Abstract
The robustness of a recently developed diagnostic microarray for influenza, the MChip, was evaluated with 16 historic subtype H1N1 influenza A viruses (A/H1N1), including A/Brevig Mission/1/1918. The matrix gene segments from all 16 viruses were successfully detected on the array. An artificial neural network trained with temporally related A/H1N1 viruses identified A/Brevig Mission/1/1918 as influenza virus A/H1N1 with 94% probability.
The rapidly evolving nature of the influenza virus presents unique challenges to public health officials (5, 11). Year-round global surveillance is necessary to ensure that vaccines contain virus strains capable of protecting the population from this deadly disease. The threat presented by newly emerging virus subtypes requires a rapid response to identify and treat infected patients and to take precautions to protect their families and other close contacts. Currently, a complete determination of subtype and genotype for influenza viruses often requires cell culture and/or immunofluorescence methods. A rapid point-of-care diagnostic for subtyping could provide the information needed to make decisions about patient management and influenza control measures.
Previous work in our lab demonstrated the utility of a low-density DNA microarray, the MChip, that targets only the matrix (M) gene segment of the influenza virus genome as an effective tool in the identification of influenza virus subtypes (1). The MChip utilizes only 15 short oligonucleotide capture and label pairs, which effectively discriminate influenza A viruses of the H1N1 (A/H1N1), H3N2, and H5N1 subtypes (1, 2). In designing the MChip, publicly available sequence information was used to locate conserved regions in the M gene segment for viruses that circulated between the years 1998 and 2005 (7). Several sequences were chosen to provide discrimination between the different subtypes of influenza virus, while others were selected to broadly react with a wide range of influenza A virus subtypes. In the assay, the viral sample is amplified, hybridized to the microarray, and scanned. Virus subtype information can be inferred by analyzing the relative fluorescence signal intensity for each of the array sequences, either by visual inspection or with an automated pattern recognition program (1, 2).
The MChip and associated assay have been demonstrated to be highly effective for both clinical samples and emerging H5N1 viruses. In a comparative study of MChip, QuickVue A+B, reverse transcription-PCR, and viral culture using over 100 clinical samples obtained by a variety of sampling methods, the MChip exhibited 98% clinical sensitivity and 98% clinical specificity as referenced to viral culture (8). The MChip has now been evaluated with over 100 avian A/H5N1 virus samples collected during the time period from 2000 to 2007. While results from only a fraction of the entire set have been published (2), the sensitivity and specificity are over 95% with no false positives.
Sequences on the microarray were designed using recently circulating viruses; therefore, we tested the robustness of the MChip with respect to detecting remotely related and novel viruses by evaluating the assay performance with historic viruses. Sixteen H1N1 viruses of human, swine, and avian origins were chosen for evaluation, based on the availability of sample and sequence data. The viruses, which are summarized in Table 1, circulated from 88 years ago to 26 years ago, and all were isolates from cell culture or egg passage. The virus samples included the recently reconstructed pandemic “Spanish Flu” strain of 1918 (10).
TABLE 1.
Sample set of historic A/H1N1 viruses tested
| Sample | Virus |
|---|---|
| 1 | A/Brevig Mission/1/1918 |
| 2 | A/swine/Iowa/1930 |
| 3 | A/WSN/1933 |
| 4 | A/Puerto Rico/8/1934a |
| 5 | A/Alaska/1935a |
| 6 | A/Galenby/1937a |
| 7 | A/Bellamy/1942a |
| 8 | A/Iowa/1943a |
| 9 | A/Fort Monmouth/1/1947a |
| 10 | A/Fort Warren/1/1950a |
| 11 | A/Missouri/301/1952a |
| 12 | A/Tasmania/30/1954a |
| 13 | A/England/19/1955a |
| 14 | A/Scotland/38/1956a |
| 15 | A/Prussia/1957a |
| 16 | A/turkey/Kansas/4880/1980 |
Training set for the artificial neural network.
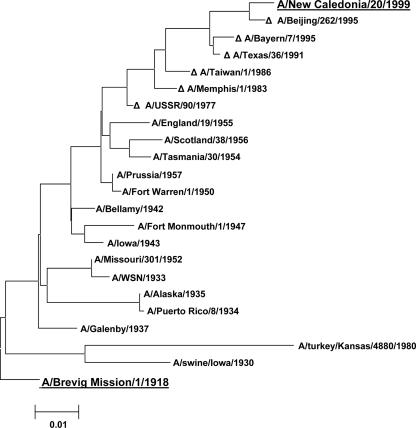
The genetic diversity of the historic H1N1s and their relationship to modern H1N1 viruses was evaluated by aligning the M gene segment sequences of the 16 H1N1 samples examined in this study to the reported sequences of recent H1N1 vaccine strains, including A/New Caledonia/20/1999. A phylogenetic tree for the M gene segment sequences is shown in Fig. 1. As can be observed in the figure, there has been a steady trend away from the 1918 pandemic virus, with the A/New Caledonia/20/1999 M gene segment considerably different from its 1918 predecessor.
FIG. 1.
Neighbor-joining tree of M gene segment sequences of historic subtype H1N1 strains aligned with those of several recent vaccine strains (denoted by Δ) since the reemergence of H1N1 with A/USSR/90/77. A/Brevig Mission/1/1918 and A/New Caledonia/20/99 are underlined and in grey to emphasize the distance between these two strains.
Experimental procedures.
Low-density microarrays were prepared as previously described (1, 7). The viruses were obtained by reconstitution of lyophilized egg-passaged virus stored at the Centers for Disease Control and Prevention (CDC) Influenza Division in Atlanta, GA. A/Brevig Mission/1/1918 viral RNA was generously provided by Terrence Tumpey of the CDC. A/Puerto Rico/8/1934 was purchased from the American Type Culture Collection (ATCC, Bethesda, MD). All experimental details have been given elsewhere (1, 2, 7, 9). Briefly, influenza virus RNA was extracted, reverse transcribed with universal influenza A virus gene-specific primers (12) that contained a T7 promoter, amplified by PCR, transcribed back to RNA, and then fragmented. The fragmented RNA was hybridized to the microarray as previously described (1, 2). After hybridization, the slides were washed and fluorescence images obtained. The images were processed as described in reference 1. Each sample was analyzed twice from the point of RNA transcription to ensure correct interpretation.
As described elsewhere (2), for automated image interpretation, a probabilistic neural network (PNN) analysis was performed using Weka (Waikato Environment for Knowledge Analysis) version 3.4.8 using the multilayer perceptron function with feed-forward back propagation of error (4). The neural network was trained with 25 images from known viruses for each of three subtypes (H1N1, H3N2, and H5N1) and 25 images from influenza virus A-negative samples. During training, 20% of the samples from each subtype were selected at random for validation. The trained neural network was used to analyze the test data or the query set (samples 1 to 3 and 16 in Table 1).
Results and discussion.
The ability of the MChip to detect viral RNA relies on the amplification of genetic material through reverse transcription-PCR (1, 7, 9). The highly conserved 3′ and 5′ ends of the influenza A virus genome provide reliable regions for PCR primers (3). Gene specificity can be gained by extending the primers into the gene segment a short distance (6, 12). The universal influenza A virus gene-specific primers developed by Zou (12) successfully amplified the M gene segment for all of the historic viruses used in this study. In all cases, DNA was amplified and detectable on an ethidium bromide-stained agarose gel without appreciable additional bands, i.e., other influenza A virus genes.
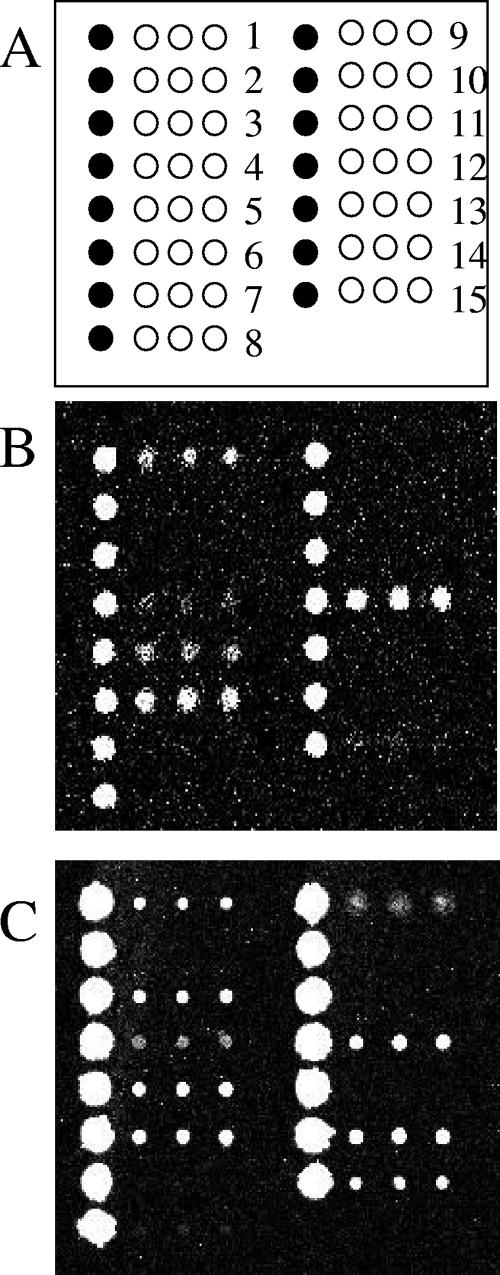
Figure 2 shows the microarray fluorescence images for a modern H1N1 virus and A/Brevig Mission/1/1918. The sequences previously identified (see reference 1) as broadly reactive to influenza A virus (numbers 1, 4, 5, 6, and 15) exhibited good signal intensities (referred to hereafter as “hit”) for all of the older viruses. Additionally, the sequence that was previously identified as H1N1 specific (sequence 12) was observed for all of the samples except lab strain A/WSN/1933. Interestingly, the three sequences designed to be sensitive to H5N1 avian influenza viruses, sequences 3, 9, and 14, hit for several older H1N1 viruses. A closer look at the samples that exhibited signal for sequence 14 revealed that two were isolates from nonhuman sources (A/swine/Iowa/1930 and A/turkey/Kansas/4480/1980); one was the lab strain A/WSN/1933; one was the 1918 isolate A/Brevig Mission/1/1918; and one was from 1952, A/Missouri/301/1952. Positive signals on sequences 3, 9, and 14, which were previously only observed with H5N1 avian influenza viruses, may be evidence of the avian ancestry of H1N1 viruses or of the slow evolution of the H5N1 strain to a more human-adapted virus.
FIG. 2.
Comparison of modern H1N1 virus to the 1918 “Spanish Flu” strain. (A) Layout of MChip with 15 sequences (open circles) spotted in triplicate next to a column of control spots (filled circles). (B) Fluorescence image from a New Caledonia-like A/H1N1-infected clinical sample obtained after 2000 (from the Colorado Department of Public Health and Environment). Brighter spots indicate higher fluorescence intensity. (C) Fluorescence image from A/Brevig Mission/1/1918.
Automated image analysis.
Our group has previously employed an artificial neural network for pattern recognition on the fluorescence images to accurately subtype unknown virus samples (1). This pattern recognition method assigns subtypes based on the relative intensities of fluorescence signals for all of the sequences after thorough training using a set of images from “known” viruses. In this work, a PNN provided the additional benefit of confidence values for the assignment.
When the PNN was trained using only modern viruses (collected after 2000), the homogeneity of the H1N1 training data prevented the accurate identification of historic H1N1 viruses, with significant misses of A/WSN/1933, A/Brevig Mission/1/1918, and both swine and turkey viruses. In order to perform subtype determination with the PNN, it was necessary to train with a similar set of known viruses, in this case historic H1N1 viruses. As expected, the PNN was able to accurately assign subtypes for a set of historic viruses when trained with a set of temporally related and modern viruses. In addition, the PNN retained the ability to identify modern viruses. Specifically, for H1N1 representation in the PNN, 12 of the historic viruses were used with 25 modern H1N1 viruses for training, and 4 historic viruses were tested as unknowns (the training and query sets are given in Table 1). The PNN correctly assigned the 1918 virus as A/H1N1, with a 94% probability, and A/WSN/1933 as A/H1N1, with 96% probability. Interestingly, the improved training provided an accurate subtype assignment for both A/swine/Iowa/1930 (70% probability) and A/turkey/Kansas/4480/1980 (97% probability).
In summary, the MChip assay and associated artificial neural network robustly detected a range of historic viruses dating back to 1918. This study demonstrated the reliability of the conserved M genes for the inference of antigenic subtype in the absence of a recent reassortment event. Studies are under way to determine the time required for the M genes to adapt and correctly report the antigenic subtype in the case where the M genes originate from a different virus than the hemagglutinin and neuraminidase genes during a reassortment event.
Acknowledgments
We gratefully acknowledge funding from NIH/NIAID (grant U01AI0 56528).
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Dawson, E. D., C. L. Moore, J. A. Smagala, D. M. Dankbar, M. Mehlmann, M. B. Townsend, C. B. Smith, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2006. MChip: a tool for influenza surveillance. Anal. Chem. 78:7610-7615. [DOI] [PubMed] [Google Scholar]
- 2.Dawson, E. D., C. L. Moore, J. A. Smagala, D. M. Dankbar, M. Mehlmann, M. B. Townsend, C. B. Smith, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2007. Identification of A/H5N1 influenza viruses using a single gene diagnostic microarray. Anal. Chem. 79:378-384. [DOI] [PubMed] [Google Scholar]
- 3.Desselberger, U., V. R. Racaniello, J. J. Zazra, and P. Palese. 1980. The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene 8:315-328. [DOI] [PubMed] [Google Scholar]
- 4.Frank, E., M. Hall, L. Trigg, G. Holmes, and I. H. Witten. 2004. Data mining in bioinformatics using Weka. Bioinformatics 20:2479-2481. [DOI] [PubMed] [Google Scholar]
- 5.Hay, A. J., V. Gregory, A. R. Douglas, and Y. P. Lin. 2001. The evolution of human influenza viruses. Philos. Trans. R. Soc. Lond. B 356:1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 7.Mehlmann, M., E. D. Dawson, M. B. Townsend, J. A. Smagala, C. L. Moore, C. B. Smith, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2006. Robust sequence selection method used to develop the FluChip diagnostic microarray for influenza virus. J. Clin. Microbiol. 44:2857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehlmann, M., A. B. Bonner, J. V. Williams, D. M. Dankbar, C. L. Moore, R. D. Kuchta, A. B. Podsiad, J. D. Tamerius, E. D. Dawson, and K. L. Rowlen. 2007. Comparison of the MChip to viral culture, reverse transcription-PCR, and the QuickVue influenza A+B test for rapid diagnosis of influenza. J. Clin. Microbiol. 45:1234-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Townsend, M. B., E. D. Dawson, M. Mehlmann, J. A. Smagala, D. M. Dankbar, C. L. Moore, C. B. Smith, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2006. Experimental evaluation of the FluChip diagnostic microarray for influenza virus surveillance. J. Clin. Microbiol. 44:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. Garcia-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77-80. [DOI] [PubMed] [Google Scholar]
- 11.Webster, R. G., S. M. Wright, M. R. Castrucci, W. J. Bean, and Y. Kawaoka. 1993. Influenza—a model of an emerging virus disease. Intervirology 35:16-25. [DOI] [PubMed] [Google Scholar]
- 12.Zou, S. 1997. A practical approach to genetic screening for influenza virus variants. J. Clin. Microbiol. 35:2623-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]