Abstract
Pulsed-field gel electrophoresis was used to determine genetic diversities of multiple nontypeable Haemophilus influenzae isolates from throat and ear specimens of eight children with otitis media. From five children, all ear and throat isolates were identical. The bacterial populations in these specimens showed less diversity than populations in throat isolates of healthy children.
Nontypeable Haemophilus influenzae (NTHi) are gram-negative coccobacilli that naturally colonize the pharynges of humans, with carriage rates between 20 and 80% (1, 6, 8). In addition to colonizing asymptomatically, H. influenzae causes upper and lower respiratory tract infections, such as sinusitis, otitis media (OM), and bronchitis.
OM is a common infection of the middle-ear space, experienced by 50 to 85% of children by 3 years of age (3, 17). In children, between 30 and 52% of episodes have been attributed to infection with NTHi (4). Previous studies (9, 11, 13, 19) examining single NTHi colonies from each specimen have shown that middle-ear isolates are both genotypically and phenotypically similar to throat isolates obtained from children with OM. Recent studies from our laboratory examining multiple NTHi colonies from each specimen suggest that throat isolates of healthy children attending day care are genetically very diverse; pulsed-field gel electrophoresis (PFGE) analysis found that between 37 and 43% of children were colonized by more than one strain of NTHi (5, 16) and that the PFGE patterns of strains varied dramatically from week to week (16).
The present study was designed to examine the genetic diversity of colonization and disease-causing NTHi isolates. Simultaneous pharyngeal and middle-ear specimens were collected from six children in Bardstown, KY, and from two children in Pittsburgh, PA, with acute OM; children P6, K8, and K21 had isolates obtained from both ears. A minimum of 5 and no more than 10 H. influenzae isolates were obtained from each specimen. All isolates were suspended in ∼1 ml of sterile skim milk and stored at −80°C.
Putative H. influenzae isolates were confirmed by growth on brain heart infusion agar (Difco, Detroit, MI) supplemented with X factor (hemin), V factor (NAD), or both (Sigma-Aldrich, St. Louis, MO); by porphyrin testing; and by an isolate's failure to hemolyze horse erythrocytes as determined using the methods of Kilian (12). In addition, each isolate was tested by colony blot analysis with the monoclonal antibody 7F3 for the presence of this epitope of P6 surface antigen (14) and by DNA dot blot hybridization for the immunoglobulin A1 protease gene to distinguish suspected H. influenzae isolates from nonhemolytic Haemophilus haemolyticus (7, 12). All isolates in this study bound to the 7F3 antibody and hybridized with the iga probe, indicating that they are H. influenzae.
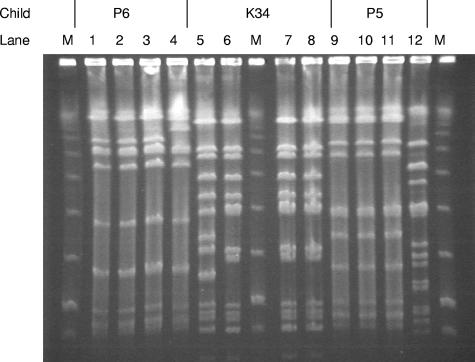
To examine genetic diversity, all study isolates were characterized by PFGE as previously described (15, 16). The PFGE patterns were first analyzed visually using the criteria of Tenover et al. (18) to determine band differences (Table 1). In five of the eight children, the nasopharyngeal and middle-ear isolates from each child were identical. Children K34, P6, and P5, however, each had one isolate that was discordant with the other isolates (Fig. 1). The unique isolates K34N2.6 and P6ME2.9 differed from the other isolates from children K34 and P6 by three and two bands, respectively. Based on Tenover et al.'s criteria (18), these isolates would be categorized as closely related to the other isolates within the respective child. The fact that they are, however, genetically distinguishable suggests that the observed diversity is the result of gain or loss of restriction enzyme sites through microbial evolution in the NTHi populations within these children. Isolate P5N2.8 differed by greater than seven bands from the other isolates from child P5 and is considered genetically unique by Tenover et al.'s criteria. The banding patterns of the isolates from two children from Bardstown, KY, K21 and K34, displayed the same PFGE pattern (with the exception of isolate K34N2.6, previously discussed). No epidemiological data are available to determine if possible bacterial sharing of strains between these two children could have occurred.
TABLE 1.
Visual and computer analyses of PFGE patterns of middle-ear and nasopharyngeal isolates from children with acute OM
| Child | No. of identical strains from indicated location/no. of strains tested by:
|
|||||
|---|---|---|---|---|---|---|
| Visual analysis
|
BioNumerics analysis
|
|||||
| Nasopharynx | Middle ear | Throat and middle ear | Nasopharynx | Middle ear | Throat and middle ear | |
| P5 | 9/10a | 10/10 | 19/20 | 9/10 | 10/10 | 19/20 |
| P6 | 10/10 | 19/20b | 29/30 | 10/10 | 19/20 | 29/30 |
| K7 | 10/10 | 8/8 | 18/18 | 10/10 | 8/8 | 18/18 |
| K8 | 10/10 | 18/18 | 28/28 | 10/10 | 18/18 | 28/28 |
| K15 | 10/10 | 10/10 | 20/20 | 10/10 | 10/10 | 20/20 |
| K17 | 8/8 | 5/5 | 13/13 | 8/8 | 5/5 | 13/13 |
| K21 | 10/10 | 20/20 | 30/30 | 10/10 | 20/20 | 30/30 |
| K34 | 9/10c | 10/10 | 19/20 | 9/10 | 10/10 | 19/20 |
Isolate P5N2.8 showed more than seven band differences from the other isolates from child P5.
Isolate P6ME2.9 showed a two-band difference from the other isolates from child P6.
Isolate K34N2.6 showed a three-band difference from the other isolates from child K34.
FIG. 1.
PFGE patterns of isolates from three children, P6 (lanes 1 to 4), K34 (lanes 5 to 8), and P5 (lanes 9 to 12), with OM. Isolates P6ME2.9 (lane 4), K34N2.6 (lane 5), and P5N2.8 (lane 12) each differ from the other isolates collected from P6, K34, and P5, respectively. Lanes M contain the size marker.
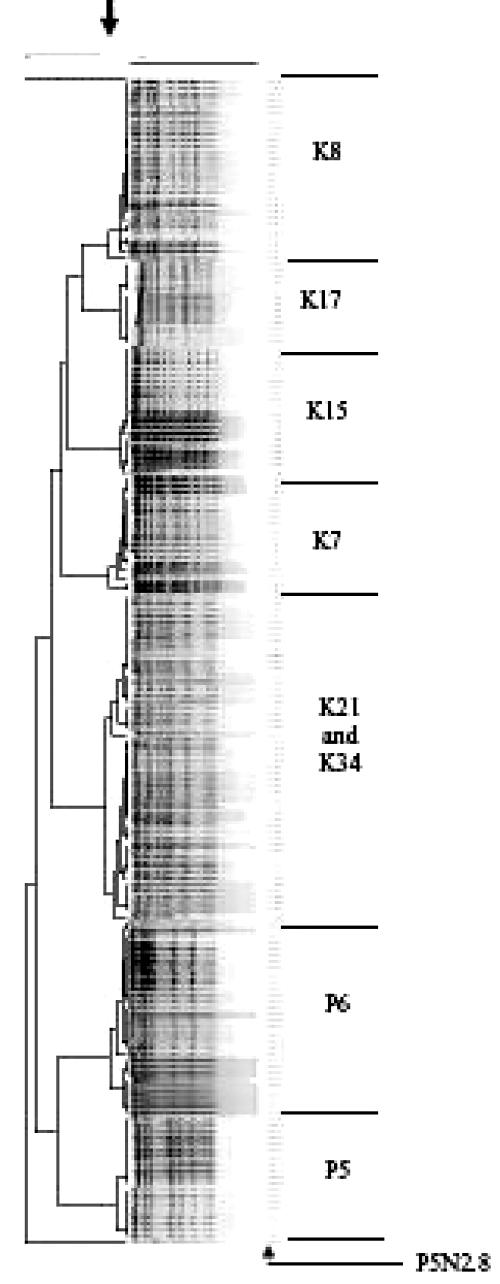
The PFGE patterns were then analyzed by the BioNumerics gel analysis software (analysis run at an optimization of 1.2% and a position tolerance of 1.2%) (Applied Math, Kortrijk, Belgium). Isolates with a similarity value of 85% or greater were considered to be identical (5). The BioNumerics results were similar to those from the visual analysis (Fig. 2). Again, in five of the eight children, the nasopharyngeal and middle-ear isolates within a child were identical. The BioNumerics analysis showed that isolates K34N2.6 and P6ME2.9 are closely related and that isolate P5N2.8 is genetically unique from the other isolates collected from each respective child.
FIG. 2.
Dendrogram (generated using BioNumerics software with the optimization setting at 1.2% and the position tolerance setting at 1.2%) of isolates digested with SmaI from children with OM. Isolates with a similarity value of greater than 85% (indicated by the arrow at the top) were considered identical.
The genetic diversity observed in throat strains from children with OM in this study was then compared to that observed in throat isolates collected from healthy children attending day care centers in Farjo et al.'s study (5). To ensure comparability, only children from Farjo et al.'s study with more than five H. influenzae (and not H. haemolyticus) isolates collected (n = 22) were included in the analysis. A diversity index was calculated for each child based on the formula derived by Hunter and Gaston (10). The following formula was utilized:
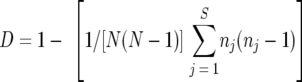 |
where D is the diversity index, N is the total number of isolates within a child, j is the PFGE type within a child, S is the total number of types described, and n is the number of isolates of that PFGE type. For this analysis, isolates with one or more band differences from the other isolates within a child were considered genetically unique PFGE types. An average diversity index was calculated for each study, and a Student t test was performed to compare the means. The results showed that two randomly chosen throat isolates from a child with OM had a 5% chance of being different, whereas two randomly chosen throat isolates from a healthy child had a 30.5% chance of being different. The throat isolates from children with OM showed significantly less diversity than those from healthy children (P = 2.37 × 10−7).
Because the children with OM may have been treated with antibiotics prior to specimen collection (but complete antibiotic histories of these children were not available to us), two steps were taken to assess the potential effect of antibiotic use on the diversity of NTHi isolates. As a first step, beta-lactamase production in the isolates was assessed. Nitrocefin disks (Remel, Lenexa, KS) were used according to the manufacturer's protocol to determine beta-lactamase production by the isolates. In seven of the eight children with OM, the beta-lactamase phenotypes were concordant among all of the isolates from each child; all of the isolates from children P6, K7, and K15 were beta-lactamase negative, and all of the isolates from children K8, K21, K17, and K34, were beta-lactamase positive. From child P5, all of the isolates, with the exception of isolate P5N2.8, were beta-lactamase negative. Isolate P5N2.8 showed beta-lactamase production. As previously discussed, the PFGE pattern from isolate P5N2.8 differed by more than seven bands, indicating that it is a genetically unique strain.
The second step to address the potential effect of antibiotics on NTHi genetic diversity relied on reanalysis of data from the previously reported study of healthy children in day care centers that included an antibiotic history of each child (2, 5). A subset of that study's population, 22 children with at least five NTHi isolates collected, were included in the analysis. Sixteen of the 22 children had no history of antibiotic use in the previous 2 weeks. Of those, nine (56%) were colonized with more than one NTHi strain. Of the six children exposed to antibiotics in the 2 weeks prior to isolate collection (four received beta-lactam-containing antibiotics, one received trimethoprim sulfamethoxazole, and two received unknown antibiotics), four (66%) were colonized with multiple strains of NTHi, indicating that antibiotic use did not cause low strain diversity in healthy children (P = 0.523 by Fisher's exact test). Thus, antibiotic use was unlikely to be the cause of the low genetic diversity observed in the pharyngeal strains of the children with OM.
The observed reduction in diversity of NTHi strains isolated from the middle ears and pharynges of children with OM compared to that of strains from pharynges of healthy children implies a selection bias favoring the persistence of pathogenic NTHi strains in the pharynges of children with OM. The commonly accepted origin of NTHi OM is as follows: NTHi colonizes the nasopharynx and migrates through the eustachian tube to the middle ear, where it persists, multiplies, and elicits an inflammatory response leading to symptomatic disease. The microbial and host factors contributing to NTHi migration into the middle-ear space are not clearly understood. Based on this general concept of OM pathogenesis and the new data suggesting a reduction in the genetic diversity of NTHi in the pharynges of children with OM, two potential models for selection arise. First, pathogenic strains could be positively selected in the pharynx by unknown mechanisms and then migrate to the middle ear, where they multiply and cause symptomatic disease. In this model, a selective force in the throat would drive the selection of pathogenic NTHi over commensal NTHi. In a second model, pathogenic NTHi strains among the large population of commensal NTHi strains first migrate to the middle ear (normally considered a sterile space), where they multiply and cause disease and then reseed the throat, where they become the predominant NTHi strains in the pharyngeal flora.
In summary, the results indicate a reduction in the genetic diversity of NTHi isolates in children with OM. Further exploration of the complex ecology between isolates and of isolates within the middle ear and the pharynx could provide valuable new information to allow us to further understand the pathogenesis of NTHi OM.
Acknowledgments
This study was supported by an award, R01-DC05840, from the National Institute on Deafness and Other Communication Disorders to J.R.G.
We thank Timothy Murphy for providing monoclonal antibody 7F3 and Ellen Wald and Stan Block for providing the H. influenzae strains.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Aniansson, G., B. Alm, B. Andersson, P. Larsson, O. Nylen, H. Peterson, P. Ringer, M. Svanborg, and C. Svanborg. 1992. Nasopharyngeal colonization during the first year of life. J. Infect. Dis. 165:S38-S42. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa-Cesnik, C., R. S. Farjo, M. Patel, J. Gilsdorf, S. I. McCoy, M. M. Pettigrew, C. Marrs, and B. Foxman. 2006. Predictors for Haemophilus influenzae colonization, antibiotic resistance, and for sharing an identical isolate among children attending 16 licensed day-care centers in Michigan. Pediatr. Infect. Dis. J. 25:219-223. [DOI] [PubMed] [Google Scholar]
- 3.Casselbrant, M. L., E. M. Mandel, P. A. Fall, H. E. Rockette, M. Kurs-Lasky, C. D. Bluestone, and R. E. Ferrell. 1999. The heritability of otitis media: a twin and triplet study. JAMA 282:2125-2130. [DOI] [PubMed] [Google Scholar]
- 4.Eskola, J., and T. Kilpi. 2000. Potential of bacterial vaccines in the prevention of acute otitis media. Pediatr. Infect. Dis. 19:72-80. [DOI] [PubMed] [Google Scholar]
- 5.Farjo, R. S., B. Foxman, M. J. Patel, L. Zhang, M. M. Pettigrew, S. I. McCoy, C. F. Marrs, and J. R. Gilsdorf. 2004. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr. Infect. Dis. J. 23:41-46. [DOI] [PubMed] [Google Scholar]
- 6.Fontanals, D., R. Bou, I. Pons, I. Sanfeliu, A. Dominguez, V. Pineda, J. Renau, C. Munoz, C. Latorre, and F. Sanches. 2000. Prevalence of Haemophilus influenzae carriers in the Catalan preschool population. Eur. J. Clin. Microbiol. Infect. Dis. 19:301-304. [DOI] [PubMed] [Google Scholar]
- 7.Fung, W. W., C. A. O'Dwyer, S. Sinha, A. L. Brauer, T. F. Murphy, J. S. Kroll, and R. R. Langford. 2006. Presence of copper- and zinc-containing superoxide dismutase in commensal Haemophilus haemolyticus isolates can be used as a marker to discriminate them from nontypeable H. influenzae isolates. J. Clin. Microbiol. 44:4222-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harabuchi, Y., H. Faden, N. Yamanaka, L. Duffy, J. Wolf, and D. Krystofik. 1994. Nasopharyngeal colonization with non-typeable Haemophilus influenzae and recurrent otitis media. J. Infect. Dis. 170:862-866. [DOI] [PubMed] [Google Scholar]
- 9.Hotomi, M., N. Yamanaka, D. S. Billal, A. Sakai, K. Yamauchi, M. Suzumoto, S. Takei, N. Yasui, S. Moriyama, and K. Kuki. 2004. Genotyping of Streptococcus pneumoniae and Haemophilus influenzae isolated from paired middle ear fluid and nasopharynx by pulsed-field gel electrophoresis. ORL J. Otorhinolaryngol. Relat. Spec. 66:233-240. [DOI] [PubMed] [Google Scholar]
- 10.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaieda, S., H. Yano, N. Okitsu, Y. Hosaka, R. Okamoto, M. Inoue, and H. Takahashi. 2005. Investigation about the homogeneity of nasopharyngeal microflora at the different location of nasopharynx of children with acute otitis media. Int. J. Pediatr. Otorhinolaryngol. 69:959-963. [DOI] [PubMed] [Google Scholar]
- 12.Killian, M. 2005. Haemophilus, p. 883-904. In D. J. Brenner, N. R. Krieg, J. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 13.Murphy, T. F., J. M. Bernstein, D. M. Dryja, A. A. Campagnari, and M. A. Apicella. 1987. Outer membrane protein and lipooligosaccharide analysis of paired nasopharyngeal and middle ear isolates in otitis media due to nontypable Haemophilus influenzae: pathogenic and epidemiological observations. J. Infect. Dis. 156:723-731. [DOI] [PubMed] [Google Scholar]
- 14.Murphy, T. F., A. L. Brauer, S. Sethi, M. Kilian, X. Cai, and A. Lesse. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 195:81-89. [DOI] [PubMed] [Google Scholar]
- 15.Pettigrew, M. M., B. Foxman, Z. Ecevit, C. F. Marrs, and J. Gilsdorf. 2002. Use of pulsed-field gel electrophoresis, enterobacterial repetitive intergenic consensus typing, and automated ribotyping to assess genomic variability among strains of nontypeable Haemophilus influenzae. J. Clin. Microbiol. 40:660-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.St. Sauver, J., C. F. Marrs, B. Foxman, R. Somsel, R. Madera, and J. R. Gilsdorf. 2000. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg. Infect. Dis. 6:622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teele, D. W., J. O. Klein, and B. Rosner. 1989. Epidemiology of otitis media during the first seven years of life in children in greater Boston; a prospective, cohort study. J. Infect. Dis. 160:83-94. [DOI] [PubMed] [Google Scholar]
- 18.Tenover, F. C., R. D. Arbeit, R. V. Goering, A. P. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villasensor-Sierra, A., and J. I. Santos. 1999. Outer membrane protein profiles of paired nasopharyngeal and middle ear isolates of nontypable Haemophilus influenzae from Mexican children with acute otitis media. Clin. Infect. Dis. 28:267-273. [DOI] [PubMed] [Google Scholar]