Abstract
A clinical study was designed to study Streptococcus pneumoniae isolates recovered from a community hospital in Japan from April 2001 to November 2002. A total of 73 isolates were defined as derived from inpatient, outpatient, and hospital staff groups. The MIC results showed that 20 strains (27.4%) were susceptible to penicillin G, 39 strains (53.4%) had intermediate resistance, and 14 strains (19.2%) had full resistance. Low susceptibility to macrolides was also detected: 32.9%, 32.9%, and 34.2% of all strains were resistant to erythromycin, clarithromycin, and azithromycin, respectively. Thirty strains (41%) were resistant to at least two different kinds of antibiotics. Nineteen disparate serotypes were detected besides two nontypeable strains, and the predominant serotypes were 19F and 23F. Pulsed-field gel electrophoresis (PFGE) pattern A was dominant in the serotype 19F group; this pattern was similar to that of the international clone Taiwan 19F. A total of 10 different patterns were detected in the 23F group and were distinguishable from those of the international clones Spain 23F and Taiwan 23F. Pattern b strains were identified in the same ward, and pattern d strains were found both in patients with nosocomial pneumococcal infections (NPI) and in outpatients. In conclusion, drug-resistant S. pneumoniae was spreading rapidly, especially isolates of the serotype 19F and 23F groups. PFGE data revealed interpatient transmission and suggested that there might be some association between NPI patient strains and outpatient strains.
Streptococcus pneumoniae is the most common pathogen of respiratory diseases, often leading to pneumonia, pharyngitis, sinusitis, otitis media, septicemia, and meningitis (1). Many studies have reported S. pneumoniae as one of the leading causes of community-acquired pneumonia (CAP) worldwide (2, 6, 19, 31, 32, 41) as well as in Japan (13). On the other hand, nosocomial pneumococcal infections (NPI) result in longer hospitalizations and higher mortality rates because of increased antimicrobial resistance induced after long-term antibiotic therapies (7, 23, 34, 35). S. pneumoniae also plays a very important role in nosocomial infections of the pulmonary tract and blood, which are more and more widely recognized (3, 8, 29, 34, 35). Age, previous hospitalization, and previous antimicrobial therapy have been thought to contribute to penicillin-resistant pneumococcal infection in nosocomial outbreaks (18, 27). Previous studies conducted to determine the antimicrobial susceptibilities of isolates from NPI found that a high percentage of pneumococcal pathogens had multidrug resistance, including resistance to new quinolones (4, 5, 28, 39). Masaki et al. reported a possible relationship between the pulsed-field gel electrophoresis (PFGE) patterns of Moraxella catarrhalis isolates from hospital- and community-acquired respiratory infections in a community hospital (20). Yet there still is not sufficient evidence to determine whether a similar pattern exists for S. pneumoniae.
Since pneumonia ranks as the fourth leading cause of death in Japan, it remains a very high priority to investigate the antimicrobial susceptibilities of S. pneumoniae isolates recovered from CAP or NPI and their genetic relatedness, in order to detect potential transmission routes. In the present study, antimicrobial susceptibilities were determined by examining MICs as well as the serotypes for all the isolates. Because PFGE has been reported as a suitable method for confirming interstrain genetic relatedness (16, 17, 21), it was used in this study to analyze and compare the molecular profiles of isolates from CAP and NPI patients.
MATERIALS AND METHODS
Setting.
Tagami Hospital, a community hospital affiliated with Nagasaki University, is located in Nagasaki, Japan. There are 180 beds, 5 floors, and 9 wards in Tagami Hospital, including the outpatient department (first floor), 41 surgery beds (second floor, wards 2W and 2E), 53 internal medicine beds (third floor, wards 3W and 3E), and long-term-care wards with 86 beds (fourth floor, 4W and 4E, 42 beds; fifth floor, 5W and 5E, 43 beds).
Participants.
Altogether, 51 patients and 5 hospital staff members were included in this research from April 2001 to November 2002 (19 months). Of 56 participants, 22 were female and 34 were male (ratio, 0.65), and the mean age was 67.6 years (range, 17 to 97). Patients' clinical records were reviewed. Forty-one patients (80.4%) had chronic obstructive pulmonary diseases, six patients were diagnosed with acute bronchitis, three patients were diagnosed with CAP, and only one patient had adenoiditis/pharyngitis. In this study, five hospital staff members took full responsibility for the medical treatment and had the highest frequency of contact with the recruited patients.
Nosocomial pneumococcal infection was defined according to the CDC recommendations (9): briefly, as pneumococcal infection that was demonstrated ≥72 h after admission, excluding those patients who were suspected of having pneumococcal diseases present or in incubation at admission. Pneumonia was diagnosed if there was an appearance of a new abnormal shadow and likely infiltration on a chest roentgenogram and if at least two of the following clinical and laboratory findings were present: fever (temperature, >37.8°C), cough, production of purulent sputum, dyspnea, and leukocytosis (leukocyte count, >10,000/ml).
Bacterial strains.
A total of 73 isolates of pneumococci were recovered from 56 adult participants. The sources of the isolates were as follows: sputum (n = 62), blood (n = 1), bronchoalveolar lavage fluid (n = 1), nasal cavity (n = 1), and pharynx (n = 8). Culture plates were incubated overnight under a 5% CO2 atmosphere, and optochin sensitivity and bile solubility tests were performed to confirm S. pneumoniae.
Serotyping and antimicrobial susceptibility test.
Isolates were serotyped by capsular swelling (Quellung reaction) observed microscopically after suspension in pneumococcal typing antisera (Statens Serum Institut, Copenhagen, Denmark).
MICs were determined by an agar dilution method according to CLSI (formerly NCCLS) guidelines (25). Serial twofold dilutions of each antibiotic (ranging from 0.008 to 128 μg/ml) were prepared. Mueller-Hinton agar supplemented with 5% horse blood was used as the culture medium. Approximately 0.01 ml (105 CFU/ml) of a bacterial suspension of each isolate was inoculated onto antibiotic-containing agar and incubated overnight at 37°C. The MIC of each antibiotic was defined as the lowest concentration that prevented visible bacterial growth. All isolates were tested for susceptibility to the following nine antibiotics: penicillin G (PCG) (Meiji Seika Kaisha, Tokyo, Japan), ceftriaxone (CTRX) (Chugai Pharmaceutical Co., Tokyo, Japan), cefditoren (CDTR) (Meiji Seika Kaisha), imipenem (IPM) (Banyu Pharmaceutical Co., Tokyo, Japan), erythromycin (EM) (Dainippon Pharmaceutical Co., Osaka, Japan), clarithromycin (CAM) (Taisho Pharmaceutical Co., Tokyo, Japan), azithromycin (AZM) (Pfizer Japan Inc., Tokyo, Japan), levofloxacin (LVFX) (Daiichi Pharmaceutical Co., Tokyo, Japan), and vancomycin (VCM) (Shionogi Co., Osaka, Japan). Penicillin-susceptible S. pneumoniae (PSSP) was defined as strains for which the penicillin MICs were <0.125; penicillin-intermediate S. pneumoniae (PISP) and penicillin-resistant S. pneumoniae (PRSP) were defined by MICs of ≥0.125 and ≤1 and by MICs of ≥2, respectively.
PFGE.
PFGE was performed on all 73 strains. The Spanish multidrug-resistant serotype 23F clone (ATCC 700669), the Taiwan multidrug-resistant serotype 19F clone (ATCC 700905), and the Taiwan multidrug-resistant serotype 23F clone (ATCC 700906) were used as reference standards. Strains were grown overnight in brain heart infusion broth, and PFGE of SmaI chromosomal digests was performed as described previously (42). DNA banding patterns were interpreted according to the criteria of Tenover et al. (37): a difference of more than three bands in the profile was needed to distinguish between PFGE types.
RESULTS
Antimicrobial susceptibility test.
The MICs for the 73 strains are shown in Table 1. According to the CLSI breakpoint criteria, 20 strains (27.4%) appeared susceptible to PCG (MIC, ≤0.063) (PSSP), 39 strains (53.4%) showed intermediate resistance (MIC, 0.125 to 1) (PISP), and 14 strains (19.2%) showed full resistance (MIC, ≥2) (PRSP). Low susceptibility to macrolides was also detected—32.9%, 32.9%, and 34.2% of all strains were resistant to EM, CAM, and AZM, respectively—while the strains showed relatively high susceptibility to all the cephems. Surprisingly, all isolates were fully susceptible to IPM and VCM. In addition, as many as 7 (9.6%) strains were resistant to LVFX.
TABLE 1.
Distribution of MICs for 73 strains of S. pneumoniaea
| Antibiotic | No. of isolates for which the MIC (μg/ml) is:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.004 | 0.008 | 0.016 | 0.032 | 0.063 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ≥128 | |
| PCG | 6 | 7 | 7 | 2 | 13 | 7 | 17* | 12 | 2 | |||||||
| CTRX | 2 | 5 | 1 | 3 | 16 | 18 | 18 | 3* | 4 | 3 | ||||||
| CDTR | 2 | 4 | 2 | 15 | 15 | 16 | 12 | 1* | 6 | |||||||
| IPM | 1 | 2 | 12 | 9 | 6 | 19 | 16 | 7 | 1 | * | ||||||
| EM | 2 | 24 | 12 | 3 | 8* | 2 | 3 | 3 | 1 | 1 | 1 | 13 | ||||
| CAM | 1 | 3 | 26 | 12 | 5 | 2* | 4 | 4 | 1 | 1 | 2 | 12 | ||||
| AZM | 6 | 21 | 10 | 4 | 4 | 3* | 1 | 4 | 1 | 3 | 1 | 15 | ||||
| LVFX | 7 | 33 | 26* | 4 | 1 | 1 | 1 | |||||||||
| VCM | 1 | 5 | 24 | 40 | 3 | * | ||||||||||
Asterisks indicate the breakpoint for each antibiotic according to CLSI criteria.
Serotyping.
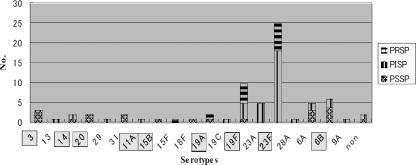
Aside from two nontypeable strains, 19 disparate serotypes were detected (Fig. 1), mainly serotypes 3 (4.1%), 6A (6.8%), 23A (6.8%), 6B (8.2%), and the predominant serotypes 19F (10 strains [13.7%]) and 23F (25 strains [34.2%]). Within the inpatient group, 17 of 23 strains (73.9%) belonged to serotypes 19F and 23F, compared to 38.1% for the outpatient group (P < 0.05 by the chi-square test). Additionally, only two strains of 19F, but no 23F strains, were detected in the staff group. Furthermore, of 73 strains, 52 (71.2%) were found to be covered by a 23-valent polysaccharide vaccine (PS23).
FIG. 1.
Serotype distribution plotted against penicillin susceptibility. Boxed serotypes were covered by PS23.
Characteristics of multidrug-resistant strains.
Thirty strains (41%) were found to be resistant to at least two different antibiotics examined in the present study, in which strains were described as PSSP, PISP, or PRSP according to their susceptibilities to penicillin (Table 2). Macrolides elicited the highest resistance: 83.3% of strains were resistant to EM, CAM, or AZM, or to all three. Seven strains (23.3%) appeared to be simultaneously resistant to penicillin and macrolides. One serotype15F strain was uniquely resistant to penicillin, cephems, and macrolides as well as fluoroquinolone, indicating a very wide spectrum of antimicrobial resistance. Seven and 11 strains in the serotype 19F and 23F groups, respectively, were multidrug resistant. In the serotype 19F group, PSSP and PISP strains showed the same pattern of resistance to all the macrolides, while PRSP strains expressed variance. Although the serotype 23F group showed results similar to those seen with the 19F group, five strains were proven to be resistant to cephems, whereas no such resistance has been found in the serotype 19F group yet (Table 2).
TABLE 2.
Characteristics of multidrug-resistant strains and details of serotype 19F and 23F strains
| Strain | Serotype | Resistancea to the following antibiotic:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PCG | CTRX | CDTR | IPM | EM | CAM | AZM | LVFX | VCM | ||
| PSSP (n = 8) | ||||||||||
| S-1 | 14 | O | O | O | ||||||
| S-2 | 3 | O | O | O | ||||||
| S-3 | Nontypeable | O | O | |||||||
| S-4 | 19F | O | O | O | ||||||
| S-5 | 6B | O | O | O | ||||||
| S-6 | 6B | O | O | O | ||||||
| S-7 | 6B | O | O | O | ||||||
| S-8 | 3 | O | O | O | O | |||||
| PISP (n = 11) | ||||||||||
| I-1 | 23F | O | O | O | ||||||
| I-2 | 19F | O | O | O | ||||||
| I-3 | 23A | O | O | O | O | |||||
| I-4 | 23A | O | O | O | ||||||
| I-5 | Nontypeable | O | O | O | ||||||
| I-6 | 23F | O | O | |||||||
| I-7 | 6B | O | O | O | ||||||
| I-8 | 23F | O | O | |||||||
| I-9 | 23F | O | O | O | ||||||
| I-10 | 23F | O | O | O | ||||||
| I-11 | 19F | O | O | O | ||||||
| PRSP (n = 11) | ||||||||||
| R-1 | 19F | O | O | O | O | |||||
| R-2 | 23F | O | O | O | ||||||
| R-3 | 23F | O | O | O | ||||||
| R-4 | 23F | O | O | O | ||||||
| R-5 | 23F | O | O | O | ||||||
| R-6 | 19F | O | O | O | O | |||||
| R-7 | 23F | O | O | O | O | |||||
| R-8 | 23F | O | O | |||||||
| R-9 | 19F | O | O | |||||||
| R-10 | 15F | O | O | O | O | O | O | O | ||
| R-11 | 19F | O | O | O | O | |||||
Designated by “O”.
Molecular characteristics by PFGE.
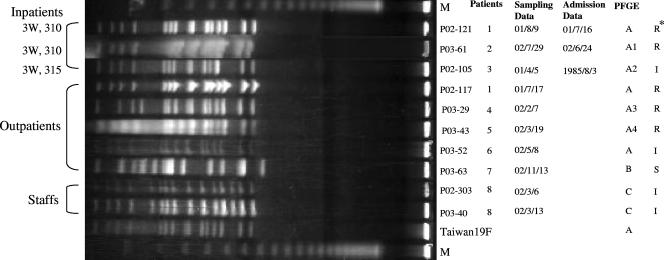
PFGE was performed on all 73 strains, and the DNA patterns showed tremendous diversity (data not shown). In the serotype 19F group, three DNA patterns were detected among 10 strains (Fig. 2). Seven strains showed pattern A, which was similar to the pattern of the internationally spreading Taiwan 19F clone; two strains had pattern C; and only one strain showed pattern B. Three of the pattern A strains were from inpatients who were regarded as NPI patients and resided in different wards, and the others were from outpatients. The strains with pattern C were both from the same member of the hospital staff. The pattern B strain, isolated from an outpatient, was the only PSSP strain in the serotype 19F group.
FIG. 2.
PFGE results for SmaI-digested DNA from serotype 19F strains and a representative of the multidrug-resistant clone Taiwan 19F. M, molecular size marker; I, PISP; R, PRSP. Asterisk indicates PCG susceptibility.
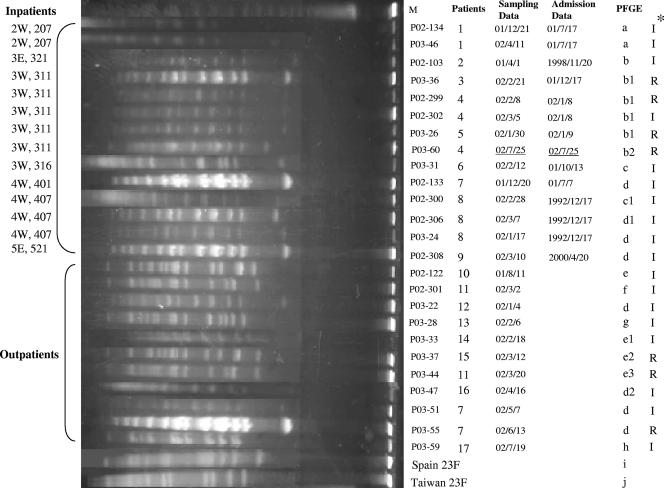
In the serotype 23F group, including predominant DNA patterns b and d, a total of 10 different patterns were detected (Fig. 3). All six pattern b strains were from inpatients residing in the same ward except for one patient, and three of these strains were isolated from the same patient on different dates. Furthermore, except for P03-60, all these strains belonged to NPI patients. As many as eight strains showed pattern d; of these, four were from inpatients defined as having NPI and the others were from outpatients. Strains P02-306 and P03-24 were isolated from the same inpatient, while P03-51 and P03-55 were from the same outpatient. The internationally spreading clones Spain 23F and Taiwan 23F each showed a unique pattern that had no relatedness to the patterns of any of the 23F clinical isolates in this study.
FIG. 3.
PFGE results for SmaI-digested DNA from serotype 23F strains and the multidrug-resistant clones Spain 23F and Taiwan 23F. M, molecular size marker; I, PISP; R, PRSP. Asterisk indicates PCG susceptibility.
DISCUSSION
Drug-resistant S. pneumoniae has spread widely in the world since the 1970s, with the increased resistance expanding to a wider spectrum of antibiotics (1, 12, 15, 22, 33). In this study, we found that 72.6% and 83.3% of all strains were not susceptible to penicillin and macrolides, respectively, numbers that were very close to the findings of previous studies (14, 30). One possible cause of the increase in drug resistance is long-term antimicrobial therapy (15). To date it has been thought that the priority for clinicians is to choose the appropriate chemotherapy for patients, usually with underlying diseases. The strains in this study showed the most susceptibility to IPM and VCM, a finding consistent with the results of a previous national survey in Japan (30). These findings suggested that the application of IPM and VCM might sometimes be effective in empirical therapy of pneumococcal infection with the prescription of β-lactam antibiotics.
We also found that the multidrug-resistant strains were relatively concentrated in serotypes 3, 14, 6B, 19F, 23A, and 23F. This suggested that strains of these serotypes may acquire drug resistance more easily than strains of other serotypes. Fortunately, these serotypes were covered by PS23, except for the serotype 23A strain. The vaccine coverage rate was 71.2% in this study, which indicated that preventive use of pneumococcal vaccine would be an effective measure to reduce morbidity and mortality in cases of pneumococcal infection, especially in the elderly.
As is well known, the pbp1a, pbp1b, pbp2x, pbp2a, pbp2b, and pbp3 genes encode penicillin-binding proteins (PBPs), and altered pbp1a, pbp2x, and pbp2b genes play a very important role in inducing penicillin resistance in S. pneumoniae by decreasing the affinity for β-lactam antibiotics (10, 24, 40). On the other hand, it has been reported that the erm(B) gene, which encodes the 23S rRNA methylase, and the mef(A) gene, which is assumed to encode the efflux pump system, are the predominant mechanisms of macrolide resistance (36, 38). No analyses of these genes as an alternative application of PFGE for detecting the transmission route were performed in this study. Although in the field of molecular epidemiology, the incidence of mutation in these genes often gives us a hint for explaining the interlinks in the genetic transmission of a pathogen, which is useful for explaining the modification and spread of antibiotic resistance characteristics, PFGE is thought to be one of the most discriminating fingerprinting methods for verifying interstrain genetic relatedness, and in comparison to other frequently used methods such as amplified fragment length polymorphism or multilocus sequence typing, PFGE uniquely analyzes the whole chromosomal DNA and is comparatively cost-effective (11, 26).
The PFGE patterns of all 73 strains showed significant diversity based on their serotypes (data not shown). Since serotype 19F and 23F strains constituted the majority, molecular analysis focused on such strains. We found genetic relatedness for most of the serotype 19F strains isolated from either NPI patients or outpatients, and the pattern was undistinguishable from that of the Taiwan 19F clone. Our previous national survey had already demonstrated that the Taiwan19F clone was spreading widely in Japan (30), which suggested that the causative pathogen spreading between NPI patients and outpatients might have originated from the same parent strain. A previous study indicated that there was some association between hospital- and community-acquired respiratory infections caused by Moraxella catarrhalis (20). In this study, we detected pattern A in the serotype 19F group in both NPI patients and outpatients, as well as pattern d in the serotype 23F group, indicating that some association existed between NPI patients and outpatients even in pneumococcal infection.
There was an NPI outbreak caused by pattern b strains in ward 3W, 311, which proved that the pathogen can spread from patient to patient under crowded conditions. Although strains P03-31 and P02-300 and strains P02-306 and P02-308 were isolated from NPI patients on separate floors, they expressed the same PFGE patterns, c and d, respectively. Why did strains isolated from different backgrounds express similar molecular characteristics? One possible reason is that the pathogen may be spread by patients or hospital staff members. We found some evidence to support the assumption of interpatient transmission. It was a pity that we failed to prove direct transmission between hospital staff members and patients in this study. Since PFGE is the suitable fingerprinting method, it appears to be necessary to use a larger sample size and a more accurate research design in a future study.
In conclusion, the prevalence of drug-resistant S. pneumoniae was still high, and the predominant serotypes were 19F and 23F. PFGE data demonstrated that the Taiwan 19F clone was spreading widely and that the Spain 23F and Taiwan 23F clones were not spreading in Japan, but some unique clones of 23F were. The data also suggested that there was some association between isolates from NPI patients and those from outpatients. Finally, we demonstrated possible NPI transmission from patient to patient due to crowded conditions. Further work should be done to expose the possible transmission route between hospital staff members and patients.
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Appelbaum, P. C. 2002. Resistance among Streptococcus pneumoniae: implications for drug selection. Clin. Infect. Dis. 34:1613-1620. [DOI] [PubMed] [Google Scholar]
- 2.Bates, J. H., G. D. Campbell, A. L. Barron, G. A. McCracken, P. N. Morgan, E. B. Moses, and C. M. Davis. 1992. Microbial etiology of acute pneumonia in hospitalized patients. Chest 101:1005-1012. [DOI] [PubMed] [Google Scholar]
- 3.Bouza, E., V. Pintado, S. Rivera, R. Blazquez, P. Munoz, E. Cercenado, E. Loza, M. Rodriguez-Creixems, and S. Moreno. 2005. Nosocomial bloodstream infections caused by Streptococcus pneumoniae. Clin. Microbiol. Infect. 11:919-924. [DOI] [PubMed] [Google Scholar]
- 4.Carter, R. J., G. Sorenson, R. Heffernan, J. A. Kiehlbauch, J. S. Kornblum, R. J. Leggiadro, L. J. Nixon, W. A. Wertheim, C. G. Whitney, and M. Layton. 2005. Failure to control an outbreak of multidrug-resistant Streptococcus pneumoniae in a long-term-care facility: emergence and ongoing transmission of a fluoroquinolone-resistant strain. Infect. Control Hosp. Epidemiol. 26:248-255. [DOI] [PubMed] [Google Scholar]
- 5.de Galan, B. E., P. M. van Tilburg, M. Sluijter, S. J. Mol, R. de Groot, P. W. Hermans, and A. R. Jansz. 1999. Hospital-related outbreak of infection with multidrug-resistant Streptococcus pneumoniae in the Netherlands. J. Hosp. Infect. 42:185-192. [DOI] [PubMed] [Google Scholar]
- 6.Fang, G. D., M. Fine, J. Orloff, D. Arisumi, V. L. Yu, W. Kapoor, J. T. Grayston, S. P. Wang, R. Kohler, R. R. Muder, et al. 1990. New and emerging etiologies for community-acquired pneumonia with implications for therapy. A prospective multicenter study of 359 cases. Medicine (Baltimore) 69:307-316. [DOI] [PubMed] [Google Scholar]
- 7.File, T. M., Jr. 2004. Streptococcus pneumoniae and community-acquired pneumonia: a cause for concern. Am. J. Med. 117(Suppl. 3A):39S-50S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folkersen, B. H., T. Hojbjerg, T. R. Urth, and H. C. Schonheyder. 2001. Nosocomial infections with penicillin-resistant Streptococcus pneumoniae. Four clusters with pneumococci serotype 9V. Ugeskr. Laeger 163:2362-2365. (In Danish.) [PubMed] [Google Scholar]
- 9.Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1988. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control 16:128-140. [DOI] [PubMed] [Google Scholar]
- 10.Hakenbeck, R., M. Tarpay, and A. Tomasz. 1980. Multiple changes of penicillin-binding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 17:364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermans, P. W., M. Sluijter, T. Hoogenboezem, H. Heersma, A. van Belkum, and R. de Groot. 1995. Comparative study of five different DNA fingerprint techniques for molecular typing of Streptococcus pneumoniae strains. J. Clin. Microbiol. 33:1606-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue, M., N. Y. Lee, S. W. Hong, K. Lee, and D. Felmingham. 2004. PROTEKT 1999-2000: a multicentre study of the antibiotic susceptibility of respiratory tract pathogens in Hong Kong, Japan and South Korea. Int. J. Antimicrob. Agents 23:44-51. [DOI] [PubMed] [Google Scholar]
- 13.Ishida, T., T. Hashimoto, M. Arita, I. Ito, and M. Osawa. 1998. Etiology of community-acquired pneumonia in hospitalized patients: a 3-year prospective study in Japan. Chest 114:1588-1593. [DOI] [PubMed] [Google Scholar]
- 14.Kasahara, K., K. Maeda, K. Mikasa, K. Uno, K. Takahashi, M. Konishi, E. Yoshimoto, K. Murakawa, E. Kita, and H. Kimura. 2005. Clonal dissemination of macrolide-resistant and penicillin-susceptible serotype 3 and penicillin-resistant Taiwan 19F-14 and 23F-15 Streptococcus pneumoniae isolates in Japan: a pilot surveillance study. J. Clin. Microbiol. 43:1640-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klugman, K. P., D. E. Low, J. Metlay, J. C. Pechere, and K. Weiss. 2004. Community-acquired pneumonia: new management strategies for evolving pathogens and antimicrobial susceptibilities. Int. J. Antimicrob. Agents 24:411-422. [DOI] [PubMed] [Google Scholar]
- 16.Lefevre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefevre, J. C., A. M. Gasc, J. Lemozy, A. M. Sicard, and G. Faucon. 1994. Pulsed field gel electrophoresis for molecular epidemiology of penicillin resistant Streptococcus pneumoniae strains. Pathol. Biol. (Paris) 42:547-552. [PubMed] [Google Scholar]
- 18.Little, J. R., J. Miller, and M. G. Kahn. 1998. Nosocomial penicillin-resistant pneumococcal infections at a Midwestern university hospital. Infect. Control Hosp. Epidemiol. 19:782-784. [DOI] [PubMed] [Google Scholar]
- 19.Marrie, T. J., H. Durant, and L. Yates. 1989. Community-acquired pneumonia requiring hospitalization: 5-year prospective study. Rev. Infect. Dis. 11:586-599. [DOI] [PubMed] [Google Scholar]
- 20.Masaki, H., N. Asoh, K. Kawazoe, K. Watanabe, T. Onizuka, S. Shimogama, T. Yamaryo, H. Watanabe, K. Oishi, and T. Nagatake. 2003. Possible relationship of PFGE patterns of Moraxella catarrhalis between hospital- and community-acquired respiratory infections in a community hospital. Microbiol. Immunol. 47:379-385. [DOI] [PubMed] [Google Scholar]
- 21.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mera, R. M., L. A. Miller, J. J. Daniels, J. G. Weil, and A. R. White. 2005. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States over a 10-year period: Alexander Project. Diagn. Microbiol. Infect. Dis. 51:195-200. [DOI] [PubMed] [Google Scholar]
- 23.Miyashita, N., H. Fukano, K. Mouri, M. Fukuda, K. Yoshida, Y. Kobashi, Y. Niki, and M. Oka. 2005. Community-acquired pneumonia in Japan: a prospective ambulatory and hospitalized patient study. J. Med. Microbiol. 54:395-400. [DOI] [PubMed] [Google Scholar]
- 24.Munoz, R., C. G. Dowson, M. Daniels, T. J. Coffey, C. Martin, R. Hakenbeck, and B. G. Spratt. 1992. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 6:2461-2465. [DOI] [PubMed] [Google Scholar]
- 25.NCCLS. 1998. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. NCCLS, Wayne, PA.
- 26.Neeleman, C., C. H. Klaassen, H. A. de Valk, M. T. de Ruiter, and J. W. Mouton. 2004. Amplified fragment length polymorphism fingerprinting is an effective technique to distinguish Streptococcus pneumoniae from other streptococci and an efficient alternative to pulsed-field gel electrophoresis for molecular typing of pneumococci. J. Clin. Microbiol. 42:369-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paradisi, F., and G. Corti. 1998. Is Streptococcus pneumoniae a nosocomially acquired pathogen? Infect. Control Hosp. Epidemiol. 19:578-580. [DOI] [PubMed] [Google Scholar]
- 28.Paradisi, F., G. Corti, and R. Cinelli. 2001. Streptococcus pneumoniae as an agent of nosocomial infection: treatment in the era of penicillin-resistant strains. Clin. Microbiol. Infect. 7(Suppl. 4):34-42. [DOI] [PubMed] [Google Scholar]
- 29.Pfaller, M. A., R. N. Jones, S. A. Marshall, M. B. Edmond, R. P. Wenzel, et al. 1997. Nosocomial streptococcal blood stream infections in the SCOPE Program: species occurrence and antimicrobial resistance. Diagn. Microbiol. Infect. Dis. 29:259-263. [DOI] [PubMed] [Google Scholar]
- 30.Qin, L., H. Watanabe, H. Yoshimine, H. Guio, K. Watanabe, K. Kawakami, A. Iwagaki, H. Nagai, H. Goto, T. Kuriyama, Y. Fukuchi, T. Matsushima, S. Kudoh, K. Shimada, K. Matsumoto, T. Nagatake, T. Mizota, and K. Oishi. 2006. Antimicrobial susceptibility and serotype distribution of Streptococcus pneumoniae isolated from patients with community-acquired pneumonia and molecular analysis of multidrug-resistant serotype 19F and 23F strains in Japan. Epidemiol. Infect. 134:1188-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rello, J., R. Rodriguez, P. Jubert, B. Alvarez, et al. 1996. Severe community-acquired pneumonia in the elderly: epidemiology and prognosis. Clin. Infect. Dis. 23:723-728. [DOI] [PubMed] [Google Scholar]
- 32.Restrepo, M. I., J. H. Jorgensen, E. M. Mortensen, and A. Anzueto. 2001. Severe community-acquired pneumonia: current outcomes, epidemiology, etiology, and therapy. Curr. Opin. Infect. Dis. 14:703-709. [DOI] [PubMed] [Google Scholar]
- 33.Song, J. H., S. I. Jung, K. S. Ko, N. Y. Kim, J. S. Son, H. H. Chang, H. K. Ki, W. S. Oh, J. Y. Suh, K. R. Peck, N. Y. Lee, Y. Yang, Q. Lu, A. Chongthaleong, C. H. Chiu, M. K. Lalitha, J. Perera, T. T. Yee, G. Kumarasinghe, F. Jamal, A. Kamarulzaman, N. Parasakthi, P. H. Van, C. Carlos, T. So, T. K. Ng, and A. Shibl. 2004. High prevalence of antimicrobial resistance among clinical Streptococcus pneumoniae isolates in Asia (an ANSORP study). Antimicrob. Agents Chemother. 48:2101-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sopena, N., and M. Sabria. 2005. Multicenter study of hospital-acquired pneumonia in non-ICU patients. Chest 127:213-219. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian, D., J. A. Sandoe, V. Keer, and M. H. Wilcox. 2003. Rapid spread of penicillin-resistant Streptococcus pneumoniae among high-risk hospital inpatients and the role of molecular typing in outbreak confirmation. J. Hosp. Infect. 54:99-103. [DOI] [PubMed] [Google Scholar]
- 36.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss, K., C. Restieri, R. Gauthier, M. Laverdiere, A. McGeer, R. J. Davidson, L. Kilburn, D. J. Bast, J. de Azavedo, and D. E. Low. 2001. A nosocomial outbreak of fluoroquinolone-resistant Streptococcus pneumoniae. Clin. Infect. Dis. 33:517-522. [DOI] [PubMed] [Google Scholar]
- 40.Williamson, R., R. Hakenbeck, and A. Tomasz. 1980. In vivo interaction of beta-lactam antibiotics with the penicillin-binding proteins of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 18:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodhead, M. A., J. T. Macfarlane, J. S. McCracken, D. H. Rose, and R. G. Finch. 1987. Prospective study of the aetiology and outcome of pneumonia in the community. Lancet i:671-674. [DOI] [PubMed] [Google Scholar]
- 42.Yano, H., M. Suetake, A. Kuga, K. Irinoda, R. Okamoto, T. Kobayashi, and M. Inoue. 2000. Pulsed-field gel electrophoresis analysis of nasopharyngeal flora in children attending a day care center. J. Clin. Microbiol. 38:625-629. [DOI] [PMC free article] [PubMed] [Google Scholar]