Abstract
Neisseria gonorrhoeae multiantigen sequence typing (NG-MAST) is a highly discriminatory molecular typing procedure that provides precise and unambiguous strain characterization. Since molecular typing can complement contact tracing for reconstructing gonorrhea sexual networks, the concordance between the NG-MAST genotypes of pairs of N. gonorrhoeae isolates from recent sexual contacts was examined. Among 72 pairs of gonococci from recent sexual contacts, the genotypes of each pair were concordant in 65 cases (90.3%). In two further pairs, the isolates from sexual contacts differed by only a single nonsynonymous substitution in the porin gene, and in both of these pairs, the isolates were the same by opa typing. The other five nonconcordant pairs of isolates were clearly different strains. opa typing data were available for 51 of the pairs of isolates from sexual contacts, and concordant opa types were obtained in 38 cases (74.5%). NG-MAST should therefore be better than opa typing at identifying recent sexual contacts and has the important advantage over opa typing of being a more precise method of strain characterization.
Typing of Neisseria gonorrhoeae isolates has provided a valuable adjunct to contact tracing for reconstructing sexual networks (3, 14) and for identifying individuals predicted to be in the same sexual network (2, 6). Phenotypic typing methods for gonococci—for example, the combination of auxotype and serovar—lack sufficient discrimination for this purpose, but a number of more-discriminatory molecular methods have been developed. Of these, pulsed-field gel electrophoresis and opa typing are considered to be the most highly discriminatory but have the disadvantage that they require the comparison of complex DNA fragment patterns on agarose or acrylamide gels (10, 12). More recently there has been a move toward the use of digital data for molecular typing, since this is more suitable for the development of molecular-typing Internet databases and the unambiguous comparison of new isolates with those already deposited in the database. In gonococci, the sequences of the porin (por) gene and fragments of both por and the transferrin-binding protein subunit B gene (tbpB) have been evaluated for molecular typing (9, 11, 13). These genes show high levels of sequence diversity among strains; they are believed to evolve rapidly, because their gene products are surface exposed and are targets of the host immune response to infection. The combination of the sequences at por and tbpB have been used to develop N. gonorrhoeae multiantigen sequence typing (NG-MAST). In this procedure each unique sequence at each locus is given a different allele number, so that a strain is defined unambiguously by two digits, corresponding to the allele numbers at por and tbpB. Each different two-digit number is assigned to a distinct sequence type (ST) that is used to describe the strain (9).
Rapid diversification of the loci used to characterize gonococci provides very high levels of discrimination between strains, but too high rates could lead to slight differences in the genotypes of isolates recovered from recent sexual contacts. We therefore used NG-MAST to examine the concordance between the genotypes of isolates of N. gonorrhoeae recovered from recent sexual contacts, in order to assess its use in supporting traditional contact-tracing methods, and compared the concordance with that obtained using opa typing.
MATERIALS AND METHODS
Gonococcal isolates were recovered from patients attending the genitourinary clinic at the Royal Hallamshire Hospital in Sheffield, United Kingdom, between March 1995 and December 1998. An additional set of isolates came from patients attending the same clinic between May 1999 and April 2000. The great majority of patients in Sheffield with gonorrhea are diagnosed and managed by this clinic, and most of those diagnosed elsewhere are referred to the clinic for management. Patients are interviewed at their first visit to the clinic, and behavioral data and information on sexual contacts during the last 3 months are recorded. Patients are encouraged to ask their sexual contacts to visit the clinic (and if necessary, clinic staff) and, with permission from the index case patient, to inform the contacts of their potential exposure to gonorrhea. Components of the sexual network were reconstructed using the contact-tracing data, and the recorded dates at which specimens were obtained from named sexual contacts were used to identify pairs of gonococcal isolates from recent, mutually named sexual contacts.
N. gonorrhoeae was grown on single-strength Difco GC medium base (Becton Dickinson, Oxford, United Kingdom) supplemented with 1% Vitox (Oxoid, Basingstoke, United Kingdom) and incubated at 37°C under 5% CO2. Chromosomal DNA was prepared and NG-MAST carried out as described by Martin et al. (9). Previously identified alleles and STs were obtained by interrogating the NG-MAST website (www.ng-mast.net); previously unrecognized alleles and allele combinations were submitted to the website in order to obtain the new allele and ST numbers. opa typing was performed as described by O'Rourke et al. (10) using TaqI. Isolates were considered to have the same opa type if the DNA fragment patterns were indistinguishable. Genotypic diversity was measured using Simpson's index of diversity (5) with its 95% confidence limit (4).
RESULTS
Identification of recent sexual contacts.
Exhaustive contact tracing of individuals with gonorrhea in Sheffield allowed the reconstruction of components of the sexual network. These data were used to identify named sexual contacts where the N. gonorrhoeae isolates were available for molecular typing. We then evaluated the concordance between the genotypes of the gonococcal isolates recovered from pairs of individuals in Sheffield who were most clearly identified as recent sexual contacts. Two criteria were used to identify recent sexual contacts. Each of the contacts must have named the other as a sexual contact (mutual naming), and the gonococcal isolates from the contacts must have been from specimens taken at the clinic within 1 month of each other. The average time between recovery of isolates from all 72 mutually named sexual contacts was 6.1 days (range, 0 to 29 days), and 72.2% of the pairs of isolates were recovered within a week of each other.
Analysis of gonococci from recent sexual contacts.
Between March 1995 and December 1998, there were 56 pairs of sexual contacts who met the above criteria, and 51 of the 56 pairs of gonococcal isolates from these contacts (91.1%) were indistinguishable by NG-MAST. Two of the five nonconcordant pairs had the same tpbB allele and differed at only a single nonsynonymous nucleotide site in por, which altered an amino acid in either loop 5 or loop 7 of the Por protein. Both of these pairs had indistinguishable opa types. For 51 of the 56 pairs of isolates from sexual contacts, there were opa-typing data; 38 of 51 pairs (74.5%) had indistinguishable opa types.
During this period, two major strains were identified in Sheffield using opa typing (7). All 167 isolates recovered between December 1995 and November 1996 were characterized by NG-MAST and confirmed the presence of the two major strains, which corresponded to ST12 and ST261 (16.2% and 26.3% of the isolates, respectively). Several of the pairs of sexual contacts were infected with ST12 or ST261, and it is possible that for some sexual contacts, infection with these prevalent strains had not been passed from one contact to the other, but the same strain had been independently acquired from additional sexual contacts. We therefore excluded pairs of sexual contacts who were both infected with either ST12 or ST261 and reexamined the proportion of sexual contacts infected with the same strain. Of the remaining 32 pairs of sexual contacts, 27 (84.4%) were infected with the same strain. Two of the nonconcordant pairs in this subset were those that differed at a single nonsynonymous nucleotide site, and if these pairs are both considered to be infections with the same strain, the concordance increases from 84.4% to 90.6%.
Isolates were also available from contact tracing carried out in Sheffield between May 1999 and April 2000. In this period, ST12 and ST261 were absent or rare (only 1 of the 119 isolates recovered during this period was ST12, and none was ST261), and the gonococcal population was significantly more diverse than in the earlier sampling period. Simpson's index of diversity was 0.90 (confidence interval, 0.86, 0.93) for the earlier period and 0.96 (confidence interval, 0.95, 0.98) for the later period. Of the 16 pairs of isolates from mutually named recent sexual contacts (according to the criteria described above) recovered in this later period, 14 were concordant in genotype by NG-MAST. If the data from these two periods are combined, there was concordance between the NG-MAST genotypes for 65/72 (90.3%) pairs of isolates from recent sexual contacts (or 93.1% concordance if isolates with single nonsynonymous substitutions in por are considered to be the same strain).
Analysis of the sexual-network components that include nonconcordant strains.
Excluding the two pairs of isolates that differed at a single site within por, five pairs of isolates were nonconcordant. Four pairs were infected with completely different strains, having multiple differences in both tbpB and por. The fifth pair had the same tbpB allele and three nonsynonymous differences in por, changing amino acids in loops 5 and 6. The isolates in the latter pair had different opa types and were also considered to be completely different strains.
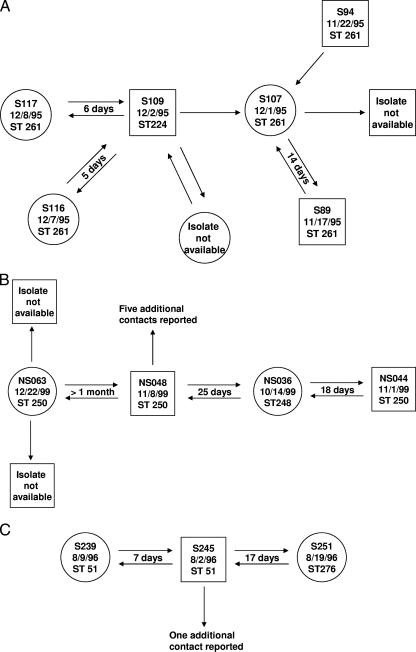
The components of the sexual network that included these five nonconcordant pairs were examined. Four of the pairs were from two sexual-network components. One involved a male (S109) infected with ST224 who named four female contacts, all within a month (Fig. 1A). Isolates were available from three of these contacts, and all were ST261. The different isolates from the male and two of the female contacts contributed two of the five nonconcordant pairs of strains. The isolate from the third female contact (S107) was not included, because there was not mutual naming. Isolates from two additional male contacts of this female were both also ST261. It is assumed that during the 1-month period in which these isolates were recovered, the male at the center of the network (S109) had been infected with ST224 and also ST261, the former infection possibly obtained from the additional contact (isolate not available) or additional undisclosed contacts.
FIG. 1.
Sexual network components including recent contacts with nonconcordant gonococcal genotypes. Panels A, B, and C show data for three network components. Circles (females) or squares (males) represent identified sexual contacts. Where an isolate was available, the information within the circle or square shows the patient number, the date of the clinic visit, and the ST. Additional contacts that are not in a circle or square were reported but unidentified. Each arrow indicates that an individual identified the other as a contact. Two arrows between individuals denotes mutual naming. The period between the visits of the contacts to the clinic are indicated.
The other component that contributed two nonconcordant pairs (Fig. 1B) included six identified individuals. Isolates were available from four, and all were ST250, except for the isolate from a female sexual contact (NS036). The isolates from this female and her two recent male contacts represented the two nonconcordant pairs. The fifth nonconcordant pair was from a small-network component (Fig. 1C) of one male who reported three female sexual contacts; one female was infected with the same strain as the male (ST51), one with a different strain (ST276), and no isolate was available from the third. Although we consider ST51 and ST276 to be different strains, they do share the same tbpB allele. However, the por alleles differ at three nucleotides, spread over a 135-bp region, indicating that for these isolates to be minor variants of the same strain, either three independent mutations in por or a single recombinational replacement would have had to occur between sexual contacts. We therefore prefer the cautious view that they are different strains.
DISCUSSION
For individuals who become infected with gonorrhea and infect a regular sexual contact, with no additional sexual contacts, there should usually be concordance between the genotypes of the isolates from the two individuals. Lack of concordance could occur either if there was incorrect or incomplete reporting of sexual contacts, if genetic variation indexed by the typing procedure evolved extremely rapidly, or if the initially infected individual had a mixed infection. In many cases, the situation is more complex, and individuals with gonorrhea may have had multiple recent sexual contacts. In such cases, different isolates may be recovered from mutually named recent sexual contacts as a result of additional infections from other sexual contacts.
Constructing gonorrhea sexual networks is difficult, because contact tracing is time-consuming and usually incomplete (1), and molecular typing can be combined with contact tracing to build a more complete picture of the sexual network (3, 14). A major requirement for using molecular typing for this purpose is that the typing method should be highly discriminatory, so that many different strains can be shown to be circulating in the community, and yet isolates from recent sexual contacts should usually be indistinguishable. NG-MAST is one of the most convenient and discriminatory of the available molecular-typing methods for N. gonorrhoeae, but except for an analysis of 10 pairs of isolates from sexual contacts that were preselected as being indistinguishable by phenotypic and molecular methods (9), as far as we are aware, there is no information on the degree of concordance between the genotypes of isolates from known sexual contacts obtained using this method.
In this study, the degree of concordance determined by using NG-MAST and sexual-contact tracing data was 90.3 to 93.1%; the higher value included the two pairs of isolates that differed at a single nonsynonymous site within por as the same strain. Concordance was higher for NG-MAST than for opa typing, presumably because variation in the opa gene repertoire occurs more rapidly than variation in por and tbpB. Two examples were found of changes in an ST between sexual contacts due to single nonsynonymous nucleotide substitutions in por, which had presumably occurred in one of the sexual contacts as a result of selection imposed by the host immune response, consistent with the fact that in both cases these substitutions alter an amino acid within a predicted surface-exposed loop in the porin protein.
The other examples of nonconcordance were due to completely different strains in sexual contacts, presumably due to additional infections from other sexual contacts or to mixed infection. Under such circumstances, nonconcordance is inevitable with any molecular typing method. Previous studies have shown that mixed gonococcal infections occur not infrequently but are difficult to detect after culturing, presumably due to overgrowth of one of the strains (8). Nonconcordance has been observed using other molecular-typing procedures. For example, 2/17 pairs of isolates from sexual contacts in Baltimore were nonconcordant by opa typing, and 1 of these 17 pairs was nonconcordant by using por sequences (13). The degree of concordance with NG-MAST in this larger study was therefore similar to that observed for these other molecular-typing procedures. NG-MAST therefore provides a molecular-typing procedure that is well suited as an adjunct to contact tracing for reconstructing sexual networks and for this purpose appears to be slightly better than opa typing, since concordance was higher. Perhaps more importantly, NG-MAST is easier to perform than opa typing, and comparisons between isolates are much simpler, because sequence data are precise and unambiguous.
Acknowledgments
This work was funded by the Wellcome Trust. B.G.S. is a Wellcome Trust Principal Research Fellow.
We thank Paul Zadik for help with the study.
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Bell, G., H. Ward, S. Day, A. C. Ghani, U. Goan, E. Claydon, and G. R. Kinghorn. 1998. Partner notification for gonorrhoea: a comparative study with a provincial and a metropolitan UK clinic. Sex. Transm. Infect. 74:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choudhury, B., C. L. Risley, A. C. Ghani, C. J. Bishop, H. Ward, K. A. Fenton, C. A. Ison, and B. G. Spratt. 2006. Identification of individuals with gonorrhoea within sexual networks: a population-based study. Lancet 368:139-146. [DOI] [PubMed] [Google Scholar]
- 3.Ghani, A. C., C. A. Ison, H. Ward, G. P. Garnett, G. Bell, G. R. Kinghorn, J. Weber, and S. Day. 1996. Sexual contact networks in the transmission of sexually transmitted diseases. An analysis of gonorrhea cases in Sheffield, UK. Sex. Transm. Dis. 23:498-503. [DOI] [PubMed] [Google Scholar]
- 4.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolader, M. E., N. H. Dukers, A. K. van der Bij, M. Dierdorp, J. S. Fennema, R. A. Coutinho, and S. M. Bruisten. 2006. Molecular epidemiology of Neisseria gonorrhoeae in Amsterdam, The Netherlands, shows distinct heterosexual and homosexual networks. J. Clin. Microbiol. 44:2689-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin, I. M., A. Ghani, G. Bell, G. Kinghorn, and C. A. Ison. 2003. Persistence of two genotypes of Neisseria gonorrhoeae during transmission. J. Clin. Microbiol. 41:5609-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin, I. M., and C. A. Ison. 2003. Detection of mixed infection of Neisseria gonorrhoeae. Sex. Transm. Infect. 79:56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin, I. M., C. A. Ison, D. M. Aanensen, K. A. Fenton, and B. G. Spratt. 2004. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J. Infect. Dis. 189:1497-1505. [DOI] [PubMed] [Google Scholar]
- 10.O'Rourke, M., C. A. Ison, A. M. Renton, and B. G. Spratt. 1995. Opa-typing: a high-resolution tool for studying the epidemiology of gonorrhoea. Mol. Microbiol. 17:865-875. [DOI] [PubMed] [Google Scholar]
- 11.Unemo, M., P. Olcén, J. Albert, and H. Fredlund. 2003. Comparison of serologic and genetic porB-based typing of Neisseria gonorrhoeae: consequences for future characterization. J. Clin. Microbiol. 41:4141-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Looveren, M., C. A. Ison, M. Ieven, P. Vandamme, I. M. Martin, K. Vermeulen, A. Renton, and H. Goossens. 1999. Evaluation of the discriminatory power of typing methods for Neisseria gonorrhoeae. J. Clin. Microbiol. 37:2183-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viscidi, R. P., J. C. Demma, J. Gu, and J. Zenilman. 2000. Comparison of sequencing of the por gene and typing of the opa gene for discrimination of Neisseria gonorrhoeae strains from sexual contacts. J. Clin. Microbiol. 38:4430-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward, H., C. A. Ison, S. E. Day, I. Martin, A. C. Ghani, G. P. Garnett, G. Bell, G. Kinghorn, and J. N. Weber. 2000. A prospective social and molecular investigation of gonococcal transmission. Lancet 356:1812-1817. [DOI] [PubMed] [Google Scholar]