Abstract
Bile acids are surface-active steroid compounds with toxic effects for bacteria. Recently, the isolation and characterization of a bacterium, Pseudomonas sp. strain Chol1, growing with bile acids as the carbon and energy source was reported. In this study, initial reactions of the aerobic degradation pathway for the bile acid cholate were investigated on the biochemical and genetic level in strain Chol1. These reactions comprised A-ring oxidation, activation with coenzyme A (CoA), and β-oxidation of the acyl side chain with the C19-steroid dihydroxyandrostadienedione as the end product. A-ring oxidizing enzyme activities leading to Δ1,4-3-ketocholyl-CoA were detected in cell extracts and confirmed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Cholate activation with CoA was demonstrated in cell extracts and confirmed with a chemically synthesized standard by LC-MS/MS. A transposon mutant with a block in oxidation of the acyl side chain accumulated a steroid compound in culture supernatants which was identified as 7α,12α-dihydroxy-3-oxopregna-1,4-diene-20-carboxylate (DHOPDC) by nuclear magnetic resonance spectroscopy. The interrupted gene was identified as encoding a putative acyl-CoA-dehydrogenase (ACAD). DHOPDC activation with CoA in cell extracts of strain Chol1 was detected by LC-MS/MS. The growth defect of the transposon mutant could be complemented by the wild-type ACAD gene located on the plasmid pBBR1MCS-5. Based on these results, the initiating reactions of the cholate degradation pathway leading from cholate to dihydroxyandrostadienedione could be reconstructed. In addition, the first bacterial gene encoding an enzyme for a specific reaction step in side chain degradation of steroid compounds was identified, and it showed a high degree of similarity to genes in other steroid-degrading bacteria.
Steroid compounds occur in eukaryotic organisms and fulfill various biological functions, such as serving as membrane components (e.g., cholesterol and phytosterols) or acting as hormones (e.g., testosterone and estradiol). Bile acids are surface-active steroid compounds that support the digestion of water-insoluble nutrients (18). The so-called primary bile acids cholate and chenodeoxycholate are synthesized in the liver, conjugated to glycine or taurine, stored in the gall bladder, and released into the duodenum as necessary. During their passage through the intestinal tract, a part of the bile acids is taken up and reused in the enterohepatic cycle (see reference 17 and references therein). The other part is excreted with urine or feces (12, 28).
In general, steroids occur in bacteria only as rare exceptions (3). Nevertheless, bacteria are capable of transforming eukaryotic steroid compounds, and they also are used as carbon and energy sources. In the intestinal tract, bacteria transform the primary bile acids cholate and chenodeoxycholate into the secondary bile acids deoxycholate and lithocholate, respectively, via reductive dehydroxylation (22, 29). Microbial bile acid transformation is also relevant for biotechnological production of steroid drugs and hormones (4, 23, 25). When bile acids are released into the environment, they can be degraded completely by numerous gram-positive and -negative bacteria (12).
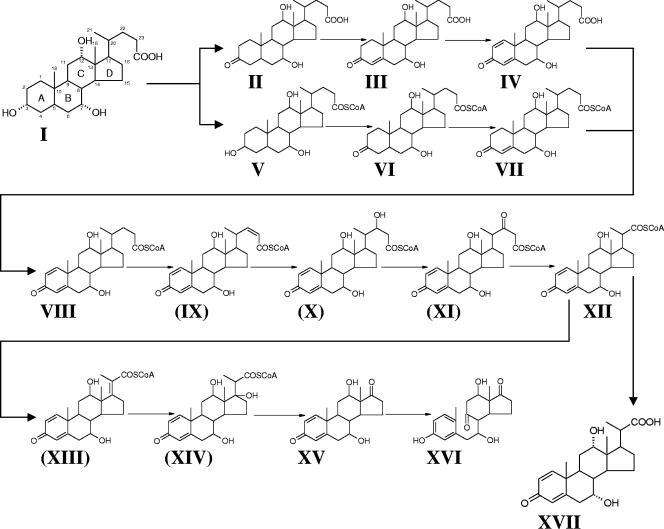
Recently, we reported the isolation and characterization of Pseudomonas sp. strain Chol1, which is able to grow aerobically with cholate and other bile acids as the sole source of carbon and energy and anaerobically with nitrate as an electron acceptor (28). Under anoxic conditions, strain Chol1 grows by transformation of cholate to 7,12-dihydroxy-1,4-androstadiene-3,17-dione (DHADD) as the end product. Under oxic conditions, strain Chol1 converts DHADD further to 3,7,12-trihydroxy-9,10-seco-1,3,5(10)-androstatriene-9,17-dione (THSATD) by an oxygenase-dependent reaction. The secosteroid THSATD is then degraded to CO2. The degradation pathway in strain Chol1 thus follows the unifying scheme proposed by Hayakawa (12). The conversion of cholate into DHADD proceeds via A-ring oxidation and degradation of the acyl side chain (Fig. 1). While the enzymatic reactions involved in A-ring oxidation have been studied in great detail since the 1960s (26, 32), the degradation of steroid side chains has been studied mainly on a phenomenological level by extracting and identifying degradation intermediates from whole steroid-transforming cultures (23). Only recently, genes in Rhodococcus sp. strain RHA1 have been proposed to be involved in β-oxidation of the side chain of steroids because their transcription was up-regulated during growth with cholesterol (34).
FIG. 1.
Proposed degradation pathway for cholate (compound I) in Pseudomonas sp. strain Chol1. The following compounds have been experimentally detected in this study: II, 3-ketocholate; III, Δ4-3-ketocholate (P1); IV, Δ1,4-3-ketocholate (P2); V, cholyl-CoA (P5); VI, 3-ketocholyl-CoA; VII, Δ4-3-ketocholyl-CoA; VIII, Δ1,4-3-ketocholyl-CoA; XII, DHOPDC-CoA (P6); and XVII, DHOPDC (P4). As the position of the double bond in the A-ring has not yet been identified, compounds III and VII could also be Δ1-3-ketocholate and Δ1-3-ketocholyl-CoA, respectively. Compounds XV (DHADD) and XVI (THSATD) were identified in a previous study (28). The compounds indicated by numbers in brackets (IX, X, XI, XIII, and XIV) have not been detected so far. DHOPDC (XVII) is a dead-end metabolite in the mutant strain R1.
Growth with or in the presence of bile acids is a challenge for bacteria because, as surface-active detergents, bile acids are toxic for bacteria (1, 13). Enteric bacteria growing in the presence of bile acids possess energy-requiring resistance mechanisms, such as diffusion barriers and efflux pumps (11). We have shown that cholate is toxic for strain Chol1 and that this strain requires an intact outer membrane and energy-dependent protection mechanisms to survive exposure to cholate (28). In addition, we found also that the degradation intermediates DHADD and THSATD are toxic. As strain Chol1 is apparently confronted with toxic compounds during cholate degradation, we investigated whether strain Chol1 possesses additional adaptive strategies for growth with cholate as a carbon and energy source. An intriguing observation during growth with cholate is the transient accumulation of degradation intermediates in culture supernatants. In the first half of exponential growth, strain Chol1 converts cholate completely into THSATD, which accumulates in the medium and is degraded during the second half of exponential growth. We hypothesized that this extracellular accumulation represents an adaptive strategy to keep the level of the intracellular pool of toxic intermediates low (28). If this is true, the cholate degradation pathway must be strictly controlled on the levels of gene expression and/or enzyme activity. Furthermore, efflux and uptake systems for degradation intermediates must be postulated. A prerequisite to understanding the potential adaptive strategies of strain Chol1 for growth with cholate is the reconstruction of the degradation pathway in vitro, thereby identifying individual enzymatic steps with their subcellular localization and their regulation on the expression and activity level. However, although bile acid degradation is widespread among bacteria and of biotechnological importance, there is only limited knowledge about these physiological aspects of bacterial growth with bile acids. Thus, the goal of this study was to detect enzymatic steps leading from cholate to DHADD in cell extracts. In particular, we wanted to verify the presumptive β-oxidation pathway for degradation of the acyl side chain. This goal was approached using biochemical and genetic methods.
MATERIALS AND METHODS
Bacteria, media, and growth experiments.
Pseudomonas sp. strain Chol1 was grown in the phosphate-buffered mineral medium MMChol with cholate and succinate as the carbon and energy sources as described previously (28). The transposon mutant strain R1 and all other transposon mutants derived from strain Chol1 were grown under the same conditions in the presence of kanamycin (10 μg ml−1) for selection of the mini-Tn5 Km1 transposon (6). Strain R1 harboring pBBR1MCS-5 (24) and pBBR(ACAD) (where ACAD is acyl-coenzyme A [CoA]-dehydrogenase) were grown in MMChol medium with gentamicin (20 μg ml−1) and kanamycin (10 μg ml−1). Pseudoalteromonas haloplanktis strain TAC125 was grown in TYP medium (16 g of Bacto tryptone, 16 g of yeast extract, 5 g of NaCl, and 2.5 g of K2HPO4 per liter) as described elsewhere (27). The Escherichia coli strains JM109, DH5α pBBR, DH5α pBBR(ACAD), and HB101 pRK600 were grown in Luria-Bertani (LB) medium (30), supplemented with chloramphenicol (30 μg ml−1) for strain HB101 pRK600 or with gentamicin (20 μg ml−1)for strains DH5α pBBR and DH5α pBBR(ACAD). Growth experiments with Pseudomonas strains Chol1, R1, R1 pBBR, and R1 pBBR(ACAD) were performed as described previously (28).
Conjugational plasmid transfer and transposon mutagenesis.
Conjugational plasmid transfer into Pseudomonas strains Chol1 and R1 was performed via triparental mating. For transposon mutagenesis of strain Chol1, the donor strain E. coli CC118λpir (14) with the suicide vector pUT(mini-Tn5Km1) (6) and the helper strain E. coli HB101 pRK600 (14) were used. The donor strain was grown overnight in LB medium containing kanamycin (10 μg ml−1). The recipient strain Chol1 was grown in LB medium, and the helper strain HB101 pRK600 was grown in LB medium with chloramphenicol (30 μg ml−1) overnight. After determination of the optical density at 600 nm, all strains were washed in prewarmed LB medium to remove antibiotics, combined in a total volume of 250 μl with a cell number ratio of 1 (donor):1 (helper):3 (recipient), and washed again. After resuspension in 100 μl of LB medium, cells were transferred to a filter with 2.5-cm diameter and 0.2-μm pore size (Schleicher and Schuell), placed on an LB agar plate, and incubated for 4 h at 30°C. Then, cells were washed off the filter with and finally resuspended in 5 ml of MMChol medium without a carbon source. For selecting transposon-carrying clones of strain Chol1, cells were spread on MMChol agar plates containing kanamycin (10 μg ml−1) and 12 mM succinate as a carbon source.
For conjugational transfers of pBBR and pBBR(ACAD) into strain R1, the donor strains DH5α pBBR and DH5α pBBR(ACAD), the helper strain HB101 pRK600, and the recipient strain R1 were grown overnight in LB medium containing gentamicin (20 μg ml−1), chloramphenicol (30 μg ml−1), and kanamycin (10 μg ml−1), respectively. The mating was performed as described above with the following modifications: the different strains were combined in 2.5 ml of LB medium, washed with LB medium, and finally resuspended in 2 ml of LB medium. For selecting plasmid-carrying strain R1 clones, cells were spread on LB medium agar plates with kanamycin (10 μg ml−1) and gentamicin (20 μg ml−1).
Molecular biological methods.
Genomic DNA of strain R1 was isolated (Puregene DNA Isolation Kit; Gentra), partially digested with 125 mU of Sau3AI at 37°C for 1 h, and separated on a 1% agarose gel. Fragments of 2 to 4 kb were purified from the gel (Qiaquick Gel Extraction Kit; QIAGEN) and ligated into the BamH1 restriction site of pUCP18 (35). These plasmids were transformed into competent E. coli strain JM109 cells (Promega). Positive clones were identified by blue-white screening on LB medium containing kanamycin (10 μg ml−1) as described previously (30).
Primers used for PCR are listed in Table 1. Arbitrarily primed PCRs with primer pairs 1 and 3 (initial PCR) and 2 and 4 (nested PCR) were performed as described previously (7) with the following modifications: for the initial PCR, 5% dimethyl sulfoxide (DMSO) was added; for nested PCR, 5% DMSO and 1.5 mM MgCl2 were added; only 1 μl instead of 5 μl of PCR product was transferred as template DNA for nested PCR. For amplification of the ACAD gene from strain Chol1 with paired primers 5 and 6, the following PCR protocol was used: 2 min of initial denaturing at 93°C, followed by 35 cycles of 30 s of denaturing at 93°C, 30 s of annealing at 60.4°C, and 1.5 min of elongation at 72°C, followed by 5 min of final elongation at 72°C. The PCR mixture contained 5% DMSO. PCR products were purified from gels as described above and ligated into the cloning vector pCR2.1 (TA Cloning Kit; Invitrogen) which was transformed into E. coli strain InvαF′ (Invitrogen). After plasmid isolation (Qiaprep Spin Miniprep Kit; QIAGEN), the ACAD gene was excised as an SalI-BamH1 fragment and ligated into pBBR1MCS-5 (see above) which was transformed into E. coli strain DH5α. Custom DNA sequencing was performed by GATC in Konstanz, Germany.
TABLE 1.
Primers used in this study
| Primer no. | Name | Tm (°C)a | Sequence (5′ to 3′) |
|---|---|---|---|
| 1 | Tn5rev | 63.7 | GTACCGAGCTCGAATTCGGCC |
| 2 | ACADrev | 67.6 | GTGGTGCGCCGGCGATGTTGG |
| 3 | ARB1 | 71.9 | GGCACGCGTCGACTAGTACNNNNNNNNNGATAT |
| 4 | ARB2 | 61.0 | GGCACGCGTCGACTAGTAC |
| 5 | ACADBamH1rev | 66.3 | GGCAAGTATCGGATCCGGAATCTC |
| 6 | ACADsal1fw | 65.4 | GTACCTGATGTCGACATATGTTCGTTGATC |
Thermal denaturation midpoint temperature.
Preparation of cell suspensions and cell extracts.
Cells of strains Chol1 and R1 in late exponential growth phase were harvested via centrifugation at 9,000 × g for 10 min at 4°C. For cell suspension experiments, cells were washed with MMChol medium without a carbon source, finally resuspended in filter-sterilized culture supernatants, and further processed as described previously (28). For preparation of cell extracts, cells were washed in 50 mM potassium phosphate buffer (pH 7.0), finally resuspended in a small volume of this buffer, and broken by three to five passages through a cooled French press (Aminco) at 138 MPa. Homogenates were centrifuged at 17,900 × g for 10 min at 4°C to separate the cell extract from cell debris. Cell extracts were further separated into cytosol and membrane fractions by ultracentrifugation at 100,000 × g for 60 min at 4°C. The membrane fraction was washed with the aforementioned buffer and submitted to a further ultracentrifugation. All cell extracts were immediately used for enzyme assays or stored at −20°C.
Detection of enzyme activities.
Enzyme assays for cholate oxidation contained 50 mM Tris-HCl (pH 8.0), 1 mM NAD(P)+ or K3Fe[CN]6, and cell extract (0.6 to 0.8 mg) and were started by the addition of 1 mM cholate. Cholate-dependent reduction of NAD(P)+ and K3Fe[CN]6 was measured spectrophotometrically at 365 nm (ɛ = 3.4 mM−1 cm−1) and at 436 nm (ɛ = 0.7 mM−1 cm−1), respectively. For detecting products of cholate oxidation, samples withdrawn immediately after the reaction was started and at defined time intervals thereafter were subsequently analyzed by high-performance liquid chromatography (HPLC) (see below). For production of 3-ketocholate as a reference compound, 0.3 U of α,β-hydroxysteroid dehydrogenase from Pseudomonas testosteroni (Sigma) was employed instead of cell extract. Enzyme assays for acylate-CoA ligase activities contained 50 mM morpholinepropanesulfonic acid buffer (pH 7.8), 1 mM CoA, 1 mM ATP, 2.5 mM MgCl2, and cell extract (0.6 to 0.8 mg) and were started by the addition of 1 mM cholate or of ca. 300 μM 7α,12α-dihydroxy-3-oxopregna-1,4-diene-20-carboxylate (DHOPDC). Samples withdrawn immediately after the reaction was started and at defined time intervals thereafter were subsequently analyzed by HPLC or LC-tandem mass spectrometry (LC-MS/MS) (see below).
HPLC.
All steroid compounds were analyzed with a reversed-phase HPLC system equipped with UV/visible light diode array detector as described previously (28). The flow rate in all HPLC methods was 1 ml min−1. For quantification of cholate, an isocratic method with 70% K-Na-phosphate buffer (10 mM; pH 7.1) (eluent A) and 30% acetonitrile (eluent B) was used. For detection of steroid degradation intermediates and CoA esters, a gradient method was applied, starting with 20% eluent B for 2 min, rising to 90% eluent B within 9 min, and returning to 20% eluent B within 1 min, followed by an equilibration of 4 min.
Spectroscopic methods.
UV/visible light spectra were acquired either with a diode array detector during HPLC analysis (see above) or with a double-beam spectrophotometer (Uvikon 930; Kontron Instruments). Infrared spectra were acquired with a Fourier transform infrared spectrometer (FTS 70; Bio-Rad) with a resolution of one wave number. Samples were dissolved in methanol and measured in a CaF2 cuvette (diameter, 0.1 mm).
Mass spectrometric analysis of CoA esters was performed on an LTQ Orbitrap LC-MS/MS instrument (Thermo Scientific) using electrospray in the positive-ion mode. Chromatographic separation was done using an HTS PAL autosampler, together with a Rheos 2200 HPLC and a CC 125/2 Nucleosil 120-3 C18 column from Macherey Nagel. Acetic acid (0.5‰ [vol/vol] in H2O) and Ultramark 1621 (for lock masses) was added postcolumn at 20 μl min−1 using a Bischoff 2200 HPLC pump (Bischoff GmBH). The following gradient system was used: 10 mM ammonium acetate, (pH 7; eluent A) and acetonitrile (eluent B) at a flow rate 250 μl min−1, starting with 5% eluent B for 2 min, followed by a linear increase to 90% eluent B in 12 min, and then returning to initial conditions in 1 min and reequilibration for 4 min, giving a total run time of 19 min. The injection volume was 50 μl. The scan range was from 50 to 2,000 Da at a mass resolution of 100,000 at mass 400. The needle voltage was set to 4,500 V, the capillary voltage was set to 44 V, the tube lens was set to 250 V, and the capillary temperature was set to 300°C. The normalized collision energy was set to 35 V for MS/MS experiments, with an isolation width of 2 Da.
LC/MS spectra with free steroid compounds were acquired on an API4000 LC-MS/MS instrument (Applied Biosystems) using electrospray in both positive- and negative-ion modes. Chromatographic separation was done using an Agilent HP1100 Series system and a CC 125/2 Nucleosil 120-3 C18 column from Macherey Nagel. Acetic acid addition and chromatographic separation were performed as described above. The scan range was from 50 to 1,300 Da in 0.8 s for a full scan and from 100 to 1,200 for MS/MS experiments. The needle voltage was set to 4,500 V (with electrospray ionization [+ESI]) or −4500 V (without ESI [−ESI]); the declustering potential was 60 V (+ESI) or −70 V (−ESI), and the temperature was set to 350°C. Collision energy was set to 30 V (+ESI) or −30 V (−ESI) for MS/MS experiments. The wavelength simultaneously monitored was 245 nm.
Nuclear magnetic resonance (NMR) measurements were carried out with HPLC-purified DHOPDC dissolved in D2O to a final concentration of ca. 10 mM. All NMR spectra were recorded at 300 K on a Bruker DRX 600 MHz spectrometer equipped with a 5-mm TXI-H/C/N inverse triple resonance probe with actively shielded Z-gradient. The proton one-dimensional spectrum was recorded with a spectral width of 16 ppm and 32,000 complex points. Residual HDO was suppressed by presaturation during the recycle delay of 2 s. Homonuclear two-dimensional correlated spectroscopy (COSY) and rotating frame nuclear Overhauser effect spectroscopy (ROESY) experiments were recorded with 4,000 complex points in the detected dimension and 256 complex points in the indirect dimension. ROESY spin-lock was achieved with a continuous wave pulse of 1 kHz field strength and a duration of 250 ms. The heteronuclear single-quantum coherence (HSQC) and heteronuclear multiple bond correlation (HMBC) spectra were recorded with 1,000 and 4,000 complex points, respectively, in the proton dimension and 256 complex points in the 13C dimension. The spectral width in the carbon dimension was 130 ppm and 185 ppm, respectively. All spectra were processed and analyzed with Bruker's Xwinnmr (version 3.6) and Topspin (version 1.3) software. Usually, zero-filling was applied to double the number of real points in each dimension. Chemical shifts were referenced to the HDO resonance at 4.7 ppm. Chemical shift assignments for 13C were determined indirectly from HSQC and HMBC spectra.
Preparation of DHOPDC.
Culture supernatants of strain R1 were acidified (pH 1 to 2) with 5 M HCl and extracted twice with dichloromethane. After dichloromethane was evaporated and the solid matter was dissolved in a small volume of distilled water, DHOPDC was purified by HPLC. DHOPDC-containing HPLC fractions were acidified and extracted twice with ethylacetate. After a drying step with MgSO4 and evaporation with ethylacetate, DHOPDC was dissolved in solvents appropriate for further analysis.
Synthesis of cholyl-CoA.
Cholyl-CoA was synthesized via a mixed anhydride procedure as described previously (31). Formation of 14.2 mg of cholyl-CoA from 30 mg of sodium cholate (Fluka) and 50 mg of CoA (trilithium salt from yeast; Sigma) was confirmed by LC-MS/MS.
Nucleotide sequence accession number.
The sequence of the ACAD gene found in this study was deposited in the GenBank database under accession number EF530217.
RESULTS
In vitro detection of A-ring oxidizing reactions.
To reconstruct the cholate degradation pathway of Pseudomonas sp. strain Chol1 in vitro, we searched for A-ring oxidizing enzyme activities in cell extracts. Cholate-dependent reduction of NAD+ and of K3Fe(CN)6 with specific activities of 86 ± 3 mU mg of protein−1 and of 69 ± 4 mU mg of protein−1 were detected in the cytosolic fraction of cholate-grown cells. HPLC analysis of assays containing NAD+ revealed that cholate was converted to a product that eluted 0.4 min earlier than cholate. This product coeluted with a compound that was formed in a reference reaction mixture with commercial α,β-hydroxysteroid dehydrogenase which is known to oxidize cholate to 3-ketocholate in an NAD+-dependent reaction. The UV spectra of the products formed in the cytosolic fraction and in the reference reaction were identical. These results confirmed that cholate was oxidized to 3-ketocholate by a soluble enzyme of strain Chol1. Cholate-dependent reduction of NADP+ was detected in cell extracts of cholate-grown cells. In this reaction, in addition to the products formed in the NAD+-dependent reaction, another product was formed that has not been identified so far.
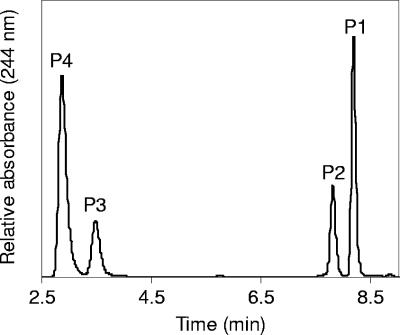
HPLC analysis of assays containing K3Fe(CN)6 showed that cholate was transformed into two products, P1 and P2 (Fig. 2), that eluted earlier than cholate (retention time, 9 min) and showed UV spectra with a maximum at 245 nm, which is characteristic for steroid compounds with a Δ1/Δ4-3-keto or Δ1,4-3-keto structure (28). These two products had the same UV spectra and coeluted with two peaks that accumulated in the culture supernatants of the mutant strain R1 (see below). LC-MS analysis revealed deprotonated molecular ions ([M-H]−) with m/z 403.0 and m/z 401.3 for P1 and P2, respectively. In MS/MS analysis, major fragments (F−) with m/z 123.1 and m/z 121.1 were generated from P1 and P2, respectively. The fragment with m/z 121.1 has been shown to originate from an A-ring with Δ1,4-3-keto structure (28). Accordingly, the m/z 123.1 fragment of P1 would correspond to an A-ring with a Δ1- or a Δ4-3-keto structure. In summary, the UV and MS data indicate that P1 and P2 were Δ1/Δ4-3-ketocholate (mass, 404 Da) and Δ1,4-3-ketocholate (mass, 402 Da), respectively. Thus, we could detect all reactions leading to A-ring oxidation of cholate in cell extracts of strain Chol1 (Fig. 1). NAD+ served as the physiological electron acceptor for oxidation of the 3-hydroxy group, and the addition of the artificial electron acceptor K3Fe(CN)6 enhanced unsaturation of the A-ring. When no exogenous electron acceptors were added, endogenous electron acceptors within the cell extract enabled a slight formation of 3-ketocholate and Δ1/Δ4-3-ketocholate (Fig. 3) and very low formation of Δ1,4-3-ketocholate (not shown). No cholate-dependent electron acceptor reductions or electron acceptor-dependent cholate transformations were detectable in heat-treated (10 min at 80°C) cell extracts or in cell extracts of succinate-grown cells.
FIG. 2.
HPLC chromatogram of a culture supernatant of Pseudomonas sp. strain R1 grown with succinate in the presence of cholate. The products P4, P3, P2, and P1 were identified as DHOPDC, its Δ1 or Δ4 monoene derivative, Δ1,4-3-ketocholate, and its Δ1 or Δ4 monoene derivative, respectively.
FIG. 3.
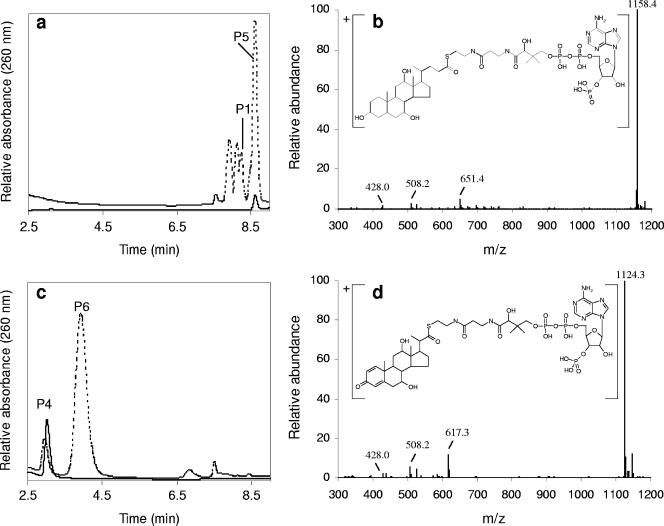
Analysis of bile acid CoA ester formation. (a) Overlaid HPLC chromatograms of a sample from a cholate-CoA ligase assay (dotted trace) and of chemically synthesized cholyl-CoA (solid trace). P5 was identified as cholyl-CoA. P1 represents the monoene derivative of Δ1,4-3-ketocholate (Fig. 2). (b) Mass spectrum and chemical structure of ([M+H]+) of cholyl-CoA (P5). (c) Overlaid chromatograms of a DHOPDC-CoA ligase assay (dotted trace) and a control assay in which cell extract had been omitted (solid trace). P6 was identified as DHOPDC-CoA. P4 represents DHOPDC (Fig. 2). (d) Mass spectrum and chemical structure of ([M+H]+) of DHOPDC-CoA (P6).
In vitro detection of cholate-CoA ligase activity.
CoA activation of cholate is the prerequisite for the postulated β-oxidation of the acyl side chain. We searched for cholate-CoA ligase activity in cell extracts of cholate-grown cells by HPLC analysis. We detected the cholate-, CoA-, ATP-, and cell extract-dependent formation of a product, P5, which showed UV spectra similar to CoA. In addition, the UV spectrum of P5 showed a slight shoulder at 233 nm, which is characteristic for CoA esters (31). P5 had the same UV absorption spectrum and retention time in HPLC analysis as chemically synthesized cholyl-CoA (Fig. 3a). LC-MS analysis of synthesized cholyl-CoA and of P5 revealed ions with m/z 1,158.4 ([M+H]+) (Fig. 3b) for both compounds. From this precursor ion, a specific fragment (F+) with m/z 651.4 was generated consisting of cholate attached to the pantetheine residue of CoA (5, 9). Furthermore, two CoA-specific fragments (F+) with m/z 428.0 (5, 9) and m/z 508.2 were generated, corresponding to 3-phospho-AMP and 3-phospho-ADP, respectively. These spectroscopic data confirmed the in vitro formation of cholyl-CoA with a molecular mass of 1,157.4 Da for the uncharged molecule (C45H74N7O20P3S). Under the assay conditions applied, the formation of cholyl-CoA occurred instantaneously within the first minute after the start of the reaction. Cholate-CoA ligase activity was localized in the cytosolic fraction and not present in cell extracts of succinate-grown cells.
In addition to cholyl-CoA, three products with UV absorption spectra similar to CoA were formed in the cholate ligase reaction assay (Fig. 3a). A small peak forming a shoulder at the beginning of the cholyl-CoA peak showed an ion ([M+H]+) with m/z 1,156.4, and two peaks eluting before the monoene derivative of Δ1,4-3-ketocholate (P1) showed ions with m/z 1,154.4 and m/z 1,152.4. From these precursor ions specific fragments (F+) with m/z 649.4, 647.4, and 645.4, respectively, were generated, corresponding to the aforementioned specific fragment with m/z 651.4 for cholyl-CoA. These spectroscopic data indicated that the three products correspond to CoA esters of 3-ketocholate (mass, 1,155.4 Da), Δ1/Δ4-3-ketocholate (mass, 1,153.4 Da), and Δ1,4-3-ketocholate (mass, 1,151.4 Da), respectively.
Characterization of the transposon mutant strain R1.
To identify genes involved in cholate degradation, strain Chol1 was subjected to random mutagenesis by insertion of the transposon mini-Tn5 Km1 as described in Materials and Methods. Of about 700 transposon mutants that were screened, four mutants were obtained that could not grow with cholate as a sole source of carbon and energy. These mutants grew with succinate in the presence of cholate. During growth with succinate, three mutants transformed cholate into four unknown compounds, while with one mutant the cholate concentration remained unchanged. HPLC analysis of culture supernatants of the cholate-transforming mutants revealed an identical product pattern for all three strains. One of these transposon mutants, strain R1, was analyzed further. The four accumulating transformation products eluted as two pairs after about 5 min (P4 and P3) and after about 8 min (P2 and P1) (Fig. 2). The UV spectra of all four products had an absorption maximum at 245 nm, indicative of steroid compounds with an oxidized A-ring as described above. For further analysis, the supernatant was analyzed by LC-MS/MS. The identification of P1 and P2 as Δ1/Δ4-3-ketocholate and Δ1,4-3-ketocholate has been described above. LC-MS analysis of the two other products revealed ions ([M-H]−) with m/z 373.0 for P4 and m/z 375.0 for P3. In MS/MS analysis, the aforementioned major fragments (F−) with m/z 121.1 and m/z 123.3 were found for P4 and P3, respectively, indicating that these two products differed in the number of double bonds in the A-ring. The further identification of P4 is described below.
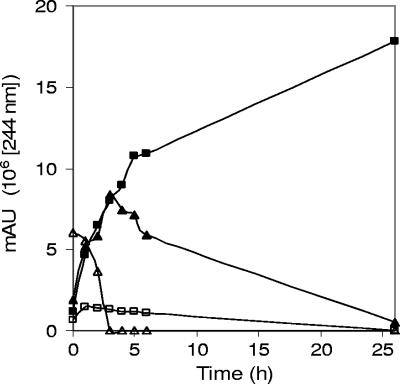
To investigate whether the four accumulating compounds were dead-end metabolites or whether they were further converted to common end products, dense cell suspensions of strain R1 were incubated with supernatants from succinate-grown cultures which had converted cholate completely into the four products P4, P3, P2, and P1. These experiments showed that P3, P2, and P1 were eventually all converted into P4 (Fig. 4). P4 itself was not further transformed or degraded by strain R1. In contrast, the wild-type strain Chol1 could grow in culture supernatants of strain R1, thereby degrading all four accumulated steroid compounds (not shown). To investigate whether strain R1 could utilize DHADD, cell suspensions of strain R1 were incubated in filter-sterilized supernatants of strain Chol1 grown anaerobically with cholate, as described previously (28). In these experiments, strain R1 converted DHADD completely into THSATD, which was later degraded as well (not shown). These physiological experiments showed that the cholate degradation pathway in strain R1 must be interrupted at a step between A-ring oxidation and DHADD hydroxylation.
FIG. 4.
Cell suspension experiment with Pseudomonas sp. strain R1 in culture supernatant of strain R1 after growth with succinate in the presence of cholate. Conversion of Δ1/Δ4-3-ketocholate (▵), Δ1,4-3-ketocholate (▴), and the monoene derivative of DHOPDC (□) into DHOPDC (▪). AU, arbitrary units.
Identification and CoA activation of DHOPDC (P4).
The final accumulating steroid compound in culture supernatants of strain R1, P4, was purified by preparative HPLC and characterized by infrared (IR) and NMR spectroscopy. IR spectroscopy of P4 in methanol revealed a signal at 1,659 cm−1, which is indicative of the 3-keto group as described previously (28). Further IR signals of P4 in methanol at 1,716 cm−1, 1,725 cm−1, and 1,742 cm−1 disappeared if the sample was alkalized, indicating the presence of a carboxyl group (15).
Table 2 shows the complete chemical shift assignment of P4. Proton chemical shift assignment was straightforward for most of the molecule from the COSY spectrum since all protons form an interconnected spin system. The resonance of the H2 proton at 6.26 ppm could be correlated via a 4J coupling to H4, which in turn shows 4J coupling to the allylic protons H6β of the B-ring. From here on, all protons, except the two methyl groups at positions 18 and 19, are linked via 2J or 3J couplings leading to mostly strong COSY correlations. Our assignment was greatly facilitated by the HSQC spectrum's yielding consistent assignments for the diunsaturated, oxygenated A-ring, the two hydroxylated CH-groups at positions 7 and 12, and the aliphatic chain attached to C-17. The HSQC data also allowed unambiguous identification of the diastereotopic methylene protons at positions 6, 11, 15, and 16, especially since there was significant spectral overlap of CH2-15 and −16 with each other and with H9 and H14. The methyl groups 18 and 19 were connected to rings A/B and C/D, respectively, by HMBCs. All chemical shifts of quarternary carbons were obtained by HMBC as well and are consistent with the structure of DHOPDC. Stereospecific assignments of methylene protons 6, 11, and 15 as well as the stereochemistry of the hydroxylated CH groups were obtained from the ROESY spectrum which, in total, shows a pattern of through-space contacts totally consistent with the presented assignment. Our chemical shift assignment of DHOPDC isolated from Pseudomonas sp. strain Chol1 agrees well with limited 1H chemical shifts of this compound published earlier (33). This structure fits the molecular mass of 373.0 Da determined for [M-H]− by LC-MS analysis (see above). P3 with a mass of 375.0 Da for the [M-H]− would thus be the monoene derivative of DHOPDC.
TABLE 2.
1H and 13C chemical shifts of DHOPDC (P4)a
| Atom no. | Shifts of DHOPDC from the present study (ppm)
|
δH of the DHOPDC methyl ester from a previous study (ppm)b | |
|---|---|---|---|
| δC | δH | ||
| 1 | 160.6 | 7.36 | 7.06 |
| 2 | 125.6 | 6.26 | 6.30 |
| 3 | 188.8 | ||
| 4 | 125.1 | 6.13 | 6.20 |
| 5 | 171.3 | ||
| 6 | 40.2 | 2.81 (β)/2.48 (α) | |
| 7 | 69.7 | 4.08 | 4.10 |
| 8 | 39.0 | 1.86 | |
| 9 | 38.5 | 1.71 | |
| 10 | 44.1 | ||
| 11 | 29.5 | 2.00 (β)/1.82 (α) | |
| 12 | 72.0 | 3.99 | 3.96 |
| 13 | 46.2 | ||
| 14 | 41.7 | 1.68 | |
| 15 | 22.6 | 1.67 (β)/1.25 (α) | |
| 16 | 25.8 | 1.69/1.28 | |
| 17 | 43.9 | 1.99 | |
| 18 | 11.8 | 0.76 | 0.81 |
| 19 | 17.2 | 1.21 | 1.25 |
| 20 | 42.9 | 2.36 | |
| 21 | 15.6 | 1.15 | 1.27 |
| 22 | 183.0 | ||
The next expected enzymatic step in DHOPDC degradation would be activation with CoA. In cell extracts of strain Chol1, we detected the CoA-, ATP-, and DHOPDC-dependent formation of the product P6 (Fig. 3c) which showed a UV absorption spectrum similar to CoA. LC-MS/MS analysis of P6 revealed an ion ([M+H]+) with m/z 1,124.3 (Fig. 3d). From this precursor ion a fragment (F+) with m/z 617.3, corresponding to DHOPDC attached to the pantetheine part of CoA, and the two aforementioned specific CoA fragments (F+) with m/z 508.2 and 428.0 were generated. These spectroscopic data confirmed the in vitro formation of DHOPDC-CoA with a molecular mass of 1,123 Da for the uncharged molecule (C43H64N7O20P3S).
Identification and complementation of the inactivated gene in strain R1.
The complete sequence of the inactivated gene in strain R1 was identified in two steps. In the first step, we screened a gene library of strain R1 in E. coli strain JM109 for kanamycin-resistant clones. We could identify a 972-bp fragment of a gene that was interrupted by the mini-Tn5Km1 transposon. The transposon was flanked by the characteristic 9-bp duplications (2). The sequence of the interrupted gene lacked a start codon although a stop codon was found. Comparison of the deduced amino acid sequence showed the highest similarity to an ACAD of P. haloplanktis strain TAC125. The predicted protein of strain TAC125 had a length of 389 amino acids (aa) while the predicted protein of strain Chol1 had a length of 324 aa, suggesting that 65 aa were missing at the N terminus. In the second step, we employed arbitrarily primed PCR to obtain the missing part of the sequence. With this method, we obtained a 328-bp DNA fragment encoding the missing 195 bp including the start codon. Furthermore, this fragment had a 38-bp overlapping part with the cloned fragment of strain R1 and another 95 bp upstream of the start codon. Thus, the complete gene was 1,170 bp long, and the transposon was inserted after bp 957 in strain R1. The deduced amino acid sequence of the complete gene showed 77% sequence identity to the ACAD of P. haloplanktis strain TAC125 (Fig. 5).
FIG. 5.
Alignment of the amino acid sequences of the putative ACAD gene in Pseudomonas sp. strain Chol1 (Acad_Chol1) and its homologous ACAD gene in P. haloplanktis TAC 125 (PSHAa0888) performed with the software DNASTAR. Conserved residues are in boldface.
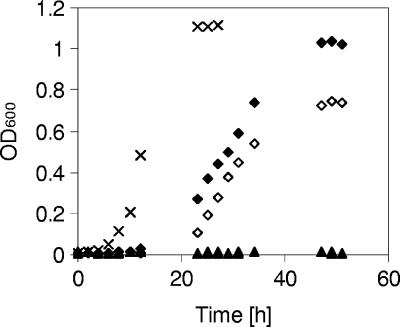
Complementation of strain R1 with the intact ACAD gene from strain Chol1 contained on the plasmid pBBR1MCS-5 showed that the complemented strain was able to grow with cholate while the vector control strain did not grow (Fig. 6). Selection with gentamicin was not necessary to maintain the complementing effect of pBBRMCS-5(ACAD).
FIG. 6.
Growth of Pseudomonas sp. strains Chol1 (×), R1 pBBR (triangles), and R1 pBBR(ACAD) (diamonds) with 2 mM cholate. Strains R1 pBBR and R1 pBBR(ACAD) were incubated in the presence of kanamycin without (closed symbols) or with gentamicin (open symbols). Open triangles overlap with closed triangles.
Other, similar deduced amino acid sequences were found in other bacteria such as Comamonas testosteroni strain KF-1 (69% sequence identity) and Rhodococcus sp. strain RHA1 (50% sequence identity) as well as several Nocardia and Mycobacterium strains (49 to 50% sequence identities). Members of these three genera are known to degrade steroid compounds (19, 23, 34) while, to our knowledge, degradation of steroids has not been investigated for P. haloplanktis. We tested P. haloplanktis strain TAC125 for cholate degradation and found that it could grow with cholate as a carbon and energy source. HPLC analysis of supernatant samples collected during growth with cholate revealed the transient accumulation of degradation intermediates also found with strain Chol1 (not shown).
DISCUSSION
The goal of our study was the in vitro reconstruction of initial reactions in the metabolic pathway for degradation of the bile acid cholate in Pseudomonas sp. strain Chol1, namely A-ring oxidation, CoA-activation, and β-oxidation of the acyl side chain. The shortest route for conversion of cholate into DHADD involves 10 steroid intermediates of which we detected five, namely cholyl-CoA and the free acids and CoA esters of 3-ketocholate, Δ1/Δ4-3-ketocholate, Δ1,4-3-ketocholate, and DHOPDC. The identification of DHOPDC as an intermediate of β-oxidation strongly supports the presumptive structures of the three intermediates preceding and the two intermediates following DHOPDC-CoA formation. Based on these results, we propose a pathway for the initial reactions of cholate degradation in Pseudomonas sp. strain Chol1 as shown in Fig. 1.
All enzyme activities involved in oxidation of the A-ring with Δ1,4-3-ketocholate as the final product were induced by cholate and localized in the cytosolic fraction. While NAD+ was found to be the physiological electron acceptor for oxidation of cholate to 3-ketocholate in strain Chol1, electron acceptors for further oxidation of the A-ring are not yet known. Often, 3-ketosteroid Δ-dehydrogenases are flavoproteins (8, 32), suggesting that they may transfer electrons to quinones or cytochromes. CoA activation of cholate was catalyzed by an inducible cholate-CoA ligase localized in the cytosolic fraction. As Δ1,4-3-ketocholate, DHOPDC, and their respective monoene derivatives accumulated transiently outside the cell, CoA activation of these acids must be possible as well to enable their further degradation. In agreement with this assumption, we have detected a CoA ligase activity for DHOPDC. To our knowledge, this enzyme activity has not been detected before. So far, we do not know whether A-ring oxidation occurs with only free acids or with CoA esters. However, the rapid formation of CoA esters with oxidized A-rings suggested that A-ring oxidation can also occur on the CoA ester level. The extracellular accumulation of degradation intermediates with monoene and diene structures in the A-ring demonstrates the complexity of the cholate degradation pathway because it implies parallel β-oxidation pathways for monoene and diene derivatives of 3-ketocholate. Furthermore, the conversion of monoenes into dienes must be possible at many if not all steps of β-oxidation. It is not known whether there are specific enzymes for β-oxidation of monoene and diene derivatives of degradation intermediates.
So far, we have not detected the three degradation intermediates between Δ1,4-3-ketocholyl-CoA and DHOPDC-CoA. Although the respective reaction steps must take place in strain R1, the corresponding intermediates did not accumulate in the medium. We also could not detect further degradation of Δ1,4-3-ketocholyl-CoA in cell extracts of strain Chol1. A possible explanation could be that these reactions are catalyzed by a multienzyme complex from which no intermediates are released. The functional assembly of enzymes and coenzymes involved in these reaction steps might have been destroyed during the preparation of cell extracts. The same might apply to the reactions leading from DHOPDC-CoA to DHADD because we could not detect any of the respective degradation intermediates in culture supernatants or in cell extracts.
The transposon-inactivated gene in strain R1 was found to encode a putative ACAD. A defect in such an enzyme delivers a plausible explanation for the accumulation of DHOPDC because an ACAD could catalyze the presumptive next degradation step with DHOPDC-CoA as a substrate. We assume that upon DHOPDC-CoA accumulation in strain R1, the CoA ester is cleaved, and DHOPDC is released into the medium. To our knowledge, we have identified the first bacterial gene encoding an enzyme that is responsible for a specific reaction step in side chain degradation of steroids. Comparison of the deduced amino acid sequence with ACADs of known function did not allow a classification of our ACAD into the existing categories (10), suggesting that we may have found a new type of ACAD. The ACAD gene found in strain Chol1 had exactly the same length as and a high similarity to a putative ACAD (gene number PSHAa0888) from P. haloplanktis TAC125, which we found to grow with cholate, too. A high similarity was also found to a putative ACAD encoded by gene number 5369 in the genome of Comamonas testosteroni strain KF-1 (http://genome.ornl.gov/microbial/ctes/17jul06/ctes.html). This gene is part of a cluster of genes (5363 to 5382) for putative proteins involved in β-oxidation with homology to genes found in P. haloplanktis TAC125. Intriguingly, this gene cluster is located just between two clusters of genes (5298 to 5314 and 5384 to 5393) that have been shown to be involved in steroid degradation in C. testosteroni strain TA441. The first gene cluster starts with tesB and encodes genes involved in further degradation of steroid skeleton after formation of the secosteroid intermediate (21). The second gene cluster starts with tesG and encodes genes involved in A-ring oxidation, hydroxylation of the B-ring, and hydroxylation of the aromatic A-ring (19, 20). A similar association of genes involved in steroid degradation with genes encoding putative β-oxidation enzymes including PSHAa0888 is also present in the genome of P. haloplanktis strain TAC125 and in Rhodococcus sp. strain RHA1 (34). It will be interesting to investigate whether these putative proteins are involved in β-oxidation of acyl side chains of steroid compounds. The discovery of such genes would be relevant for biotechnology because mutants accumulating steroids with partly oxidized side chains are used for the synthesis of corticosteroids (16). Attempts to detect further degradation steps in vitro and the search for further genes involved in bile acid degradation are ongoing in our laboratory.
Acknowledgments
The authors thank Antje Karst for technical assistance and Bernhard Schink for continuous support.
This work was funded by the Deutsche Forschungsgemeinschaft (projects PH71/2-1 and B9 in CRC454) and by the University of Konstanz (AFF project 58/03).
Footnotes
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625-651. [DOI] [PubMed] [Google Scholar]
- 2.Berg, D. 1989. Transposon Tn5, p. 185-210. In D. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, DC.
- 3.Bode, H. B., B. Zeggel, B. Silakowski, S. C. Wenzel, H. Reichenbach, and R. Müller. 2003. Steroid biosynthesis in prokaryotes: identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the myxobacterium Stigmatella aurantiaca. Mol. Microbiol. 47:471-481. [DOI] [PubMed] [Google Scholar]
- 4.Bortolini, O., A. Medici, and S. Poli. 1997. Biotransformations on steroid nucleus of bile acids. Steroids 62:564-577. [DOI] [PubMed] [Google Scholar]
- 5.Dalluge, J. J., S. Gort, R. Hobson, O. Selifonova, F. Amore, and R. Gokarn. 2002. Separation and identification of organic acid-coenzyme A thioesters using liquid chromatography/electrospray ionization-mass spectrometry. Anal. Bioanal. Chem. 374:835-840. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinosa-Urgel, M., A. Salido, and J. L. Ramos. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182:2363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Florin, C., T. Kohler, M. Grandguillot, and P. Plesiat. 1996. Comamonas testosteroni 3-ketosteroid-Δ4(5α)-dehydrogenase: gene and protein characterization. J. Bacteriol. 178:3322-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan-Schreier, H., J. G. Okun, D. Kohlmueller, C.-D. Langhans, V. Peters, H. J. ten Brink, N. M. Verhoeven, C. Jakobs, A. Voelkl, and G. F. Hoffmann. 2005. Measurement of bile acid CoA esters by high-performance liquid chromatography-electrospray ionisation tandem mass spectrometry (HPLC-ESI-MS/MS). J. Mass. Spectrom. 40:882-889. [DOI] [PubMed] [Google Scholar]
- 10.Ghisla, S., and C. Thorpe. 2004. Acyl-CoA dehydrogenases. Eur. J. Biochem. 271:494-508. [DOI] [PubMed] [Google Scholar]
- 11.Gunn, J. S. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2:907-913. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa, S. 1982. Microbial transformation of bile acids. A unified scheme for bile acid degradation, and hydroxylation of bile acids. Z. Allg. Mikrobiol. 22:309-326. [DOI] [PubMed] [Google Scholar]
- 13.Helenius, A., and K. Simons. 1975. Solubilization of membranes by detergents. Biochim. Biophys. Acta 415:29-79. [DOI] [PubMed] [Google Scholar]
- 14.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hesse, M., H. Meier, and B. Zeeh. 1995. Spektroskopische methoden in der organischen chemie, 5th ed. Thieme, Stuttgart, Germany.
- 16.Hill, F. F., J. Schindler, R. D. Schmid, R. Wagner, and W. Voelter. 1982. Microbial conversion of sterols. Appl. Microbiol. Biotechnol. 15:25-32. [Google Scholar]
- 17.Hofmann, A. F. 1999. Bile acids: the good, the bad, and the ugly. News Physiol. Sci. 14:24-29. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann, A. F., and K. J. Mysels. 1988. Bile salts as biological surfactants. Colloids Surf. 30:145-173. [Google Scholar]
- 19.Horinouchi, M., T. Hayashi, T. Yamamoto, and T. Kudo. 2003. A new bacterial steroid degradation gene cluster in Comamonas testosteroni TA441 which consists of aromatic-compound degradation genes for secosteroids and 3-ketosteroid dehydrogenase genes. Appl. Environ. Microbiol. 69:4421-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horinouchi, M., T. Hayashi, and T. Kudo. 2004. The genes encoding the hydroxylase of 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione in steroid degradation in Comamonas testosteroni TA441. J. Steroid. Biochem. Mol. Biol. 92:143-154. [DOI] [PubMed] [Google Scholar]
- 21.Horinouchi, M., T. Kurita, T. Yamamoto, E. Hatori, T. Hayashi, and T. Kudo. 2004. Steroid degradation gene cluster of Comamonas testosteroni consisting of 18 putative genes from meta-cleavage enzyme gene tesB to regulator gene tesR. Biochem. Biophys. Res. Commun. 324:597-604. [DOI] [PubMed] [Google Scholar]
- 22.Hylemon, P. B., and J. Harder. 1998. Biotransformation of monoterpenes, bile acids, and other isoprenoids in anaerobic ecosystems. FEMS Microbiol. Rev. 22:475-488. [DOI] [PubMed] [Google Scholar]
- 23.Kieslich, K. 1985. Microbial side-chain degradation of sterols. J. Basic Microbiol. 25:461-474. [DOI] [PubMed] [Google Scholar]
- 24.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 25.Mahato, S. B., and S. Garai. 1997. Advances in microbial steroid biotransformation. Steroids 62:332-345. [DOI] [PubMed] [Google Scholar]
- 26.Maser, E., G. Xiong, C. Grimm, R. Ficner, and K. Reuter. 2001. 3α-Hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni: biological significance, three-dimensional structure and gene regulation. Chem. Biol. Interact. 130-132:707-722. [DOI] [PubMed] [Google Scholar]
- 27.Medigue, C., E. Krin, G. Pascal, V. Barbe, A. Bernsel, P. N. Bertin, F. Cheung, S. Cruveiller, S. D'Amico, A. Duilio, G. Fang, G. Feller, C. Ho, S. Mangenot, G. Marino, J. Nilsson, E. Parrilli, E. P. Rocha, Z. Rouy, A. Sekowska, M. L. Tutino, D. Vallenet, G. von Heijne, and A. Danchin. 2005. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philipp, B., H. Erdbrink, M. J.-F. Suter, and B. Schink. 2006. Degradation of and sensitivity to cholate in Pseudomonas sp. strain Chol1. Arch. Microbiol. 185:192-201. [DOI] [PubMed] [Google Scholar]
- 29.Ridlon, J. M., D.-J. Kang, and P. B. Hylemon. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid. Res. 47:241-259. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Shah, P. P., and E. Staple. 1968. Synthesis of coenzyme A esters of some bile acids. Steroids 12:571-576. [DOI] [PubMed] [Google Scholar]
- 32.Talalay, P. 1965. Enzymatic mechanisms in steroid biochemistry. Annu. Rev. Biochem. 34:347-380. [DOI] [PubMed] [Google Scholar]
- 33.Tenneson, M. E., J. D. Baty, R. F. Bilton, and A. N. Mason. 1979. The degradation of cholic acid by Pseudomonas sp. NCIB 10590. Biochem. J. 184:613-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van der Geize, R., K. Yam, T. Heuser, M. H. Wilbrink, H. Hara, M. C. Anderton, E. Sim, L. Dijkhuizen, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. USA 104:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]