Abstract
Clostridium difficile binary toxin (CDT) is an actin-specific ADP-ribosyltransferase that is produced by various C. difficile isolates, including the “hypervirulent” NAP1/027 epidemic strains. In contrast to the two major toxins from C. difficile, toxin A and toxin B, little is known about the role of CDT in virulence or how C. difficile regulates its production. In this study we have shown that in addition to the cdtA and cdtB toxin structural genes, a functional cdt locus contains a third gene, here designated cdtR, which is predicted to encode a response regulator. By introducing functional binary toxin genes into cdtR+ and cdtR-negative strains of C. difficile, it was established that the CdtR protein was required for optimal expression of binary toxin. Significantly increased expression of functional binary toxin was observed in the presence of a functional cdtR gene; an internal deletion within cdtR resulted in a reduction in binary toxin production to basal levels. Strains that did not carry intact cdtAB genes or cdtAB pseudogenes also did not have cdtR, with the entire cdt locus, or CdtLoc, being replaced by a conserved 68-bp sequence. These studies have shown for the first time that binary toxin production is subject to strict regulatory control by the response regulator CdtR, which is a member of the LytTR family of response regulators and is related to the AgrA protein from Staphylococcus aureus.
Clostridium difficile is an anaerobic, gram-positive spore-forming bacterium that is recognized as the causative agent of a broad spectrum of intestinal diseases ranging from mild self-limiting diarrhea to more serious and potentially life threatening manifestations such as pseudomembranous colitis and toxic megacolon (4). Collectively, these diseases are referred to as C. difficile-associated disease (CDAD). The onset of CDAD is typically associated with the use of antibiotics or other therapeutics such as cytotoxic anticancer drugs, which disrupt the normal intestinal microflora (50). Once this colonization resistance (51) is removed, it is proposed that ingested C. difficile spores germinate, colonize the gastrointestinal tract, and elaborate toxins, which results in an acute inflammatory response and severe damage to the intestinal epithelium.
C. difficile is a significant and increasing source of morbidity and mortality and is a very significant financial burden in hospitals throughout the developed world (9, 22, 41, 59). More recently, a new, highly virulent group of strains, known as NAP1/027, has emerged. These hypervirulent strains are reported to produce significantly more toxin than other strains and are associated with more severe disease and high mortality rates (59). The hypervirulent strains are highly resistant to fluoroquinolones and are responsible for epidemics seen in Canada, the United States, and the United Kingdom and several other European countries (21, 25, 28, 48).
The major virulence factors of C. difficile are two members of the large clostridial cytotoxin family, toxin A and toxin B, which are encoded by the genes tcdA and tcdB, respectively, and are located within a five-gene locus known as the PaLoc (6). Both toxins A and B are potent monoglucosyltransferases that have been shown to target small GTP-binding proteins, including Rho, Rac, and Cdc42, which are involved in the regulation and formation of the actin cytoskeleton in eukaryotic cells (18, 19).
In addition to producing toxins A and B, some C. difficile strains, including the NAP1/027 epidemic strains, produce a third toxin, known as C. difficile binary toxin (CDT) (12, 38, 53). This toxin is an actin-specific ADP-ribosyltransferase that consists of two subunits, namely, CDTa, which is the enzymatic component, and CDTb, the binding component (3, 39). It has a high degree of amino acid sequence similarity to iota toxin from Clostridium perfringens type E strains and Clostridium spiroforme toxin, the subunits of which are reportedly interchangeable (38). CDTb binds to an as-yet-unidentified cellular receptor, leading to the internalization of CDTa into the cytosol, where it catalyzes the ADP-ribosylation of monomeric actin and the resultant disruption of the actin cytoskeleton (1, 47). Since most human C. difficile strains do not produce functional CDT, its role in the pathogenesis of CDAD is yet to be determined. Recent studies have shown that CDT production is much more prevalent in strains isolated from animals, with the CDT structural genes being found in approximately 23% to 100% of these isolates (43). However, the significance of this finding for C. difficile-mediated disease in animals is also unknown.
CDT is encoded by two genes, cdtA and cdtB, which are transcriptionally linked and are located on the C. difficile chromosome at a locus separate from the PaLoc (38). In contrast to the situation with toxin A and toxin B, very little research has been carried out to examine how the expression of this potentially important virulence factor is regulated. Here we report the identification of a functional LytTR family response regulator, which we have named CdtR, and we show that it is able to activate CDT production in C. difficile. Furthermore, our results reveal that the cdtR and the cdtAB genes are genetically linked as part of the CDT locus (CdtLoc). Finally, in CDT-negative isolates we have identified a novel 68-bp nucleotide sequence that is absent in isolates carrying the CdtLoc.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The characteristics and origins of all recombinant strains and plasmids are shown in Table 1. All bacteriological culture media were obtained from Oxoid. For mating procedures and extraction of plasmid and genomic DNAs, C. difficile strains were cultured in BHIS broth (49) supplemented with 0.1% l-cysteine HCl and 0.5% glucose or on BHIS agar with the additional supplement of 0.09% FeSO4. Cycloserine (250 μg · ml−1), cefoxitin (8 μg · ml−1), thiamphenicol (10 μg · ml−1), or erythromycin (10 μg · ml−1) (all from Sigma-Aldrich) were used for selection of recombinant strains, as required. C. difficile cultures were grown in an atmosphere of 10% H2, 10% CO2, and 80% N2 at 37°C in a Coy anaerobic chamber. Escherichia coli strains were derivatives of either DH5α, Top10, or HB101(pVS520) and were cultured aerobically at 37°C in 2× YT agar or broth (30). Recombinant E. coli cultures were supplemented with chloramphenicol (25 μg · ml−1), erythromycin (400 μg · ml−1), or tetracycline (10 μg · ml−1), as required.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics and/or origina | Source or reference |
|---|---|---|
| Strains | ||
| C. difficile | ||
| 630 | Clinical isolate, Tcr Emr, Switzerland | 13 |
| CD196 | Binary toxin-producing reference strain, France | 39 |
| VPI 10463 | Toxinotype 0 reference strain, United States | 56 |
| L253 | Toxigenic isolate | 14 |
| CD37 | Nontoxigenic isolate, United States | 49 |
| JIR8094 | 630 Ems; has a spontaneous deletion of an erm(B) gene from Tn5398 | 35 |
| KZ1623 | Nontoxigenic isolate, Japan | 32 |
| 662 | Nontoxigenic isolate, Switzerland | 60 |
| M7404 | NAP1/027 Canadian isolate, 2005 | J. Pepin |
| 661 | Toxigenic isolate, France, 1992 | M. Sebald |
| 660/2 | Toxigenic isolate, France, 1992 | M. Sebald |
| 685 | Toxigenic isolate, France, 1992 | M. Sebald |
| AM421 | Nontoxigenic isolate; Fairfield Hospital, Australia; 1980 | R. Wilkinson |
| AM662 | Nontoxigenic isolate; St. Vincents Hospital, Australia; 1982 | R. Wilkinson |
| AM1180 | Toxigenic isolate; Latrobe Valley Regional Hospital, Australia; 1985 | R. Wilkinson |
| M334581 | Toxigenic isolate; Austin Hospital, Australia; 2002 | L. Grayson |
| M336632 | Toxigenic isolate; Austin Hospital, Australia; 2002 | L. Grayson |
| M336948 | Nontoxigenic isolate; Austin Hospital, Australia; 2002 | L. Grayson |
| M345206 | Nontoxigenic isolate; Austin Hospital, Australia; 2002 | L. Grayson |
| M345300 | Toxigenic isolate; Austin Hospital, Australia; 2002 | L. Grayson |
| M361441 | Nontoxigenic isolate; Austin Hospital, Australia; 2002 | L. Grayson |
| WA7 | Toxigenic isolate; Rockingham Kwinana District Hospital, Australia; 2006 | T. Riley |
| WA9 | Toxigenic isolate; Katanning Hospital, Australia; 2006 | T. Riley |
| MMC CD1 | Toxigenic isolate; Monash Medical Centre, Australia; 2006 | G. Jenkin |
| MMC CD2 | Toxigenic isolate; Monash Medical Centre, Australia; 2006 | G. Jenkin |
| MMC CD8 | Toxigenic isolate; Monash Medical Centre, Australia; 2006 | G. Jenkin |
| E. coli | ||
| DH5α | F− φ80dlacZM15 (lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Life Technologies |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| HB101 | thi-1 hsdS20(rB− mB−) supE44 recAB ara-14 leuB5 proA2 lacY1 galK rpsL20 (Smr) xyl-5 mtl-1 | 5 |
| Plasmids | ||
| pVS520 | Tra+ Mob+ RP4 derivative, Tcr | 37 |
| pMTL9301 | E. coli-C. difficile mobilizable shuttle vector, carries pCD6 replication region, Emr | 40 |
| pMTL500E | E. coli-Clostridium shuttle vector (pAMβ1 replicon), Emr | 36 |
| pMTL9361Cm | E. coli-C. difficile mobilizable shuttle vector, carries pCD6 replication region, Tmr | G. Carter, N. Minton (unpublished) |
| pCR2.1-TOPO | E. coli PCR cloning vector, ColE1, Apr Kmr | Invitrogen |
| pT7Blue3 | E. coli PCR cloning vector, ColE1, Apr | Novagen |
| pJIR3107 | pMTL9361Cm Ω CD196 4.5-kb cdtAB fragment (SacI/SphI), CDT binary toxin expression plasmid, Tmr | Recombinant |
| pJIR3394 | pMTL9301 (FspI) Ω JIR8094 cdtR 1.13 kb (EcoRV/5maI), CdtR expression plasmid, Emr | Recombinant |
| pJIR3395 | pMTL3394 ΔcdtR (BamHI/PstI), internal deletion in cdtR, Emr | Recombinant |
Tmr, thiamphenicol (chloramphenicol) resistance; Emr, erythromycin resistance; Apr, ampicillin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Smr, streptomycin resistance.
Isolation and manipulation of nucleic acids.
Plasmid DNA was isolated from E. coli strains grown overnight in 5 ml of 2 ×YT broth or from C. difficile strains grown in 5 ml of BHIS broth, with appropriate antibiotic selection, using the QIAprep spin miniprep kit (QIAGEN) according to the manufacturer's instructions. For C. difficile plasmid preparations, cell pellets were incubated in buffer P1 (supplied with the kit) supplemented with 30 μg · ml−1 lysozyme (Sigma-Aldrich) for 30 min at 37°C, before extraction as per the manufacturer's instructions. C. difficile genomic DNA was prepared using the DNeasy tissue kit (QIAGEN) according to the manufacturer's instructions. DNA was quantified using a NanoDrop spectrophotometer (NanoDrop Technologies) and stored at −20°C. Standard methods for the digestion, modification, ligation, and analysis of plasmid and genomic DNA and PCR products were used (44). Nucleotide sequence analysis was carried out using a PRISM BigDye Terminator cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions. Sequence detection was carried out on an Applied Biosystems 3730S genetic analyzer and was performed by Micromon at Monash University. Sequences were analyzed using ContigExpress (Invitrogen).
PCR.
Oligonucleotide primer sequences are listed in Table 2. Unless otherwise stated, all PCR experiments were carried out with Taq DNA polymerase and the supplied 10× PCR buffer with magnesium according to the manufacturer's instructions (Roche). The following parameters were utilized for all PCR experiments: initial denaturation was performed at 94°C for 4 min, followed by 30 cycles consisting of a denaturation step at 94°C for 30 s, an oligonucleotide primer annealing step at 50°C for 30 s, and an extension step at 72°C. The extension time was 1 min per 1 kb of amplified DNA. A final extension step was performed at 72°C for 12 min.
TABLE 2.
Oligonucleotide primers
| Primer | Sequence (5′→3′) | Usea |
|---|---|---|
| JRP1744 | GGGAAAGAAAAGAAGCAGAAAG | Amplify internal fragment of cdtA (+) |
| JRP1745 | TGTATTTTCATTGTTTCTCCTC | Amplify internal fragment of cdtA (−) |
| JRP2342 | CCGGAATTCGCTCTATTGGACTAGACCGTTG | Amplify internal fragment of tcdA (+) |
| JRP2343 | CCGGAATTCTTAAAGTTTTTTCTATAGTATT | Amplify internal fragment of tcdA (−) |
| JRP2344 | CCGGAATTCGTTCAGTAACAGTGGATTTTTG | Amplify internal fragment of tcdB (+) |
| JRP2345 | CCGGAATTCTCAAATGGATATTCATACTGCC | Amplify internal fragment of tcdB (−) |
| JRP2423 | CCGGAATTCGGCCATATATGTCGTATATCTA | Amplify C. difficile CD196 cdtA and cdtB genes and upstream, region introduces EcoRI site (+) |
| JRP2424 | CCGGAATTCTTGTGGGGACAAATTTAAATCA | Amplify C. difficile CD196 cdtA and cdtB genes and upstream region, introduces EcoRI site (−) |
| JRP2523 | AGTTCTAGAGGAATGAAACTAAAAGAAAAAAT | Amplify internal fragment of cdtR (+) |
| JRP2524 | AGTTGCGCAGCATGCATCTAAATCTGGTAAAG | Amplify internal fragment of cdtR (−) |
| JRP3631 | GTTGCGGGGATCCTCTAGTAACGG | Amplify ΔcdtR by SOE-PCR (+) |
| JRP3632 | CTCATTTTAATATTTCTTACAACATCATTATCAAAACCTTC | Amplify ΔcdtR by SOE-PCR (−) |
| JRP3633 | TAAGAAATATTAAAATGAGTAATGGCCATATATG | Amplify ΔcdtR by SOE-PCR (+) |
| JRP3634 | GGTCAGGCTGCATCTGCAGAATTCG | Amplify ΔcdtR by SOE-PCR (−) |
| JRP3720 | CTTCTATAATTATTGTTAAATAATTCTTC | Amplify cdtR gene and upstream region (+) |
| JRP3721 | GAGACATCTCCTTTTTCTATTTATTATG | Amplify cdtR gene and upstream region (−) |
| JRP3722 | GTTGATAAATTTATTATTAACTCTCTAG | Amplify internal fragment of CD2602 (+) |
| JRP3723 | GTAAACACTAGAATAATATCTATAG | Amplify internal fragment of CD2602 (−) |
| JRP3724 | CATGTTTACAATCTCTTCTACACTC | Amplify internal fragment of trpS (+) |
| JRP3725 | CAATCACATGTCAAAGAGCATGTTGAAC | Amplify internal fragment of trpS (−) |
| JRP3726 | CTACTATTAGATATTATAACAATTGTTCC | Amplify putative cdtA promoter region (+) |
| JRP3727 | CCCTACAAATATTAGTTTAGTTAAC | Amplify putative cdtA promoter region (−) |
| JRP3728 | GCCACCTGATACTATAGATATTATTC | Amplify/sequence CD2602/trpS intervening region (+) |
| JRP3729 | AAGTTGGATTTATAGAGAGAAG | Amplify/sequence CD2602/trpS intervening region (−) |
| JRP3845 | TGCAATACTACTTACAAGGCTCCTATAGA | Amplify internal cdtA fragment in QRT-PCR (+) |
| JRP3846 | TCTTTCCCATTCTTTAGCCTTTTC | Amplify internal cdtA fragment in QRT-PCR (−) |
+, leading-strand-specific oligonucleotide primer; −, lagging-strand-specific oligonucleotide primer.
Construction of recombinant plasmids.
For the construction of the CDT expression plasmid, PCR was performed using primers JRP2423 and JRP2424 (Table 2), chromosomal DNA from strain CD196, and the Triple Mastermix PCR system (Roche). The resultant 4.5-kb fragment containing the CDT region was cloned into the EcoRV site of pT7Blue3 using the Perfectly Blunt cloning kit according to the manufacturer's instructions (Novagen). After sequencing, the 4.5-kb fragment carrying both the cdtA and cdtB genes was excised from the pT7Blue3 derivatives with SphI and SacI before cloning into the equivalent sites of the E. coli-C. difficile shuttle plasmid pMTL9361Cm, resulting in pJIR3107.
For construction of the cdtR-plasmid pJIR3394, PCR was performed using primers JRP3720 and JRP3721 (Table 2). Chromosomal DNA from strain JIR8094 was used in the Failsafe PCR system with buffer E (Epicenter Biotechnologies) to amplify the cdtR region. The PCR product was then TA cloned into pCR2.1-TOPO, according to the manufacturer's instructions (Invitrogen). The 1.13-kb cdtR fragment was excised from this plasmid with XhoI and BamHI and subcloned into the equivalent sites of pMTL500E. The insert was subsequently excised from this intermediate vector using EcoRV and SmaI and inserted into the FspI site of pMTL9301, resulting in pJIR3394.
The deletant cdtR allele was generated using splice overlap extension PCR (SOE-PCR) (15, 16). PCR was used to amplify the left and right sides of the cdtR region using primers JRP3631 with JRP3632 and JRP3633 with JRP3634, respectively (Table 2). This reaction used pJIR3394 as the template together with the Failsafe PCR system enzyme mix and buffer E. Each fragment was purified by extraction from a 1% agarose gel using the Qiaquick gel extraction kit (QIAGEN). An aliquot of each of the purified PCR products was mixed in an equimolar ratio and used as the DNA template in a second PCR with primers JRP3631 and JRP3634, which resulted in a DNA fragment with a 561-bp deletion within cdtR, which was then TA cloned into pCR2.1-TOPO (Invitrogen). The fragment was excised from pCR2.1-TOPO using BamHI and PstI, and cloned into the equivalent sites of pJIR3394, resulting in the final construct, pJIR3395.
Transfer of plasmid DNA into C. difficile by conjugation.
The conjugation procedure was carried out as previously described (40), except that HB101(pVS520) (37) was used as the E. coli donor. Bacterial pellets collected from 1-ml aliquots of an overnight E. coli donor culture were resuspended in 200-μl aliquots of the C. difficile recipient strain, which had been collected from 20-ml overnight cultures incubated at 37°C in an anaerobic chamber. This mating mixture was then spread onto a thick BHIS agar plate and incubated for 6 h or overnight at 37°C under anaerobic conditions. Bacterial growth was harvested by flooding the agar three times with 500 μl of sterile phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 1.4 mM KH2PO4, 4.3 mM Na2HPO4). Aliquots (100 μl) of the combined cell suspension were then spread onto BHIS agar supplemented with cycloserine and cefoxitin to prevent growth of the E. coli donor and with either thiamphenicol or erythromycin to select for C. difficile transconjugants carrying the appropriate plasmids. The plates were incubated under anaerobic conditions for 24 to 72 h.
Determination of the integration site of the CdtLoc.
The region flanked by CD2602 and trpS from C. difficile strain CD37 was amplified by PCR using primers JRP3722 and JRP3725 (Table 2) and the Failsafe PCR system buffer G. The sequence of the amplified product was then determined using primers JRP3722, JRP3725, JRP3728, and JRP3729 (Table 2). The CDT integration sites of all other nontoxigenic C. difficile strains were determined by PCR amplification of this region using primers JRP3728 and JRP3729 and Failsafe buffer E, followed by sequencing with primers JRP3728 and JRP3729.
Detection of CDTa and CDTb proteins by Western blotting.
Isolation of CDTa and CDTb and Western blotting were carried out as previously described (38). Proteins were precipitated at a concentration of 70% ammonium sulfate from cell-free supernatants of C. difficile cultures grown overnight in 100 ml of BHIS broth. The precipitate was collected, dissolved in 2 ml of distilled water, and dialyzed against 10 mM Tris-HCl buffer, pH 7.0, for 24 h with three changes of buffer. Proteins were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (23) and transferred by electrophoresis (44) to nitrocellulose membranes (Whatman). The membranes were incubated for 1 h in phosphate-buffered saline containing 5% dry milk powder and then incubated overnight with immunopurified antibodies specific for C. perfringens iota toxin components A (Ia) and B (Ib), which are cross-reactive with the C. difficile CDTa and CDTb proteins, respectively (38). CDTa- and CDTb-bound antibodies were then detected with peroxidase-conjugated anti-rabbit sheep antibodies (Chemicon) and the Western lightning chemiluminescence reagent kit (Perkin Elmer), according to the manufacturer's instructions.
ADP-ribosyltransferase assays.
ADP-ribosyltransferase assays were carried out essentially as before (55). In brief, precipitated supernatant protein was incubated for 1 h in a reaction solution containing 50 mM triethanolamine-HCl buffer (pH 7.5), 5 mM MgCl2, 10 mM dithiothreitol, 10 mM thymidine, and 10 μg rabbit skeletal muscle actin (Sigma-Aldrich), together with [32P]NAD (3 μCi). The entire reaction volume was then applied to a glass fiber filter (Millipore) and allowed to dry. Protein was then precipitated onto the filter by washing with ice-cold 10% (wt/vol) trichloroacetic acid, followed by three washes in ice-cold 5% (wt/vol) trichloroacetic acid and a final wash using 95% ethanol. The filters were allowed to dry before being placed in Optiphase Hisafe 3 scintillation fluid (Perkin-Elmer), and radioactivity was counted using a Tri-Carb 2100TR liquid scintillation analyzer (Packard Biosciences).
Quantitative real-time PCR (QRT-PCR) analysis of cdtA expression.
C. difficile cultures (10 ml) were grown to a turbidity of approximately 1.0, and RNA was extracted using the Ribopure Bacteria kit (Ambion Inc.) according to the manufacturer's instructions. To remove contaminating genomic DNA, the purified RNA was treated with the DNA-free kit (Ambion Inc.), according to the manufacturer's instructions. Reverse transcription reactions were then performed using 2 μg of purified RNA, 0.5 μM primer, and the Omniscript reverse transcription kit (QIAGEN) according to the manufacturer's instructions. The reaction mixtures were diluted twofold, and real-time PCR analysis was performed in a final volume of 25 μl with SYBR green PCR master mix (Applied Biosystems), 2 μl of diluted reverse transcription reaction mixture, and 120 nM primers using an Eppendorf Realplex 4 Mastercycler. Triplicate reactions were performed in multiple experiments using RNAs from three biological replicates. The data were normalized to C. difficile rpoA mRNA levels.
Nucleotide sequence accession number.
The GenBank accession number of the CD196-derived sequence determined in this study is EF581852.
RESULTS
More binary toxin is produced in a strain 630 background than in a strain CD37 background.
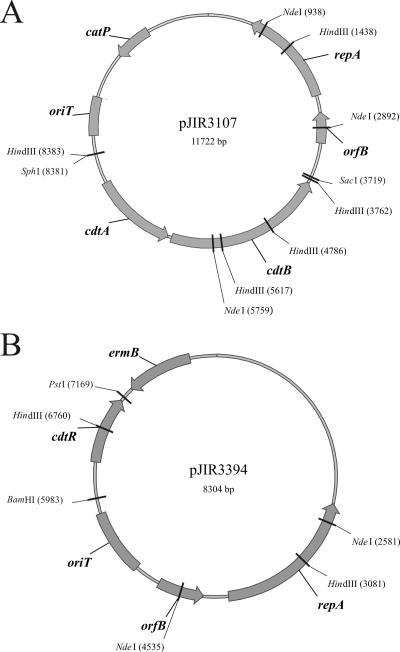
To facilitate the functional analysis of CDT in C. difficile, the E. coli-C. difficile shuttle plasmid pJIR3107 (Fig. 1A) was constructed. This plasmid encoded the complete binary toxin region of C. difficile strain CD196. The nucleotide sequence of the cloned 4.5-kb fragment was determined to see if any PCR-derived mutations had been introduced. When this sequence was compared to the CD196-derived binary toxin region in the GenBank database (accession number L76081), several differences were observed. Sequencing of two independent CDT-specific PCR products revealed that these changes were not the result of PCR-derived errors.
FIG. 1.
Relevant shuttle plasmids. Schematic representations of the CDT binary toxin expression plasmid pJIR3107 (A) and the cdtR complementation plasmid pJIR3394 (B) are shown. The position and orientation of the pCD6 plasmid replication genes (repA and orfB) are shown, as is the transfer origin (oriT), the selectable markers (ermB or catP), the CDT binary toxin genes (cdtA and cdtB), and the cdtR gene.
The most significant difference occurred within cdtA, after nucleotide position 267, where the residues ACCAGAAGA were present in the sequence determined here but not in the database sequence. The addition of these residues would lead to the insertion of three amino acids, Thr Arg Arg. Within the sequenced region, there were two other changes that would lead to a change to the resultant protein sequence. Within cdtA there was a G at position 871, not an A, resulting in a change from Asn to Ser. Within cdtB, the T at position 1720 was an A, changing the encoded amino acid from Ser to Thr. It is not known if errors in the previous CD196-derived sequence or strain variation is the reason for the differences that were observed. However, comparison of the cdtAB sequence that we obtained from CD196 with those of the sequenced NAP1/027 strain QCD-32g58 (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=120673853&view=gbwithparts) and of an additional cdtAB positive isolate, CCUG 20309 (accession number AF271719) (7), revealed that the differences observed here were present in both of these strains. This analysis showed that the CDT region that had been cloned into pJIR3107 was identical to that from the chromosome of strain CD196, as well as strains QCD-32g58 and CCUG 20309, validating the use of this plasmid in subsequent studies.
The CDT+ plasmid pJIR3107 was introduced by conjugation into JIR8094, an erythromycin-susceptible derivative of the tcdA+ tcdB+ strain 630, which does not have intact cdt genes and therefore produces toxins A and B but not binary toxin. The plasmid was also introduced into the nontoxigenic strain CD37. The integrity of the plasmids from several independently derived transconjugants was confirmed by introducing plasmid DNA into E. coli DH5α and carrying out restriction analysis.
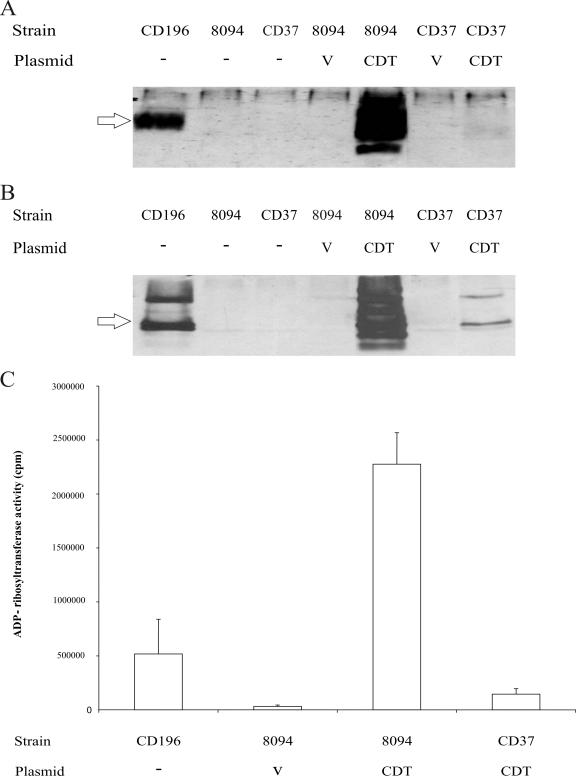
To determine if the C. difficile transconjugants produced the CDTa and CDTb proteins, Western blotting was performed. Neither JIR8094, CD37, nor derivatives of these strains carrying the vector plasmid pMTL9361Cm produced any detectable CDTa or CDTb protein (Fig. 2A and B). As expected, both CDT components could be detected in the supernatant of the reference binary toxin-producing strain CD196. The JIR8094 derivative that carried pJIR3107 produced substantial quantities of CDTa and CDTb, much more than was produced by CD196. Unexpectedly, the CD37 derivative carrying the same plasmid produced very low levels of the CDT proteins (Fig. 2A and B).
FIG. 2.
Analysis of binary toxin production by complemented strains. (A) CDTa Western immunoblot using cross-reactive Ia-specific antibodies and precipitated supernatant proteins from the strains indicated. (B) CDTb Western immunoblot using cross-reactive Ib-specific antibodies and precipitated supernatant proteins. (C) ADP-ribosyltransferase activity assay using purified rabbit skeletal muscle actin, [32P]NAD and precipitated supernatant proteins. The data are the mean values and standard deviations from three independent experiments. CD196, binary toxin-positive reference strain; JIR8094, binary toxin-negative strain; CD37, nontoxigenic strain; V, pMTL9361Cm vector plasmid; CDT, pJIR3107, which carries the wild-type cdtAB genes. The arrows indicate the 48-kDa CDTa protein and 75-kDa CDTb protein. The higher-molecular-mass products are most likely higher-order aggregates and the smaller protein bands breakdown products.
To quantitatively measure CDT production and to confirm that functional CDT binary toxin was being produced, ADP-ribosyltransferase assays were performed using ammonium sulfate-precipitated culture supernatants prepared from JIR8094 and CD37 derivatives carrying pJIR3107, together with the appropriate controls (Fig. 2C). In agreement with the Western blots, the level of ADP-ribosyltransferase activity associated with JIR8094(pJIR3107) was more than 15-fold higher than that associated with CD37(pJIR3107) and more than 4-fold higher than that associated with the CD196 reference strain (Fig. 2C). No significant ADP-ribosyltransferase activity was detected in extracts from strain JIR8094(pMTL9361Cm), as expected (Fig. 2C).
A LytTR family response regulator gene is adjacent to the CDT-encoding genes.
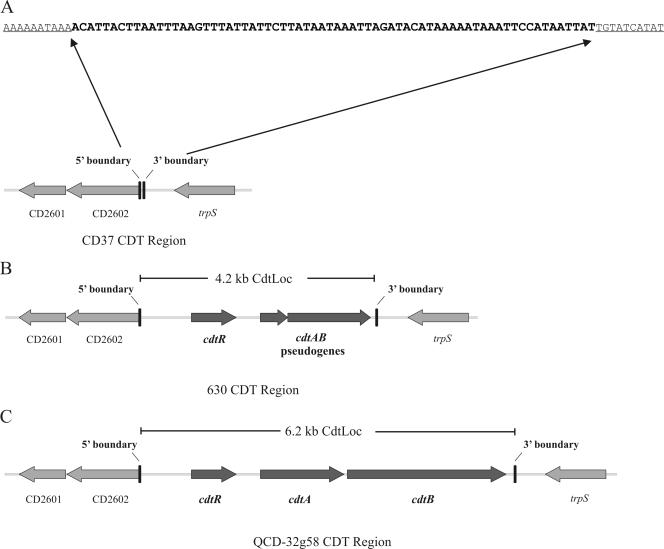
To determine if a regulatory gene was responsible for the increased levels of CDT production by JIR8094(pJIR3107), bioinformatic analysis of the CdtLoc of the strain 630 genome sequence (45) and of strain QCD-32g58 was undertaken. Note that although strain 630 does not produce CDT or have an intact cdtAB operon, it does have cdtAB pseudogenes (Fig. 3), which have accumulated several frameshift mutations and premature stop codons (45).
FIG. 3.
Schematic representation of the CDT region and flanking genes. The regions from the nontoxigenic isolate CD37 (A), the binary toxin-negative isolate strain 630 (B), and the binary toxin-positive isolate QCD-32g58 (C) are shown. The positions of the 5′ flanking genes CD2601 and CD2602, the 3′ flanking gene trpS, the response regulator gene cdtR, and the CDT binary toxin-encoding genes cdtAB, or their pseudogenes, are shown. For each variant of the CDT region the positions of the 5′ and 3′ conserved boundaries are shown, and the size of the entire CdtLoc is indicated. The unique 68-bp sequence that is present in CD37 and other nontoxigenic isolates in place of the CdtLoc is shown in bold, and the nucleotide boundaries of the CDT region that are conserved in all three variants are underlined.
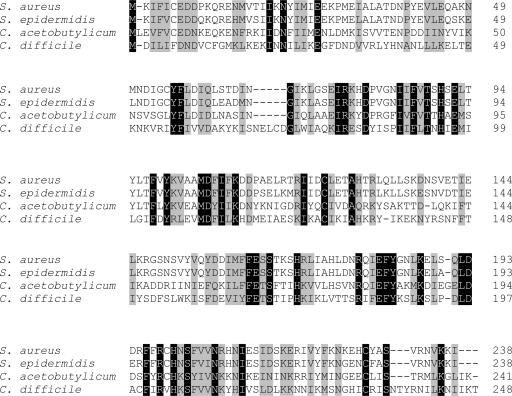
Our analysis revealed the presence of a gene (CD2603) encoding a putative LytTR family response regulator, which was located immediately upstream of the predicted cdtA pseudogene (Fig. 3). Due to its close proximity to the cdtAB genes, and in view of the results reported in this paper, we have named CD2603 the “Clostridium difficile binary toxin regulatory gene,” or cdtR. The cdtR gene was 747 bp in length and encoded a putative 30-kDa CdtR protein that had approximately 28% amino acid sequence identity to the accessory gene regulator A (AgrA) protein from Staphylococcus aureus and that had similarity to several other AgrA orthologs (Fig. 4). Unlike AgrA from S. aureus, cdtR was an orphan response regulator gene, since it did not appear to have a cognate sensor histidine kinase gene associated with it.
FIG. 4.
Alignment of CdtR orthologs. The predicted CdtR protein from C. difficile strain 630 and predicted AgrA orthologs of S. aureus (ABB17535), Staphylococcus epidermidis (AAO25552) and Clostridium acetobutylicum (AAK78066) were aligned using CLUSTALW (58). Identical amino acid residues are shaded with black, and strongly similar amino acid residues are shaded with gray.
The similar genetic organizations of the cdt locus in strains 630 and QCD-32g58 (Fig. 3) suggested that cdtR might encode a regulator of binary toxin production. The low level of CDT production by CD37(pJIR3107) might be due to the absence of cdtR or to a mutation within this gene in CD37. To investigate this hypothesis, primers JRP3720 and JRP3721 (Table 2) were used in an attempt to PCR amplify the cdtR gene region from strains JIR8094 and CD37. While a PCR product specific to cdtR was amplified using chromosomal DNA from JIR8094, no such product could be generated when CD37-derived DNA was used (data not shown). This result indicated either that cdtR was absent from CD37 or that the nucleotide sequence of the gene was significantly different from that of the gene in strain 630. PCR analysis using other primers that anneal within the cdtR coding region yielded similar results (data not shown), indicating that strain CD37 does not carry the cdtR gene.
The cdt locus was found only in toxigenic C. difficile isolates.
The cdt loci from 26 C. difficile strains, isolated over a period of at least 27 years, were investigated in more detail. Genomic DNA from each strain was subjected to PCR amplification using primers specific for the cdtR gene, the predicted promoter region of cdtA, and CD2602 and trpS, which are located immediately upstream and downstream, respectively (Fig. 3). The cdtA promoter region was analyzed in preference to an internal cdtA fragment since there are a number of isolates that have large deletions within the coding regions of cdtAB (53). Any such strains would have appeared as false negatives in cdtA-specific analysis. In addition, the major toxin genes, tcdA and tcdB, were also PCR amplified. Of these 26 strains, eight isolates did not yield amplification products for either tcdA or tcdB and therefore were classed as nontoxigenic isolates. Both the cdtR gene and the predicted cdtA promoter region were found exclusively in the remaining 18 isolates, which had both tcdA and tcdB, although all 26 strains carried both CD2602 and trpS.
To define the cdt locus, the region located between the CD2602 and trpS genes of strain CD37 was PCR amplified. A product of only 1.8 kb in size was generated (data not shown), indicating that a substantial portion of the equivalent 4.2-kb and 6.2-kb regions present in strains 630 and QCD-32g58, respectively, was absent from strain CD37. The nucleotide sequence of this 1.8-kb fragment was determined, allowing the boundaries of the cdt locus to be defined. The cdtR, cdtA, and cdtB genes, as well as some flanking sequences, were all absent in strain CD37. Instead, a unique 68-bp nucleotide sequence was found at this location (Fig. 3). This region did not have any similarity with sequences within the cdt locus of strain 630 or QCD-32g58 or with any other sequences found in the database.
To determine whether the same 68-bp region was present in the remaining seven nontoxigenic strains, PCR was used to amplify a smaller internal fragment located between the CD2602 and trpS genes. In every strain, a product of approximately 700 bp was generated, which was identical in size to that from CD37 (data not shown). Subsequent sequence analysis of each PCR product revealed the presence of the same 68-bp sequence in every nontoxigenic isolate examined (Fig. 3).
CdtR positively regulates binary toxin production in C. difficile.
To determine if CdtR regulates the expression of the cdtAB operon, the cdtR gene and a 300-bp upstream region were PCR amplified from strain JIR8094 and cloned into the shuttle plasmid pMTL9301, generating pJIR3394 (Fig. 1B), which encodes erythromycin resistance. This plasmid was transferred by conjugation into the CD37 derivative that carries the cdtA+B+ plasmid, pJIR3107. The transconjugants were isolated and maintained on medium supplemented with both thiamphenicol and erythromycin to ensure the continued presence of both plasmids, each of which was derived from a pCD6 replicon. PCR analysis of several transconjugants confirmed the presence of both cdtA and cdtR (data not shown), verifying the presence of both pJIR3107 and pJIR3394 in the resultant strain.
To provide the appropriate control for this experiment, pJIR3395 was constructed. This plasmid was a derivative of pJIR3394 that contained a large in-frame internal deletion of cdtR, spanning codons 36 to 222, designated ΔcdtR. This plasmid was transferred by conjugation into strain CD37(pJIR3107), and thiamphenicol and erythromycin resistant transconjugants were selected, maintained, and analyzed as before.
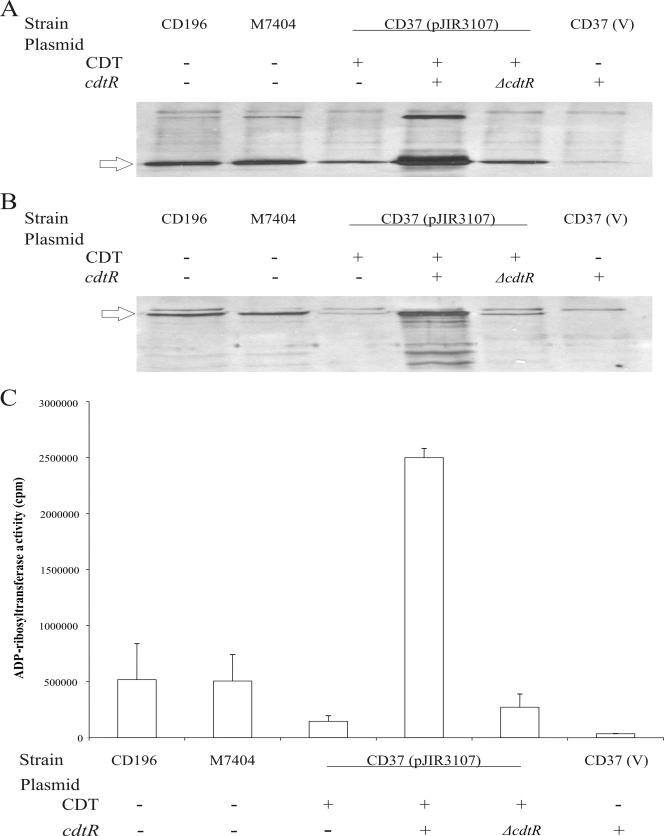
Western blot analysis was then performed on the isogenic CD37 derivatives carrying pJIR3107 (cdtA+B+) alone or pJIR3107 together with pJIR3394 (cdtR+) or pJIR3395 (ΔcdtR), as well as CD37 carrying the shuttle vector pMTL9361Cm and pJIR3394. Other controls included the CD196 binary toxin reference strain and a CDT+ NAP1/027 strain, M7404. The results showed that the presence of pJIR3394 in CD37(pJIR3107) led to a dramatic increase in the production of both CDTa and CDTb (Fig. 5A and B). By contrast, pJIR3395 had no effect on CDTa or CDTb production. As expected, a CD37 derivative carrying pMTL9361Cm and pJIR3394 did not produce any detectable CDTa or CDTb, since it did not contain the cdtA or cdtB gene. Note that CDTa and CDTb production by CD37(pJIR3107) carrying the cdtR+ plasmid appeared to be greater than that by either of the binary toxin-producing positive controls.
FIG. 5.
Analysis of binary toxin production in derivatives of CD37. (A) CDTa Western immunoblot using cross-reactive Ia specific antibodies and precipitated supernatant proteins from the strains indicated. (B) CDTb Western immunoblot using cross-reactive Ib specific antibodies and precipitated supernatant proteins. (C) ADP-ribosyltransferase activity assayed using purified rabbit skeletal muscle actin, [32P]NAD and precipitated supernatant proteins. The data are the mean values and standard deviations from three independent experiments CD196, binary toxin-positive reference strain; M7404, binary toxin-positive NAP1/027 strain; CD37, nontoxigenic strain; pJIR3107, encodes the wild-type cdtAB genes (shown as + in the CDT rows); V, pMTL9361Cm vector plasmid (shown as − in the CDT rows); pJIR3394, encodes the wild-type cdtR gene (shown as + in the cdtR rows); pJIR3395, encodes the ΔcdtR gene (shown as ΔcdtR in the cdtR rows). The arrows indicate the 48-kDa CDTa protein and 75-kDa CDTb protein.
To confirm the data obtained by Western blot analysis, ADP-ribosytransferase assays were performed on the same strains (Fig. 5C). The results showed that the functional cdtR gene results in a 17-fold increase in CDT activity. The CD37(pJIR3107)(pJIR3394) strain had approximately fivefold more activity than the positive control strains CD196 and M7404. The introduction of the internal deletion into cdtR led to an approximately ninefold reduction in ADP-ribosytransferase activity, to a level that was not significantly different from that of the isogenic negative controls. These results provide good experimental evidence that the cdtR gene encodes a positive activator of binary toxin production in C. difficile.
To verify that CdtR regulates the expression of the binary toxin genes at the transcriptional level, cdtA-specific QRT-PCR analysis was performed on RNAs extracted from strains CD37(pJIR3107)(pJIR3394) and CD37(pJIR3107)(pJIR3395). The results showed that there was a 90-fold ± 10.5-fold increase in the levels of cdtA-specific mRNA in the strain carrying the intact cdtR gene compared to the strain carrying the ΔcdtR plasmid (P < 0.0002). These data confirm that cdtR is a transcriptional regulator of the binary toxin genes.
DISCUSSION
The results presented in this paper clearly show that binary toxin production in C. difficile is positively regulated by the LytTR family response regulator, CdtR. The evidence for this conclusion comes from the observation that strain 630-derived recombinant strains carrying the binary toxin structural genes cdtA and cdtB produced large amounts of binary toxin, but the equivalent CD37 derivatives, which do not have a cdtR gene, produced approximately 15-fold less binary toxin, despite these strains carrying the same CDT-encoding plasmid. In addition, when a recombinant CD37 derivative carrying cdtA and cdtB was complemented with a functional cdtR gene, binary toxin production was activated and greater than 17-fold more binary toxin was produced.
To rule out the possibility that another plasmid-encoded factor was responsible for the increase in binary toxin production seen in the complemented strains, the intact cdtR gene was replaced with a mutant cdtR allele. This ΔcdtR gene contained an in-frame deletion that spans codons 36 to 222, resulting in the removal of most of the cdtR sequence. When complemented with ΔcdtR, CDT production was not statistically different from that of the noncomplemented parental strain, indicating that CdtR is indeed responsible for the observed up-regulation of binary toxin production.
Compared to the NAP1/027 strain M7404 and the reference CDT+ strain CD196, which both produced similar levels of binary toxin, the cdtAB-complemented strain 630 derivative and the cdtAB- and cdtR-complemented CD37 derivative had significantly more ADP-ribosyltransferase activity. This result most likely reflects the higher copy number of the plasmid-carried cdtAB genes in the recombinant strains.
The comparative genomic studies showed that there was an absolute correlation between the presence of the cdtR gene and the upstream cdtA region. Strains that did not have cdtA or a cdtA pseudogene did not carry cdtR. The term PaLoc is widely used to describe the cytotoxin gene locus of C. difficile, a locus that includes both toxin structural genes and regulatory genes. Based on the findings presented here, and in keeping with the PaLoc nomenclature, it is proposed that the term CdtLoc be used to describe the cdt locus, which is hereby defined as the region containing cdtR, cdtA, and cdtB (as shown in Fig. 3), irrespective of whether in a particular isolate cdtA and cdtB are functional genes or pseudogenes.
Of the strains examined here, only those isolates that had the tcdA and tcdB genes carried cdtR and therefore had the CdtLoc. Recently, other workers performed genomic microarray analysis that compared diverse C. difficile isolates (52), very few of which were included in our study. Of the 75 strains analyzed, 68 provided definitive data with regard to the presence or absence of cdtR. Our analysis of the microarray data appears to reinforce the findings reported here, since approximately 96% of cdtR-positive isolates also carried the cdtAB genes or, at the very least, fragments of these genes, indicating that cdtR and the cdtAB genes are genetically linked. Interestingly, we found that approximately 82% of the cdtR-negative isolates were clustered within the A−B+ clade. The remaining 18% were located in the HA1 clade and were not from disease outbreaks. Upon closer inspection of the entire A−B+ clade, we discovered that only four of these 22 isolates carried the CdtLoc. Remarkably, each of these four strains was actually genotypically classified as A+B+ and not A−B+, and they were all of animal origin (52). Analysis of the seven A−B− isolates included in the microarray study revealed that only two contained the CdtLoc. These strains were located within the HA1 and HA2 clades, respectively. The remaining five genotypically A−B− isolates were cdtR negative and were clustered within the A−B+ clade.
Other studies have identified CDT-positive but toxin A- and toxin B-negative strains, providing evidence that these loci are not always linked (10, 11). However, the observations described above, together with the results presented in this study, suggest that, with the exception of strains found within the A−B+ clade, there is a correlation between the presence of the PaLoc and the CdtLoc. More than 98% of CdtLoc-positive strains also have the PaLoc. Furthermore, it is apparent, again with the exception of A−B+ strains, that the CdtLoc is widely disseminated, with greater than 90% of the non-A−B+ clade isolates analyzed with the microarrays containing this region.
Further analysis of the site of insertion/deletion of the CdtLoc in CD37 and seven other PaLoc-negative isolates showed that the entire CdtLoc was absent in these isolates and had been replaced by a novel 68-bp sequence of unknown function. This finding is reminiscent of the analysis of the PaLoc, where nontoxigenic strains were found to harbor a unique and highly conserved 115-bp sequence instead of the PaLoc (6). As with the PaLoc, there is no evidence of transposon-, plasmid-, or bacteriophage-related genes in close proximity to the CdtLoc. Since both of these toxin loci share some important characteristics, namely, the loss of an apparently innocuous nucleotide sequence upon their acquisition and the absence of any obvious genes or nucleotide sequences related to mobile genetic elements, it is postulated that the integration of the PaLoc and the CdtLoc may have occurred through similar, but as yet unknown, mechanisms.
It is estimated that between 1.6% and 6% of all C. difficile isolates produce a functional binary toxin, including the highly virulent toxinotype III NAP1/027 epidemic strains (53). However, the role of this toxin in the pathogenesis of C. difficile infections remains to be determined. Several studies have suggested an association between binary toxin-producing strains and more severe disease and community-acquired infections (2, 29, 57). Furthermore, CDT is known to be cytotoxic to Vero cells (38) and causes fluid accumulation in the rabbit ileal loop model (10). Indirect evidence therefore does suggest that CDT plays a role in C. difficile-mediated disease. However, a recent study showed that toxin A- and toxin B-negative, CDT-positive isolates were unable to cause disease in the hamster model (10).
Many bacteria tightly control their ability to produce toxins. C. difficile regulates the expression of its major toxins, toxin A and toxin B, in response to several environmental cues, including nutritional signals, cell density, and temperature (8, 17, 20, 27). This process involves the tcdR and tcdC genes, which are encoded within the PaLoc. However, the PaLoc does not carry genes encoding a two-component signal transduction system.
Members of the LytTR response regulator family, named after the LytT and LytR response regulators from Bacillus subtilis and S. aureus, respectively, are characterized by the presence of a novel DNA-binding domain, which shows little similarity to the helix-turn-helix or winged-helix domains that are more typical of other response regulators (33). They are widely distributed in low-G+C gram-positive bacteria, such as the clostridia, and many members have a well-documented association with bacterial virulence. Some play an integral role in quorum sensing, such as the AgrA response regulator, which is the master regulator of the quorum-sensing-controlled virulence response of S. aureus (31). Others are important in the regulation of toxin production, as demonstrated by the VirR response regulator of C. perfringens, which controls the synthesis of several toxins, including perfringolysin O (26, 42, 46), while others are involved in the production of extracellular polysaccharides, such as alginate in Pseudomonas aeruginosa, which is regulated by the AlgR response regulator (24). CdtR is now another virulence-related member of this family, controlling the production of CDT in C. difficile. CdtR may activate cdtAB gene expression either directly or by a secondary regulator.
LytTR response regulators are activated by the transfer of a phosphoryl group to a conserved aspartate residue located in their receiver domain, through the action of a cognate sensor histidine kinase (54). The genes encoding these proteins often comprise an operon, as observed for both virR and agrA, which are adjacent to virS (26) and agrC (34), respectively. By contrast, the cdtR gene is an orphan response regulator; there is no sensor kinase gene located in close proximity, and the histidine kinase that presumably is responsible for the activation of CdtR is unknown. However, the integration site of the CdtLoc positions cdtR only 11 bp from the predicted start site of a sensor histidine kinase gene (CD2602), although this gene is transcribed on the opposite DNA strand (Fig. 3). CD2602 appears to be associated with another downstream LytTR response regulator (CD2601). It is possible that the acquisition of the CdtLoc might result in the differential regulation of CD2602. Current studies in this laboratory are aimed at determining whether CD2601 and CD2602 are involved in the regulation of binary toxin expression.
In conclusion, we have identified a LytTR family response regulator that activates binary toxin production in C. difficile and together with the cdtAB operon comprises a newly defined locus, the CdtLoc. The genetic boundaries of this region were defined by the comparison of different CDT+ and CDT− isolates. Key questions that remain to be resolved but which are the subject of current studies in this laboratory involve the determination of the environmental signals and signal transduction pathways that lead to the activation of CdtR and binary toxin production and the determination of the role of binary toxin production in diseases caused by this increasingly important human pathogen.
Acknowledgments
We thank R. Poon for technical assistance; J. Cheung for helpful discussions; P. Wright for assistance with the ADP-ribosyltransferase assay; G. Jenkin, J. Pepin, L. Grayson, and T. Riley for providing C. difficile isolates; N. Minton for kindly providing pMTL9301 and pMTL9361Cm; M. Popoff for the kind gift of Ia- and Ib-specific antibodies; and R. Stabler for providing access to the C. difficile genomic microarray data.
This research was funded by grant AI057637 from the National Institute of Allergy and Infectious Diseases and by Program Grant 284214 from the Australian National Health and Medical Research Council.
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Aktories, K., and A. Wegner. 1992. Mechanisms of the cytopathic action of actin-ADP-ribosylating toxins. Mol. Microbiol. 6:2905-2918. [DOI] [PubMed] [Google Scholar]
- 2.Barbut, F., B. Gariazzo, L. Bonne, V. Lalande, B. Burghoffer, R. Luiuz, and J. C. Petit. 2007. Clinical features of Clostridium difficile-associated infections and molecular characterization of strains: results of a retrospective study, 2000-2004. Infect. Control Hosp. Epidemiol. 28:131-139. [DOI] [PubMed] [Google Scholar]
- 3.Barth, H., K. Aktories, M. R. Popoff, and B. G. Stiles. 2004. Binary bacterial toxins: biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol. Mol. Biol. Rev. 68:373-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borriello, S. P. 1998. Pathogenesis of Clostridium difficile infection. J. Antimicrob. Chemother. 41(Suppl. C):13-19. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., T. Hundsberger, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29-38. [DOI] [PubMed] [Google Scholar]
- 7.Chang, S. Y., and K. P. Song. 2001. ADP-ribosylating binary toxin genes of Clostridium difficile strain CCUG 20309. DNA Seq. 12:115-120. [DOI] [PubMed] [Google Scholar]
- 8.Dupuy, B., and A. L. Sonenshein. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 27:107-120. [DOI] [PubMed] [Google Scholar]
- 9.Eggertson, L. 2004. Quebec strikes committee on Clostridium difficile. Can. Med. Assoc. J. 171:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geric, B., R. J. Carman, M. Rupnik, C. W. Genheimer, S. P. Sambol, D. M. Lyerly, D. N. Gerding, and S. Johnson. 2006. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J. Infect. Dis. 193:1143-1150. [DOI] [PubMed] [Google Scholar]
- 11.Geric, B., S. Johnson, D. N. Gerding, M. Grabnar, and M. Rupnik. 2003. Frequency of binary toxin genes among Clostridium difficile strains that do not produce large clostridial toxins. J. Clin. Microbiol. 41:5227-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goncalves, C., D. Decre, F. Barbut, B. Burghoffer, and J. C. Petit. 2004. Prevalence and characterization of a binary toxin (actin-specific ADP-ribosyltransferase) from Clostridium difficile. J. Clin. Microbiol. 42:1933-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hachler, H., B. Berger-Bachi, and F. H. Kayser. 1987. Genetic characterization of a Clostridium difficile erythromycin-clindamycin resistance determinant that is transferable to Staphylococcus aureus. Antimicrob. Agents. Chemother. 31:1039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayter, P. M., and J. W. Dale. 1984. Detection of plasmids in clinical isolates of Clostridium difficile. Microbios Lett. 27:151-156. [Google Scholar]
- 15.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 16.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 17.Hundsberger, T., V. Braun, M. Weidmann, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1997. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur. J. Biochem. 244:735-742. [DOI] [PubMed] [Google Scholar]
- 18.Just, I., J. Selzer, M. Wilm, C. von Eichel-Streiber, M. Mann, and K. Aktories. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500-503. [DOI] [PubMed] [Google Scholar]
- 19.Just, I., M. Wilm, J. Selzer, G. Rex, C. von Eichel-Streiber, M. Mann, and K. Aktories. 1995. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J. Biol. Chem. 270:13932-13936. [DOI] [PubMed] [Google Scholar]
- 20.Karlsson, S., B. Dupuy, K. Mukherjee, E. Norin, L. G. Burman, and T. Akerlund. 2003. Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect. Immun. 71:1784-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuijper, E. J., B. Coignard, and P. Tull. 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin. Microbiol. Infect. 12(Suppl. 6):2-18. [DOI] [PubMed] [Google Scholar]
- 22.Kyne, L., M. B. Hamel, R. Polavaram, and C. P. Kelly. 2002. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin. Infect. Dis. 34:346-353. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lizewski, S. E., D. S. Lundberg, and M. J. Schurr. 2002. The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect. Immun. 70:6083-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442-2449. [DOI] [PubMed] [Google Scholar]
- 26.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 27.Mani, N., D. Lyras, L. Barroso, P. Howarth, T. Wilkins, J. I. Rood, A. L. Sonenshein, and B. Dupuy. 2002. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J. Bacteriol. 184:5971-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald, L. C. 2005. Clostridium difficile: responding to a new threat from an old enemy. Infect. Control Hosp. Epidemiol. 26:672-675. [DOI] [PubMed] [Google Scholar]
- 29.McEllistrem, M. C., R. J. Carman, D. N. Gerding, C. W. Genheimer, and L. Zheng. 2005. A hospital outbreak of Clostridium difficile disease associated with isolates carrying binary toxin genes. Clin. Infect. Dis. 40:265-272. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 31.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura, S., K. Yamakawa, S. Nakashio, S. Kamiya, and S. Nishida. 1987. Correlation between susceptibility to chloramphenicol, tetracycline and clindamycin, and serogroups of Clostridium difficile. Med. Microbiol. Immunol. 176:79-82. [DOI] [PubMed] [Google Scholar]
- 33.Nikolskaya, A. N., and M. Y. Galperin. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30:2453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 35.O'Connor, J. R., D. Lyras, K. A. Farrow, V. Adams, D. R. Powell, J. Hinds, J. K. Cheung, and J. I. Rood. 2006. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 61:1335-1351. [DOI] [PubMed] [Google Scholar]
- 36.Oultram, J. D., H. Peck, J. K. Brehm, D. E. Thompson, T. J. Swinfield, and N. P. Minton. 1988. Introduction of genes for leucine biosynthesis from Clostridium pasteurianum into C. acetobutylicum by cointegrate conjugal transfer. Mol. Gen. Genet. 214:177-179. [DOI] [PubMed] [Google Scholar]
- 37.Palombo, E. A., K. Yusoff, V. A. Stanisich, V. Krishnapillai, and N. S. Willetts. 1989. Cloning and genetic analysis of tra cistrons of the Tra 2/Tra 3 region of plasmid RP1. Plasmid 22:59-69. [DOI] [PubMed] [Google Scholar]
- 38.Perelle, S., M. Gibert, P. Bourlioux, G. Corthier, and M. R. Popoff. 1997. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect. Immun. 65:1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popoff, M. R., E. J. Rubin, D. M. Gill, and P. Boquet. 1988. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect. Immun. 56:2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purdy, D., T. A. O'Keeffe, M. Elmore, M. Herbert, A. McLeod, M. Bokori-Brown, A. Ostrowski, and N. P. Minton. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46:439-452. [DOI] [PubMed] [Google Scholar]
- 41.Riley, T. V., J. P. Codde, and I. L. Rouse. 1995. Increased length of hospital stay due to Clostridium difficile associated diarrhoea. Lancet 345:455-456. [DOI] [PubMed] [Google Scholar]
- 42.Rood, J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333-360. [DOI] [PubMed] [Google Scholar]
- 43.Rupnik, M. 2007. Is Clostridium difficile-associated infection a potentially zoonotic and foodborne disease? Clin. Microbiol. Infect. 13:457-459. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd. ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 45.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 46.Shimizu, T., W. Ba-Thein, M. Tamaki, and H. Hayashi. 1994. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J. Bacteriol. 176:1616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simpson, L. L., H. Zepeda, and I. Ohishi. 1988. Partial characterization of the enzymatic activity associated with the binary toxin (type C2) produced by Clostridium botulinum. Infect. Immun. 56:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, A. 2005. Outbreak of Clostridium difficile infection in an English hospital linked to hypertoxin-producing strains in Canada and the US. Eur. Surveill. 10:E050630 2. [DOI] [PubMed] [Google Scholar]
- 49.Smith, C. J., S. M. Markowitz, and F. L. Macrina. 1981. Transferable tetracycline resistance in Clostridium difficile. Antimicrob. Agents Chemother. 19:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer, R. C. 1998. Clinical impact and associated costs of Clostridium difficile-associated disease. J. Antimicrob. Chemother. 41(Suppl. C):5-12. [DOI] [PubMed] [Google Scholar]
- 51.Spencer, R. C. 1998. The role of antimicrobial agents in the aetiology of Clostridium difficile-associated disease. J. Antimicrob. Chemother. 41(Suppl. C):21-27. [DOI] [PubMed] [Google Scholar]
- 52.Stabler, R. A., D. N. Gerding, J. G. Songer, D. Drudy, J. S. Brazier, H. T. Trinh, A. A. Witney, J. Hinds, and B. W. Wren. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stare, B. G., M. Delmee, and M. Rupnik. 2007. Variant forms of the binary toxin CDT locus and tcdC gene in Clostridium difficile strains. J. Med. Microbiol. 56:329-335. [DOI] [PubMed] [Google Scholar]
- 54.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 55.Stubbs, S., M. Rupnik, M. Gibert, J. Brazier, B. Duerden, and M. Popoff. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307-312. [DOI] [PubMed] [Google Scholar]
- 56.Sullivan, N. M., S. Pellett, and T. D. Wilkins. 1982. Purification and characterization of toxins A and B of Clostridium difficile. Infect. Immun. 35:1032-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terhes, G., E. Urban, J. Soki, K. A. Hamid, and E. Nagy. 2004. Community-acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene-positive isolates in Hungary. J. Clin. Microbiol. 42:4316-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]
- 60.Wust, J., and U. Hardegger. 1983. Transferable resistance to clindamycin, erythromycin, and tetracycline in Clostridium difficile. Antimicrob. Agents Chemother. 23:784-786. [DOI] [PMC free article] [PubMed] [Google Scholar]