an ABI dye terminator mixture, and products were separated on an ABI 3730S genetic analyzer. The DNA sequence of the P. multocida strain VP161 pcgDABC locus was determined on both strands (GenBank accession number EU089981), while the corresponding region in P. multocida strain X-73 was determined on a single strand.
Subtractive hybridization.
Subtractive hybridization was performed using a Clontech (Mountain View, CA) PCR-Select bacterial genome subtraction kit, following the manufacturer's instructions. Briefly, 1 μg of VP161 genomic (tester) DNA was digested with Sau3AI and hybridized against 5 μg of Pm70 genomic DNA (driver DNA). PCR products were cloned into pPCR2.1 (Table 1), and E. coli transformants were selected on 2YT agar containing kanamycin and ampicillin. The nucleotide sequences of the inserts from 180 clones were determined, and the functions of genes were predicted based on BLAST analysis.
Construction of a pcgC mutant and complementation.
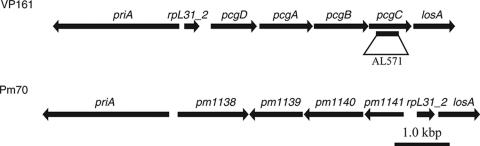
An internal fragment of the pcgC gene (Fig. 2) was amplified from VP161 genomic DNA, using the oligonucleotides BAP3356 and BAP3357 (Table 2). This PCR fragment was digested with SalI and cloned into SalI-digested pUA826 (5), generating pAL297 (Table 1). pUA826 is a λ pir-dependent vector which is unable to replicate in P. multocida and which can be mobilized from the E. coli strain SM10 λ pir. Filter matings were carried out as described previously (12), and transconjugants were selected on nutrient broth with 0.3% yeast extract containing tetracycline, streptomycin, and spectinomycin.
FIG. 2.
Genetic organization of the VP161 region involved in PCho addition to LPS. For comparison, the gene organization of the similar region in Pm70 is shown below. The section of pcgC used for insertional mutagenesis to construct the PCho mutant strain AL571 is shown by the black line below the pcgC gene and labeled AL571.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence | Description and gene |
|---|---|---|
| BAP3353 | CCAATTGGATCCATGGCTCTAAACATAACA | Forward primer for amplification of the complete pcgC gene; contains a BamHI site for cloning |
| BAP3354 | AAATTAGTCGACTTCTAGGGGAATTTTTAAGG | Reverse primer for amplification of the complete pcgC gene; contains a SalI site for cloning |
| BAP3356 | AAAACAGTCGACAAAACCTTATCGAAGCAGGA | Forward primer for amplification of an internal segment of pcgC; contains a SalI site for cloning |
| BAP3357 | TTTTATGTCGACAGCGTCTGATTTTGACCA | Reverse primer for amplification of an internal segment of pcgC; contains a SalI site for cloning |
For complementation of the pcgC mutant, an intact copy of the pcgC gene was amplified from VP161 genomic DNA using the oligonucleotides BAP3353 and BAP3354 (Table 2). The amplified fragments were digested with BamHI and SalI and cloned into BamHI/SalI-digested pAL99, generating pAL293 (Table 1). Expression of the cloned pcgC gene in pAL99 is driven by the strong, constitutively active P. multocida tpiA promoter. The pcgC mutant (AL571) was transformed with pAL293, generating the complemented strain AL829 and with pAL99 generating the control strain AL831 (Table 1).
Analysis of LPS by PAGE and immunoblotting.
Proteinase K-treated whole-cell lysates were analyzed on a Bio-Rad miniprotein gel apparatus, using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (19). LPS was then visualized by carbohydrate silver staining using a SilverSNAP stain kit II (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions. For immunoblotting, samples were transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Billerica, MA) by electroblotting. Membranes were incubated at 20°C for 16 h with a 1/800 dilution of TEPC-15, an antibody specific for phosphocholine (39), washed with Tris-buffered saline containing 5% skim milk, and incubated for 1 h at 37°C with a 1/1,000 dilution of goat anti-mouse immunoglobulin A-horseradish peroxidase conjugate (Sigma-Aldrich, St. Louis, MO). Enzyme-labeled bands were detected by chemiluminescence with an ECL Western blotting detection reagent (Amersham Pharmacia Biotech, Buckinghamshire, England) and visualized on a Fujifilm LAS-3000 (Raytest, Germany).
Competitive growth assays, direct virulence trials, and fowlicidin-1 sensitivity assays.
Competitive growth assays to assess the abilities of the pcgC mutant and the complemented strains to grow in vivo and in vitro were performed as described previously (12). The virulence of the pcgC mutant and the parent strain AL435 were determined by injection of either 40 CFU (of the pcgC mutant) or 60 CFU (of strain AL435) into the breast muscle of groups of seven 10-week-old Hy-Line Brown chickens. Birds were observed closely for signs of fowl cholera and were euthanized when deemed incapable of survival. All animal work was performed with the approval of the relevant Animal Ethics Committees. Sensitivity to the chicken antimicrobial peptide fowlicidin-1 (RVKRVWPLVIRTVIAGYNLYRAIKKK) (41) was determined by direct colony counts as described previously (12). Statistical significance of the differences in sensitivity between the parent and the mutant strains was determined by using an unpaired t test.
Purification of LPS, LPS-OH, and core OS.
Preparation of LPS and LPS-OH for small-scale analyses was done as previously described (12). For large-scale analyses, flask-grown cells (at 37°C, 200 rpm for 16 h in 2 liters of BHI broth following inoculation from chocolate agar plate-grown cells) were killed by the addition of phenol to 2%. LPS was isolated and purified as described previously (34). Briefly, cells (∼17.3 g [wet weight]) were freeze-dried, yielding ∼3.0 g, and washed with organic solvents (1× ethanol, 2× acetone, 2× light petroleum ether) to remove lipids and other lipophilic components. Washed cells (2.51 g) were extracted by the hot phenol/water method and treated with DNase and RNase at 37°C for 4 h and then proteinase K at 37°C for 4 h. Small peptides were removed by dialysis. After freeze-drying, the retentate was made up to a 2% solution in water and centrifuged at 8,000 × g for 15 min (yielding a pellet of ∼24 mg), followed by further centrifugation of the supernatant at 100,000 × g for 5 h. The pellet containing purified LPS was redissolved and freeze-dried, yielding ∼1 mg. Pellet material (1 mg) was O deacylated. The core oligosaccharide (OS) was isolated by separately treating the 8,000 × g pellet material (∼20 mg) and the LPS (∼1 mg) with 1% acetic acid (10 mg/ml; 100°C; 1.5 h), with subsequent removal of the insoluble lipid A by centrifugation (5,000 × g). The lyophilized OS sample from the 8,000 × g pellet was subsequently purified further on a Bio-Gel P-2 column.
Analytical methods, mass spectrometry, and NMR spectroscopy.
Sugars were identified as their alditol acetate derivatives, and linkage analysis was determined following methylation analysis by gas-liquid chromatography-mass spectrometry (GLC-MS) as described previously (34). Combined capillary electrophoresis-electrospray mass spectrometry (CE-ES-MS) analysis and nuclear magnetic resonance (NMR) experiments were performed as previously described (34).
RESULTS
Identification of genes involved in PCho addition to LPS.
The P. multocida strains VP161 and X-73 both express LPS with terminal PCho residues (33, 34). The genome sequence of P. multocida strain Pm70 has been determined (23), but this strain expresses LPS without PCho (35), and its genome does not contain any genes with a similarity to PCho transferases. Therefore, to identify the genes involved in PCho addition to P. multocida strain VP161 LPS, we used a subtractive hybridization of VP161 genomic DNA against Pm70 genomic DNA. We sequenced 180 putative VP161-specific DNA fragments, of which more than 60 showed either no or very limited (<15%) identity to Pm70 genes (data not shown). One of these fragments encoded two incomplete open reading frames whose predicted products were similar to the choline kinase (LicA) and the choline permease (LicB) from H. influenzae. The DNA surrounding this gene fragment was amplified using inverse PCR (25), and the nucleotide sequence was determined. Four open reading frames were identified and designated phosphocholine addition to galactose (pcg)D, pcgA, pcgB, and pcgC. A comparison of the VP161 nucleotide sequence of this region with the corresponding sequence from Pm70 showed that the PCho operon was located between the genes rpL31_2 and losA, which in Pm70 consists of a short noncoding sequence (Fig. 2). PCR analysis of the genomic region upstream of the rpL31_2 gene revealed that genes pm1138 through to pm1141 were absent (Fig. 2). Moreover, we were unable to individually amplify genes pm1138, pm1139, and pm1141 from VP161 by using low annealing temperatures, indicating that they are not located elsewhere on the genome (data not shown). The absence of these genes from strain VP161 is not unexpected as the glycosyltransferases encoded by pm1138 through pm1141 are predicted to be required for the assembly of the outer core region of the Pm70 LPS molecule but are not required for assembly of the VP161 LPS structure (Fig. 1).
All of the proteins predicted to be encoded by pcgA, pcgB, and pcgC were highly similar to proteins known to be involved in phosphocholine metabolism. PcgA was 41% identical to the choline kinase (LicA) from H. influenzae, PcgB displayed 39% identity to the LicB (putative choline permease) protein from H. influenzae, and PcgC displayed 46% identity to the CTP:phosphocholine cytidylyltransferase (LicC) from H. influenzae. PcgD showed very limited similarity to characterized bacterial proteins but was 29% identical to the human protein encoded by the fukutin gene, which is implicated in Fukuyama-type congenital muscular dystrophy (18). The last gene in the H. influenzae lic operon (LicD) encodes a phosphocholine transferase required for the transfer of the activated phosphocholine to the nascent LPS molecule (21). Although the predicted protein encoded by pcgD did not show similarity to LicD, using normal BLAST (expected [E] value of >10) analysis, it showed significant similarity to LicD after 3 PSI-BLAST iterations (E = 8 × 10−11). Furthermore, pcgD encoded a section with identity to part of the LicD domain (pfam04991; E = 0.002). Therefore, we predict that the pcgD gene encodes the PCho transferase.
Construction and complementation of a VP161 pcgC mutant.
To confirm that the pcg gene cluster was required for PCho addition to LPS, we constructed a pcgC mutant by insertional mutagenesis using the mobilizable P. multocida suicide plasmid pUA826. An internal fragment of pcgC (Fig. 2) was cloned into pUA826, generating pAL297, and this plasmid was mobilized into the VP161 strain AL435 (Tetr) by conjugation. Transconjugants were selected on tetracycline, streptomycin, and spectinomycin, and one colony was designated AL571 (Table 1). Insertion of pAL297 into pcgC was confirmed by PCR (data not shown). The mutant was then complemented with pAL293, which expresses intact pcgC, to generate AL829 (Table 1). As a control, the mutant was also transformed with the empty expression vector pAL99 to generate AL831 (Table 1).
Structure of the LPS produced by the P. multocida pcgC mutant.
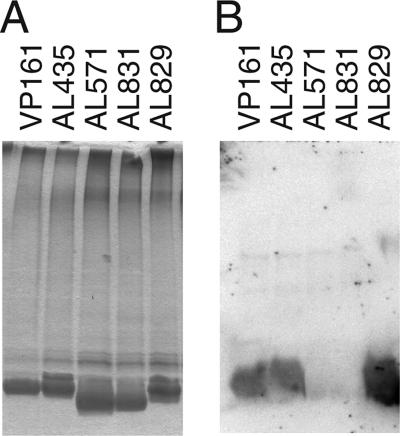
The LPS structures produced by the pcgC mutant, the parent strain AL435, and the complemented strain AL829 were assessed by SDS-PAGE, followed by silver staining and Western immunoblotting. The pcgC mutant (AL571) and the pcgC mutant harboring the vector pAL99 (AL831) expressed truncated LPS (Fig. 3A, lanes 3 and 4) and did not react with the anti-PCho TEPC-15 antibody (Fig. 3B, lanes 3 and 4). Both properties were restored in the complemented strain AL829 (Fig. 3A and B, lanes 5). O-deacylated LPS (LPS-OH) and core OS were prepared from flask-grown cells and analyzed by CE-ES-MS. Compositions consistent with the absence of PCho were inferred from the MS analysis of both the LPS-OH and OS, but a composition containing PCho was inferred from the mass spectra of both the wild-type strain and the complemented mutant (Table 3). MS analysis also revealed compositions consistent with lesser amounts of hexose in the pcgC mutant than in the parent and complemented strains, consistent with the notion that one of the terminal galactose residues was inefficiently attached in the absence of the PCho residue. 1H NMR analyses of the core OS from the mutant strains were performed which confirmed and extended the MS inferences. The characteristic signal at 3.3 ppm from the nine protons of the choline moiety was absent in the 1H NMR spectrum from the pcgC mutant. Additionally, the chemical shifts of the galactose residue were consistent with it now being a terminal residue, with upfield shifts for the H-1, H-2, H-3, and H-4 resonances compared to that of the wild-type strain (data not shown). Nuclear Overhauser effect connectivities suggested that the galactose residue linked to the 4 position of the outer core heptose residue was consistently present but there was only a minor amount of the second terminal galactose residue (linked to the 6 position of the outer core heptose), consistent with the MS analysis. Finally, methylation analysis confirmed the presence of terminally located galactose, and the identification of a 4-linked mono-substituted heptose residue and a 4,6-linked disubstituted heptose residue in an approximate 2:1 ratio confirmed the NMR inference that the galactose at the 6 position of the distal heptose residue is mainly absent in this mutant background (data not shown). These data clearly indicate that intact pcgC is required for the addition of PCho to P. multocida strain VP161 LPS. Furthermore, the lack of galactose at the 6 position of the distal heptose in most LPS glycoforms expressed by the pcgC mutant suggests that the galactose transferase required for this addition requires PCho on the galactose linked to the heptose for efficient acceptor recognition.
FIG. 3.
Phenotypes of the pcgC mutant and the complemented strains as analyzed by silver-stained PAGE (A) and Western blotting probed with TEPC-15 (anti-PCho) antibody (B). Equal amounts of proteinase K-treated whole-cell lysates (panel A) or untreated whole-cell lysates (panel B) of the following strains were run as follows: lane 1, wild-type strain VP161; lane 2, parent strain AL435; lane 3, pcgC mutant (AL571); lane 4, pcgC mutant harboring vector pAL99 (AL831); lane 5, complemented mutant (AL829).
TABLE 3.
O-deacylated LPS (LPS-OH) from P. multocida VP161 wild-type and mutant strainsa
| Strain | CE-ES-MS data
|
Proposed composition | |||||
|---|---|---|---|---|---|---|---|
| [M-4H]4− | [M-3H]3− | [M-2H]2− | Observed molecular ion | Calculated molecular ion | Relative intensity | ||
| AL435 | 743.8 | 991.9 | 1,488.6 | 2,979.1 | 2,977.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| 749.4 | 999.2 | 1,499.4 | 3,001.0 | 2,999.5 | 0.7 | 2PCho, 4Hex, 4Hep, Kdo-P, lipid A-OH | |
| 780.2 | 1,040.5 | 3,124.6 | 3,122.6 | 0.3 | 2PCho, 4Hex, 4Hep, Kdo-P-PEtn, lipid A-OH | ||
| AL571 | 620.7 | 828.0 | 1,242.0 | 2,486.5 | 2,485.3 | 1.0 | 2Hex, 4Hep, 2Kdo, lipid A-OH |
| 626.4 | 835.2 | 1,253.0 | 2,508.5 | 2,507.3 | 1.0 | 3Hex, 4Hep, Kdo-P, lipid A-OH | |
| 657.0 | 876.1 | 1,314.5 | 2,631.3 | 2,630.3 | 0.5 | 3Hex, 4Hep, Kdo-P-PEtn, lipid A-OH | |
| 666.0 | 889.2 | 2,669.3 | 2,669.4 | 0.2 | 4Hex, 4Hep, Kdo-P, lipid A-OH | ||
| AL829 | 743.7 | 992.1 | 1,488.1 | 2,978.6 | 2,977.6 | 1.0 | 2PCho, 3Hex, 4Hep, 2Kdo, lipid A-OH |
| 749.2 | 999.3 | 1,499.3 | 3,000.7 | 2,999.5 | 0.8 | 2PCho, 4Hex, 4Hep, Kdo-P, lipid A-OH | |
| 780.0 | 1,040.2 | 1,560.8 | 3,123.6 | 3,122.6 | 0.3 | 2PCho, 4Hex, 4Hep, Kdo-P-PEtn, lipid A-OH | |
| AL831 | 620.7 | 827.8 | 1,242.3 | 2,486.6 | 2,485.3 | 0.7 | 2Hex, 4Hep, 2Kdo, lipid A-OH |
| 626.3 | 835.1 | 1,253.0 | 2,508.3 | 2,507.3 | 0.7 | 3Hex, 4Hep, Kdo-P, lipid A-OH | |
| 656.9 | 876.2 | 1,314.9 | 2,631.7 | 2,630.3 | 1.0 | 3Hex, 4Hep, Kdo-P-PEtn, lipid A-OH | |
Negative-ion CE-ES-MS data and proposed composition of O-deacylated LPS (LPS-OH) from the P. multocida VP161 wild-type and mutant strains are shown. Average molecular mass units were used for the calculation of molecular weight based on the proposed composition, as follows: lipid A, 952.00; Hex [hexose], 162.15; Hep [heptose], 192.17; Kdo [3-deoxy-d-mannooctulosonate], 220.18; Kdo-P [3-deoxy-d-mannooctulosonate phosphate], 300.13; PEtn [phosphoethanolamine], 123.05; PCho, 165.05.
Virulence of the pcgC mutant.
The ability of the pcgC mutant and of the complemented mutant to grow in vivo was assessed by using competitive growth assays (Table 4). The pcgC mutant lacking PCho, AL571, was reduced in its ability to grow in chickens, following either intramuscular (i.m.) inoculation (relative competitive index [rCI] = 0.1 ± 0.1) or transmucosal intratracheal (i.t.) inoculation (rCI = 0.003 ± 0.008). The complemented mutant (AL829) showed a significant increase in rCI (rCI = 0.5 ± 0.1; P = 0.02) compared with those of both the pcgC mutant and the pcgC mutant containing empty vector. Direct challenge assays and subsequent PCR analyses of recovered bacteria showed that the PCho mutant could still cause fowl cholera in all chickens injected with AL571 (40 CFU). However, the time to establish the disease was much longer than that seen following direct challenge with the parent strain. Chickens injected with the parent strain AL435 (60 CFU) all showed clinical signs of fowl cholera within 24 h, and all birds had to be euthanized before 36 h postinfection (average time, 28 ± 6 h). In contrast, birds injected with the PCho mutant showed no signs of fowl cholera until at least 50 h postinfection, and six of the seven birds in the group showed only initial signs when the experiment was terminated at 66 h. These data indicate that the pcgC gene is involved in but not essential for the virulence of P. multocida in chickens.
TABLE 4.
Competitive growth assays of the pcgC mutant and complemented strains
| Strain | Description | rCI ± SDa | Infection route |
|---|---|---|---|
| AL571 | PCho−; pcgC mutant | 0.1 ± 0.1 | i.m. |
| AL571 | PCho−; pcgC mutant | 0.003 ± 0.008 | i.t. |
| AL829 | PCho+; pcgC mutant complemented with pcgC | 0.5 ± 0.1 | i.m. |
| AL831 | PCho−; pcgC mutant harboring pAL99 | 0.01 ± 0.01 | i.m. |
rCI values were calculated for individual animals by dividing the in vivo competitive index (CI) by the in vitro CI, and the values shown are the average rCI values for three replicate animals ± 1 standard deviation (SD). Each CI value was determined by dividing the output mutant/wild-type ratio by the input mutant/wild-type ratio.
Susceptibility of the pcgC mutant to fowlicidin-1.
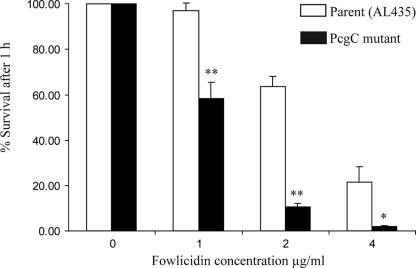
The susceptibility of the pcgC mutant to the antimicrobial peptide fowlicidin-1 was determined by assessing the survival of wild-type and mutant strains in the presence of various concentrations of fowlicidin-1 for 1 h at 37°C (Fig. 4). The pcgC mutant was significantly more susceptible to killing in the presence of fowlicidin-1 than the parent strain (AL435), with 12% survival for the mutant in the presence of 2 μg/ml fowlicidin-1 after 1 h, compared with 68% survival for the parent strain (P < 0.0001; unpaired t test). Therefore, the loss of the terminal PCho substituents of LPS results in increased sensitivity to fowlicidin-1.
FIG. 4.
Sensitivity of P. multocida strains to the action of fowlicidin-1. Bacterial survival was determined by direct colony counts after incubation with various concentrations of synthetic fowlicidin-1 for 1 h at 37°C. Numbers are the mean percentages of survival for three replicates and error bars are ± 1 standard deviation. The differences between the mean percent survival values for parent and mutant strains were statistically significant at all concentrations tested (**, P < 0.001; *, P = 0.01).
DISCUSSION
PCho is a common constituent of surface structures of mucosal and respiratory bacterial pathogens such as Neisseria and Haemophilus spp., where it plays a clear role in virulence (9, 22, 36). Terminal PCho residues have been identified on the LPS structures from the P. multocida serotype A:1 fowl cholera-causing strains X-73 and VP161 but not on the LPS of the genome-sequenced strain Pm70 (serotype F:3) (33-35). To identify the unique regions on the VP161 genome, including the genes encoding the proteins required for phosphocholine biosynthesis, we performed subtractive hybridization of the VP161 genomic DNA against Pm70 genomic DNA. A locus was identified consisting of four genes, three of which (pcgA, pcgB, and pcgC) encoded proteins with significant similarity to the H. influenzae proteins LicA, LicB, and LicC, respectively, required for the import of choline (LicB), phosphorylation of choline to phosphocholine (LicA), and activation of phosphocholine by the addition of CTP (LicC). The first gene in the operon, pcgD, encoded a protein which contained a partial LicD domain and showed identity to LicD, following a PSI-BLAST search, and we predict that this gene encodes the PCho transferase that adds the activated PCho to the galactose residues on the VP161 LPS molecule. Interestingly, the pcgD gene product has only limited similarity with any characterized bacterial proteins but shares identity with genes encoding the human fukutin proteins, which are predicted to be involved in glycosylation of neuronal α-dystroglycan and are implicated in Fukuyama congenital muscular dystrophy (18, 28). It is tempting to speculate that the human fukutin proteins may also have a role in PCho addition to α-dystroglycan, although it is possible that the protein domains which show identity are involved solely in sugar-acceptor binding.
In order to demonstrate involvement of the pcg operon in the PCho addition to P. multocida LPS, we attempted an insertional inactivation of the putative phosphocholine transferase gene, pcgD, as it was the first gene in the VP161 PCho locus, but we were unsuccessful (data not shown). Subsequently, we generated a mutant pcgC strain, which we predicted carries the CTP:phosphocholine cytidylyltransferase required to activate the phosphocholine residue prior to transfer to the LPS structure. Structural analysis of the LPS expressed by the pcgC mutant, AL571, and the lack of immunoreactivity with the TEPC-15 PCho-specific antibody demonstrated that this mutant was unable to add PCho to LPS. Moreover, complementation with an intact pcgC gene restored the ability of the bacteria to add PCho to LPS, thus confirming that pcgC is required for the PCho addition to LPS (Table 3 and Fig. 3).
To assess the mutant and complemented strains for their abilities to grow in vivo, competitive growth assays were performed using the i.m. route of infection that showed that the mutant displayed significantly reduced growth compared to that of VP161. The in vivo growth of the mutant was restored to near wild-type levels by in trans complementation with the intact pcgC (Table 4). In H. influenzae, PCho is important for adhesion and invasion of epithelial cells (36), so we anticipated that the PCho mutant might exhibit a more significant in vivo growth defect following the mucosal route of infection. However, competitive growth assays showed there were no statistically significant differences between the survival of the PCho mutant after i.m. compared with i.t. inoculation.
Competitive growth assays require both the mutant and the wild-type strains to grow within the same environment and do not assess the ability of a strain to independently colonize the host and establish disease. To determine if the PCho mutant was capable of causing fowl cholera, direct virulence trials showed that while all birds injected with the PCho mutant showed symptoms of fowl cholera, the disease progressed at a much slower rate than that caused by the parent strain. Previously we have shown that inactivation of the waaQPM gene in P. multocida results in a strain that expresses severely truncated LPS and is unable to cause disease in chickens (13). The results in this study indicate that the PCho mutant is not as attenuated as the waaQPM mutant, indicating that the addition of PCho onto the distal galactose residues in the VP161 LPS structure plays a role, but not an essential one, in the pathogenesis of fowl cholera.
In H. influenzae, PCho is found on LPS and has been shown to be important for colonization and persistence at the mucosal surface (15, 38). Conversely, PCho decoration of LPS has also been shown to increase susceptibility to complement-mediated killing in serum by the direct binding of C-reactive protein to PCho (9, 21, 38). Thus, for H. influenzae, the expression of PCho on LPS gives a selective advantage at the mucosal surface during initial colonization and invasion but is disadvantageous during systemic growth (16). For P. multocida, it is unlikely that PCho plays a significant role in susceptibility to complement-mediated killing as P. multocida strains are highly resistant to killing in serum, and this property is mediated primarily by the capsule (8). Furthermore, a waaQPM mutant which expresses no full-length LPS was no more susceptible to complement-mediated killing than the parent strain (13).
The presence of PCho residues on the LPS structure has been shown to decrease the sensitivity of H. influenzae to the human upper respiratory tract peptide cathelicidin LL-37/hCAP18 (22). To test whether PCho is important for the resistance of P. multocida to host antimicrobial peptides, we synthesized the chicken cathelicidin fowlicidin-1. This peptide is expressed in a wide range of chicken tissues including gizzard, small and large intestines, liver, kidney, trachea, and bone marrow (20) Furthermore, fowlicidin-1 has high antimicrobial activity against both gram-positive and gram-negative bacteria (41). We analyzed the sensitivities of the P. multocida parent strain and the PCho mutant to the antibacterial effects of fowlicidin-1 (Fig. 4). The PCho mutant was significantly more susceptible to fowlicidin-1 than the parent strain (Fig. 4). Thus, the presence of terminal PCho residues on the LPS mediates significant resistance to the fowlicidin-1 antimicrobial peptide, but our results do not preclude a role for other residues. It has been proposed that cationic antimicrobial peptides must interact with the negatively charged lipid A before they can cause bacterial lysis (10, 24, 32, 42). Thus, it is likely that the LPS oligosaccharide sugars play some role in shielding lipid A from the cationic peptide, thereby increasing bacterial resistance. To this end, the positively charged PCho clearly plays an important role.
In conclusion, we have identified a four-gene locus in P. multocida strain VP161 which is necessary for the PCho addition to LPS. The presence of PCho on the LPS is required for full in vivo growth of P. multocida during the infection of chickens, and the loss of the PCho residues increases the sensitivity of the bacteria to the chicken antimicrobial fowlicidin-1. However, clearly, other LPS components play a pivotal role in in vivo growth in chickens, and we are currently exploring the roles these other LPS substituents play in virulence.
Acknowledgments
We thank Marietta John, Perry Fleming, and Jacek Stupak for excellent technical assistance.
This work was funded in part by grants from the Australian Research Council, the Australian Poultry CRC, and the NRC Women in Engineering and Sciences Scholarship Program.
Footnotes
Published ahead of print on 17 August 2007.
REFERENCES
- 1.Bosch, M., R. Tarrago, M. E. Garrido, S. Campoy, A. R. F. de Henestrosa, A. M. P. de Rozas, I. Badiola, and J. Barbe. 2001. Expression of the Pasteurella multocida ompH gene is negatively regulated by the Fur protein. FEMS Microbiol. Lett. 203:35-40. [DOI] [PubMed] [Google Scholar]
- 2.Boyce, J. D., and B. Adler. 2000. The capsule is a virulence determinant in the pathogenesis of Pasteurella multocida M1404 (B:2). Infect. Immun. 68:3463-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, J. D., R. Y. C. Lo, I. Wilkie, and B. Adler. 2004. Pasteurella and Mannheimia, p. 385-396. In C. Gyles, C. Thoen, J. Prescott, and G. Songer (ed.), Pathogenesis of bacterial infections of animals. Blackwell Publishing, Ames, IA.
- 4.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 70:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardenas, M., A. R. F. de Henestrosa, S. Campoy, A. M. P. de Rozas, J. Barbe, I. Badiola, and M. Llagostera. 2001. Virulence of Pasteurella multocida recA mutants. Vet. Microbiol. 80:53-61. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter, T. E., K. P. Snipes, D. Wallis, and R. McCapes. 1988. Epidemiology and financial impact of fowl cholera in turkeys: a retrospective analysis. Avian Dis. 32:16-23. [PubMed] [Google Scholar]
- 7.Carter, G. R. 1967. Pasteurellosis: Pasteurella multocida and Pasteurella hemolytica. Adv. Vet. Sci. 11:321-379. [PubMed] [Google Scholar]
- 8.Chung, J. Y., I. Wilkie, J. D. Boyce, K. M. Townsend, A. J. Frost, M. Ghoddusi, and B. Adler. 2001. Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect. Immun. 69:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cundell, D. R., N. P. Gerard, C. Gerard, I. Idanpaan-Heikkila, and E. I. Tuomanen. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435-438. [DOI] [PubMed] [Google Scholar]
- 10.Daugelavicius, R., E. Bakiene, and D. H. Bamford. 2000. Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob. Agents Chemother. 44:2969-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, W., T. Behr, R. Hartmann, J. Peter-Katalinic, and H. Egge. 1993. Teichoic acid and lipoteichoic acid of Streptococcus pneumoniae possess identical chain structures. A reinvestigation of teichoid acid (C polysaccharide). Eur. J. Biochem. 215:851-857. [DOI] [PubMed] [Google Scholar]
- 12.Harper, M., J. D. Boyce, A. D. Cox, F. St Michael, I. Wilkie, P. Blackall, and B. Adler. 2007. Pasteurella multocida expresses two lipopolysaccharide glycoforms simultaneously, both in vitro and in vivo, but only a single form is required for virulence: identification of two acceptor specific heptosyl I transferases. Infect. Immun. 75:3885-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper, M., A. D. Cox, F. St Michael, I. W. Wilkie, J. D. Boyce, and B. Adler. 2004. A heptosyltransferase mutant of Pasteurella multocida produces a truncated lipopolysaccharide structure and is attenuated in virulence. Infect. Immun. 72:3436-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heddleston, K. L., J. E. Gallagher, and P. A. Rebers. 1972. Fowl cholera: gel diffusion precipitin test for serotyping Pasteurella multocida from avian species. Avian Dis. 16:925-936. [PubMed] [Google Scholar]
- 15.Hong, W., K. Mason, J. Jurcisek, L. Novotny, L. O. Bakaletz, and W. E. Swords. 2007. Phosphorylcholine decreases early inflammation and promotes the establishment of stable biofilm communities of nontypeable Haemophilus influenzae strain 86-028NP in a chinchilla model of otitis media. Infect. Immun. 75:958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphries, H. E., and N. J. High. 2002. The role of licA phase variation in the pathogenesis of invasive disease by Haemophilus influenzae type b. FEMS Immunol. Med. Microbiol. 34:221-230. [DOI] [PubMed] [Google Scholar]
- 17.Karlsson, C., P. E. Jansson, and U. B. Sorensen. 1998. The chemical structures of the capsular polysaccharides from Streptococcus pneumoniae types 32F and 32A. Eur. J. Biochem. 255:296-302. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, K., J. Sasaki, E. Kondo-Iida, Y. Fukuda, M. Kinoshita, Y. Sunada, Y. Nakamura, and T. Toda. 2001. Structural organization, complete genomic sequences and mutational analyses of the Fukuyama-type congenital muscular dystrophy gene, fukutin. FEBS Lett. 489:192-196. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Lynn, D. J., R. Higgs, S. Gaines, J. Tierney, T. James, A. T. Lloyd, M. A. Fares, G. Mulcahy, and C. O'Farrelly. 2004. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics 56:170-177. [DOI] [PubMed] [Google Scholar]
- 21.Lysenko, E., J. C. Richards, A. D. Cox, A. Stewart, A. Martin, M. Kapoor, and J. N. Weiser. 2000. The position of phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae affects binding and sensitivity to C-reactive protein-mediated killing. Mol. Microbiol. 35:234-245. [DOI] [PubMed] [Google Scholar]
- 22.Lysenko, E. S., J. Gould, R. Bals, J. M. Wilson, and J. N. Weiser. 2000. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect. Immun. 68:1664-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neville, F., D. Gidalevitz, G. Kale, and A. Nelson. 2006. Electrochemical screening of anti-microbial peptide LL-37 interaction with phospholipids. Bioelectrochemistry 70:205-213. [DOI] [PubMed] [Google Scholar]
- 25.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rimler, R. B., and K. R. Rhoades. 1987. Serogroup F, a new capsule serogroup of Pasteurella multocida. J. Clin. Microbiol. 25:615-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ring, A., J. N. Weiser, and E. I. Tuomanen. 1998. Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J. Clin. Investig. 102:347-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito, Y., T. Yamamoto, M. Mizuguchi, M. Kobayashi, K. Saito, K. Ohno, and M. Osawa. 2006. Altered glycosylation of alpha-dystroglycan in neurons of Fukuyama congenital muscular dystrophy brains. Brain Res. 1075:223-228. [DOI] [PubMed] [Google Scholar]
- 29.Serino, L., and M. Virji. 2002. Genetic and functional analysis of the phosphorylcholine moiety of commensal Neisseria lipopolysaccharide. Mol. Microbiol. 43:437-448. [DOI] [PubMed] [Google Scholar]
- 30.Serino, L., and M. Virji. 2000. Phosphorylcholine decoration of lipopolysaccharide differentiates commensal Neisseriae from pathogenic strains: identification of licA-type genes in commensal Neisseriae. Mol. Microbiol. 35:1550-1559. [DOI] [PubMed] [Google Scholar]
- 31.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 32.Srimal, S., N. Surolia, S. Balasubramanian, and A. Surolia. 1996. Titration calorimetric studies to elucidate the specificity of the interactions of polymyxin B with lipopolysaccharides and lipid A. Biochem. J. 315:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.St. Michael, F., J. Li, and A. D. Cox. 2005. Structural analysis of the core oligosaccharide from Pasteurella multocida strain X73. Carbohydr. Res. 340:1253-1257. [DOI] [PubMed] [Google Scholar]
- 34.St. Michael, F., J. Li, E. Vinogradov, S. Larocque, M. Harper, and A. D. Cox. 2005. Structural analysis of the lipopolysaccharide of Pasteurella multocida strain VP161: identification of both Kdo-P and Kdo-Kdo species in the lipopolysaccharide. Carbohydr. Res. 340:59-68. [DOI] [PubMed] [Google Scholar]
- 35.St. Michael, F., E. Vinogradov, J. Li, and A. D. Cox. 2005. Structural analysis of the lipopolysaccharide from Pasteurella multocida genome strain Pm70 and identification of the putative lipopolysaccharide glycosyltransferases. Glycobiology 15:323-333. [DOI] [PubMed] [Google Scholar]
- 36.Swords, W. E., B. A. Buscher, K. Ver Steeg Ii, A. Preston, W. A. Nichols, J. N. Weiser, B. W. Gibson, and M. A. Apicella. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13-27. [DOI] [PubMed] [Google Scholar]
- 37.Waltman, W. D., and A. M. Horne. 1993. Characteristics of fowl cholera diagnosed in Georgia, 1989-91. Avian Dis. 37:616-621. [PubMed] [Google Scholar]
- 38.Weiser, J. N., N. Pan, K. L. McGowan, D. Musher, A. Martin, and J. Richards. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J Exp. Med. 187:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiser, J. N., M. Shchepetov, and S. T. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65:943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkie, I. W., S. E. Grimes, D. O'Boyle, and A. J. Frost. 2000. The virulence and protective efficacy for chickens of Pasteurella multocida administered by different routes. Vet. Microbiol. 72:57-68. [DOI] [PubMed] [Google Scholar]
- 41.Xiao, Y., Y. Cai, Y. R. Bommineni, S. C. Fernando, O. Prakash, S. E. Gilliland, and G. Zhang. 2006. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 281:2858-2867. [DOI] [PubMed] [Google Scholar]
- 42.Yin, N., R. L. Marshall, S. Matheson, and P. B. Savage. 2003. Synthesis of lipid A derivatives and their interactions with polymyxin B and polymyxin B nonapeptide. J. Am. Chem. Soc. 125:2426-2435. [DOI] [PubMed] [Google Scholar]