Abstract
Molecular diagnostics based on reverse transcription (RT)-PCR are routinely complicated by the lack of stable internal controls, leading to falsely negative results. We describe a strategy to produce a stable competitive internal control (CIC) based on a Qβ phage derivative (recombinant Qβ [rQβ]) bearing primers KY78 and KY80, which are widely used in the detection of hepatitis C virus (HCV). rQβ was RNase resistant and stable at 4°C for 452 days in SM medium (0.1 M NaCl, 8 mM MgSO4·7H2O, 50 mM Tris HCl [pH 7.5], 2% gelatin) and for 125 days after lyophilization and reconstitution. rQβ performance as a CIC was evaluated. rQβ was added to HCV-positive samples, followed by RNA extraction and a CIC-HCV RT-PCR assay. This method combines RT-PCR, liquid hybridization with nonradioactive probes, and enzyme immunoanalysis. No influence of the CIC on qualitative HCV detection was observed independently of viral load, and results had high concordance with those of commercial kits. In conclusion, we describe a versatile, low-cost alternative strategy to armored RNA technology that can be adapted for detection or real-time applications of any RNA target. Moreover, the CIC reported here is an essential reagent for HCV screening in blood banks in resource-limited settings.
Molecular diagnostic tests based on PCR preceded by reverse transcription (RT-PCR) have become the method of choice for the detection of viral pathogens with RNA genomes (24). A variety of commercial assays have been developed for several RNA viruses with high clinical impact. However, in developing countries, commercially available kits tend to be expensive and hard to find, highlighting a need for developing molecular diagnostic tools using affordable technology and standardized procedures for the clinical laboratory setting in these nations.
One of the deficiencies of in-house diagnostic methods based on RT-PCR is that they generally omit reaction-specific internal controls (ICs) to monitor the extraction, RT-PCR, and detection steps of the assay with the risk of obtaining false-negative results. An appropriate control for RT-PCR-based assays should be stable, homogeneous, economical to produce, noninfectious, absent from clinical samples, and able to verify the efficiency of the procedure at every step (24, 25). Such a control could indicate if a negative result is due to the absence of virus in the sample, the presence of reaction inhibitors, or an unsuccessful nucleic acid extraction step. However, the production of controls with the above-described characteristics has been technically difficult because of the need to produce stable and RNase-resistant RNAs.
The main strategy developed to overcome this problem includes the production of an exogenous IC that is added to the sample prior to nucleic acid extraction. For these purposes, noncompetitive ICs and competitive ICs (CICs) have been described (2, 5, 6, 7, 8, 9, 14, 25, 32). In the noncompetitive IC strategy, separate primer pairs are used to detect the control and the target of interest. This approach is attractive because a noncompetitive IC could be used as a universal control for the detection of different RNA targets (6, 7). However, amplification of such a control may not accurately reflect the amplification of the target due to differences in the amplification efficiencies between them. In contrast, the CIC strategy overcomes this limitation of noncompetitive ICs. CICs hybridize to the same primers and have identical amplification efficiencies as the target nucleic acid but contain discriminating features, such as sequence variations, that allow specific targeting using hybridization probes. Currently, armored RNAs (25) are the only technology fulfilling the ideal requirements for the production of RNase-resistant controls. Armored RNA is a noninfectious coliphage MS2 derivative that is produced by assembling specific RNA sequences and viral coat proteins that are packed as MS2 pseudoviral particles and thus protected from RNase degradation (25). This technology was first applied to the development of standards for RT-PCR detection of human immunodeficiency virus (HIV) (25), hepatitis C virus (HCV) (32), West Nile virus (9), and other high-consequence animal viruses (14). Subsequently, armored RNA CICs for the screening of blood donors for HIV (8) and enteroviruses (2) were described. These CIC RNAs contain the same primer binding sites as the target RNA but have a different probe region. Armored RNAs for various RT-PCR assays are commercially available, but their cost has prevented their widespread use.
Coliphage Qβ is a member of the Leviviridae viral family, like MS2. Qβ infects Escherichia coli F-positive strains and has a 4,219-bp RNA genome that encodes a replicase and the structural proteins C, A, and A1 (18). Different Leviviridae family members have been used for the expression of heterologous epitopes. It was shown that insertions in the C-terminal region of the A1 protein did not alter phage production and assembly, and it was possible to obtain chimeric particles that expressed the pre-S1 region of hepatitis B virus (31) or the dihydrofolate reductase enzyme in the phage capsid (16). By electron microscopy analysis, it was observed that the icosahedral structure of the recombinant particles was similar to that of the wild-type phages (31).
In this article, we describe a strategy for the production of a stable CIC based on a Qβ phage derivative to be used for the molecular detection of RNA viruses. We first applied our strategy to HCV, an RNA virus that is the major causative agent of parenterally transmitted non-A, non-B hepatitis (12, 20).
The CIC described here is a recombinant Qβ phage (rQβ) that bears the same primer pair used for the detection of HCV RNA. rQβ is added to the plasma sample and is processed together with the target nucleic acid. rQβ and HCV RNAs are reverse transcribed and amplified in a single reaction, and both amplicons are differentiated by the use of specific probes. The presence of HCV RNA in plasma was detected using the CIC-HCV RT-PCR assay, a method optimized in our laboratory. This method combines RT-PCR, liquid hybridization with nonradioactive probes, and enzyme immunoanalysis (EIA).
The strategy depicted here is a versatile and low-cost alternative to armored RNA technology for the production of stable CICs and can be adapted for the detection or real-time applications of any RNA target. Moreover, this CIC could be an essential reagent for HCV screening in blood banks in resource-limited settings.
MATERIALS AND METHODS
CIC construction.
Primers QβBst98I (5′-TATCTTAAGTCGGCAGAAAGCGTCTAGCCATGGCGTGATATTGCG GCCTA-3′) and QβNsiI (5′-GACATGCATTTCCTCGCAAGCACCCTATCAGGCAGTACGACTATCACGGC-3′) (bearing KY78 and KY80 sequences, respectively) were used for amplification using pBRT7Qβ as a template (29) (Fig. 1). pBRT7Qβ was kindly supplied by Hans Weber (Universitat Zurich, Zurich, Switzerland) and is a pBR322 derivative in which the Qβ phage genome cDNA was cloned downstream of the T7 RNA polymerase promoter. PCR was carried out in a 50-μl reaction mixture containing 0.2 mM deoxynucleoside triphosphate, 30 pmol of each primer, 2.5 U of Taq polymerase (Fermentas, Burlington, Canada), 10 mM Tris-HCl (pH 9.5), 50 mM KCl, and 3 mM MgCl2. Amplification was performed under the following cycling conditions: an initial heat denaturation step at 95°C for 2 min; 35 cycles of template denaturation (94°C for 20 s), primer annealing (50°C for 20 s), and primer extension (72°C for 20 s); and a final extension step (72°C for 5 min).
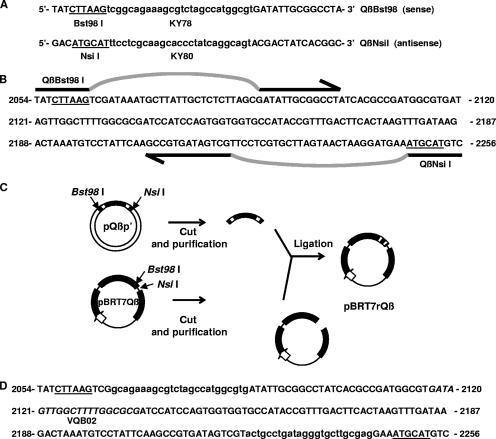
FIG. 1.
Strategy of the rQβ construction as a CIC for an HCV RT-PCR detection assay. (A) QβBst98I and QβNsiI primer sequences. (B) Strategy for KY78 and KY80 insertion into the wild-type Qβ genome. The selected region for the insertion of HCV primer binding sequences lies in the gene coding for the A1 capsid protein of Qβ (GenBank accession number M99039), where Bst98I and NsiI restriction sites are present. Primers QβBst98I and QβNsiI are indicated by arrows. According to our strategy, Primers QβBst98I and QβNsiI would hybridize pBRT7Qβ in the selected region under low-stringency PCR conditions (annealing temperature of 50°C), leading to the generation of a 200-bp chimera fragment containing KY78- and KY80-flanking Qβ sequences. (C) Construction of pBRT7rQβ. Amplicons containing heterologous sequences of KY78 and KY80 were purified and subcloned into the pGEM-T-Easy cloning vector, creating plasmid pQβp′. Each plasmid, pQβp′ and pBRT7Qβ, was digested with Bst98I and NsiI restriction enzymes. The 200-bp chimera fragment from pQβp′ was purified from the vector and ligated into pBRT7Qβ derived from the 200-bp wild-type fragment, creating pBRT7rQβ. (D) A1 region sequence of the pBRT7rQβ derivative. The correct construction of pBRT7rQβ was confirmed by sequencing of the A1 region, where heterologous primers were introduced. Note that no nucleotides were added or deleted with respect wild-type Qβ to avoid alterations in phage RNA folding and assembly. The Qβ sequence where the VQβ02 probe hybridizes is shown in italics. Wild-type Qβ sequences are shown in capital letters, and heterologous sequences (KY78 and KY80) are shown in lowercase letters. Bst98I and NsiI restriction sites are underlined.
A 200-bp fragment containing primers KY78 and KY80 (34) and flanking Qβ sequences was obtained and subcloned into pGEM-T-Easy (Promega, Madison, WI), creating pQβp′. Later, pQβp′ was digested using Bst98I and NsiI enzymes, and the 200-bp fragment was cloned into pBRT7Qβ, creating pBRT7rQβ. The entire strategy for the construction of rQβ is shown in Fig. 1 and is described in detail in Results below.
Phage lysate production.
E. coli strain BL21(DE3)/pLysS was transformed with plasmid pBRT7Qβ or pBRT7rQβ. A saturated culture was diluted and grown in LB medium containing 100 μg ampicillin/ml and 20 μg chloramphenicol/ml at 37°C until reaching an optical density (OD) at 600 nm (OD600) of 0.600. Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added, and induction was performed for 2 h at 37°C. The culture was then centrifuged for 2 min at 12,000 × g (Eppendorf microcentrifuge). Chloroform was added to supernatants, and cellular debris was eliminated by centrifugation.
Evaluation of lysate infectivity.
In order to test phage lysate infectivity, E. coli strain XL1 Blue F′, sensitive to phage Qβ, was grown in LB supplemented with tetracycline until reaching stationary phase. One hundred microliters of this culture was infected with 10 μl of lysate dilutions for 30 min in the presence of 5 mM CaCl2 and 10 mM MgCl2. Soft agar LB was added into the culture, and the mixture was spread onto petri dishes containing solid LB and incubated overnight at 37°C. Phage plaques were counted the following day.
Production of rQβ particles for use as CIC.
The production of rQβ particles for use as CIC was performed according to methods described previously by Sambrook et al. (28), with some modifications. Briefly, rQβ phage particles were obtained by transforming phage-producing plasmid pBRT7rQβ into E. coli strain BL21(DE3)/pLysS. A saturated culture was diluted and grown in LB containing 100 μg ampicillin/ml and 20 μg chloramphenicol/ml at 37°C until reaching an OD600 of 0.600. Induction was performed for 2 h after the addition of 1 mM IPTG. The culture was centrifuged at 16,000 × g for 5 min at 4°C. Supernatants were collected and warmed to room temperature. Pancreatic DNase (Sigma, Steinheim, Germany) and RNase (QIAGEN, Hilden, Germany) were added to a final concentration of 1 μg/ml and incubated overnight at room temperature. Later, solid NaCl was added to a final concentration of 1 M and was allowed to stand for 1 h on ice. Debris was removed by centrifugation at 11,000 × g for 10 min at 4°C. Supernatants were pooled, and solid polyethylene glycol 6000 (Sigma, Steinheim, Germany) was added to a final concentration of 10% (wt/vol). The mixture was incubated for at least 1 h on ice to allow bacteriophage particles to precipitate. After that, rQβ particles were recovered by centrifugation at 11,000 × g for 10 min at 4°C. The bacteriophage pellets obtained from every 500 ml of supernatant were resuspended in 8 ml of SM medium (0.1 M NaCl, 8 mM MgSO4·7H2O, 50 mM Tris HCl [pH 7.5], 2% gelatin). Phage particles were collected by centrifugation at 30,000 × g for 2 h at 4°C in a Beckman SW40 rotor. The supernatant was poured, and 1 to 2 ml of SM medium was added to each tube and incubated overnight at 4°C. The following morning, the solution was pipetted gently to ensure that all the bacteriophage particles had been resuspended. The resulting rQβ solution was kept at 4°C and constituted the CIC stock solution.
Clinical samples and RNA purification.
From February 2002 to August 2004, a total of 16 plasma samples from 16 known HCV-infected subjects were obtained from the Cátedra de Gastroenterología, Hospital Provincial del Centenario, Rosario, Argentina. An aliquot of each HCV-positive specimen was used immediately after sample collection to determine HCV viral load using Amplicor HCV Monitor 2.0 (Roche Diagnostics Corp., Indianapolis, IN) or by quantitative RT-PCR at the National Genetics Institute (LabCorp, Los Angeles, CA). Samples used in this study were obtained from patients receiving therapeutic treatment according to standardized protocols and were collected at different time points. HCV genotypes were determined by sequencing at the National Genetics Institute before therapeutic trials were started. Residual plasma was aliquoted and stored at −20°C.
RNA purification from phage lysates was performed according to an in-house method described previously (22), with the following modifications: 100 μl of phage suspension was mixed with 400 μl of a lysis solution (4 M guanidine thiocyanate, 10 mM Tris [pH 7.5], 1% β-mercaptoethanol, 1 μg of tRNA/ml) and incubated at 60°C for 10 min. RNA was precipitated with 500 μl of isopropanol at room temperature, centrifuged, washed with 70% ethanol, and resuspended in 100 μl of diethyl pyrocarbonate-H2O. All RNA samples were kept on ice (up to 2 h) until amplification.
For analysis of clinical specimens, commercial RNA purification was performed with Nuclisens isolation reagents according to the manufacturer's instructions (bioMérieux, Boxtel, The Netherlands). RNA was purified from 200 μl of plasma and eluted in 50 μl of elution buffer. All RNA recovered from each clinical sample (approximately 34 μl) was used for HCV testing using the CIC-HCV RT-PCR assay.
CIC-HCV RT-PCR assay.
The CIC-HCV RT-PCR assay system (Fig. 2) was designed with a strategy similar to those of assays previously used for the detection of other viruses (4, 10, 19). RT-PCR was carried out in 50-μl reaction mixtures containing purified RNA, 1× PCR buffer (5 mM KCl, 1 mM Tris-HCl [pH 9]), 2 mM MgCl2, 0.3 mM deoxynucleoside triphosphate, 5 mM dithiothreitol, 30 pmol of antisense biotinylated primer KY78 (B-KY78) (TIB Molbiol, Genoa, Italy), 20 U RNase inhibitor (Promega, Madison, WI), and 40 U murine leukemia virus reverse transcriptase (Promega, Madison, WI). RT reactions were carried out using a Mastercycler Personal apparatus (Eppendorf, Hamburg, Germany) at 37°C for 30 min, followed by 5 min at 95°C for murine leukemia virus reverse transcriptase inactivation. After the RT step, cDNA mixtures were placed on ice, and 50 μl of PCR mix containing 1× PCR buffer, 30 pmol of sense primer KY80 (TIB Molbiol, Genoa, Italy), 2 mM MgCl2, and 2.5 U of Taq polymerase (Fermentas, Burlington, Canada) was added to each tube. PCR mixtures were placed into the thermocycler once the temperature reached 80°C and held at 95°C for 2 min. Amplification was performed with the following profile: 5 cycles of denaturation (20 s at 95°C), primer annealing (20 s at 55°C), and DNA extension (20 s at 72°C), followed by 35 cycles of denaturation (20 s at 95°C), primer annealing (20 s at 52°C), and DNA extension (20 s at 72°C). A final DNA extension step was performed for 4 min at 72°C (Fig. 2B).
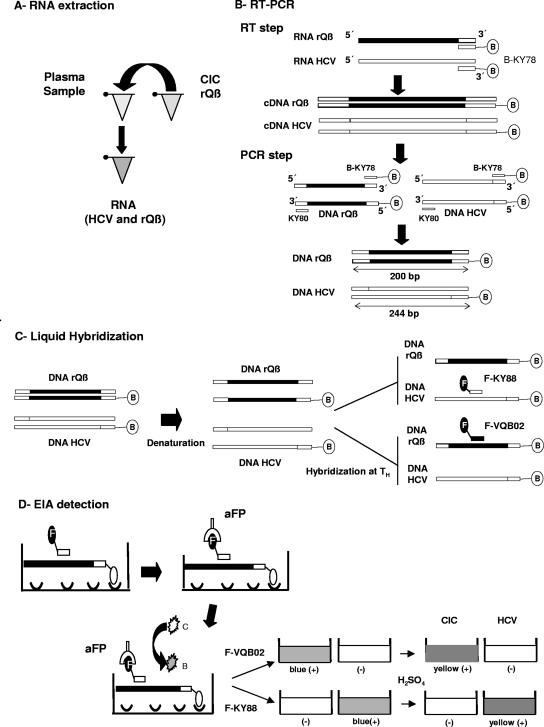
FIG. 2.
Steps of the CIC-HCV RT-PCR assay. (A) RNA extraction. A fixed amount of the CIC (rQβ) was added to the plasma sample, followed by RNA extraction. (B) RT-PCR. The CIC (rQβ) and HCV were first retrotranscribed using the antisense HCV-specific primer biotinylated in the 5′ end (B-KY78). After the RT reaction, biotinylated cDNAs were generated from both targets. Sense HCV primer (KY80) was added, and PCR amplification was performed, leading to the generation of biotinylated amplicons of 244 bp (HCV) and 200 bp (rQβ). (C) Liquid hybridization. Biotinylated amplicons were denatured and then hybridized to specific HCV (F-KY88) or rQβ (F-VQβ02) probes at the corresponding hybridization temperatures (TH). Since both probes contain fluorescein in their 5′ ends (F−), biotinylated-fluoresceinated hybrids were obtained after this procedure. (D) EIA. Biotinylated-fluoresceinated hybrids were captured in streptavidin-coated microplate wells. Colorimetric detection was performed with a monospecific anti-fluorescein antibody conjugated with horseradish peroxidase (aFP), TMB substrate, and H2O2. Reactive samples turn from colorless (C) to blue (B) by the oxidation of TMB substrate. The reaction was stopped by the addition of H2SO4, and a color change from blue to yellow occurred if the sample gave a positive result. The end-point OD was read at 450 nm in a microplate reader.
Upon completion of the amplification reaction, biotinylated amplicons were hybridized in solution with fluorescein-containing probes specific for HCV (KY88) (35) or the CIC (VQβ02 [5′-GATTGTTGGCTTTTGGCGCG-3′]). Five microliters of biotinylated PCR products was added to 120 μl of hybridization buffer (0.75 M NaCl, 75 mM sodium citrate) containing 2 pmol of the KY88 probe or 1 pmol of the VQβ02 probe. Mixtures were heated at 95°C for 5 min to denature the DNA duplex and held for 10 min at the corresponding hybridization temperature (55°C for KY88 and 65°C for VQβ02) (Fig. 2C).
For EIA, 45 μl of the hybridized products was transferred into a streptavidin-coated microplate well (Nunc, Miami, FL) for hybrid capture. Following a 1-h incubation at 37°C, unbound components were removed by extensive washing. Conjugate solution containing 1.5 U of horseradish peroxidase-conjugated anti-fluorescein antibodies (Roche, Buenos Aires, Argentina), 100 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 3% fetal calf serum was added to each microwell. The microplate was incubated for 30 min at room temperature on a microplate shaker and washed, and H2O2 and a 3,3′,5,5′-tetramethylbenzidine (TMB) solution (Wiener Laboratory, Rosario, Argentina) were added to each well. The microplate was then incubated for 30 min at room temperature to allow color development. The reaction was stopped by the addition of 1 M H2SO4, and the end-point OD was read in a microplate reader at 450 nm with a reference filter of 630 nm (Fig. 2D). Cutoff values were calculated for each experiment as two times the OD450 of the negative control. Thus, a specimen was considered to be positive or negative if its OD450 was greater or less than the cutoff value for that experiment, respectively.
rQβ stability.
The stability of the CIC stock solution was tested at −20°C, 4°C, 37°C, and 45°C. To evaluate stability at 4°C, a 10−6 dilution of the CIC stock solution was prepared in SM medium for every time point assayed. The choice of the 10−6 dilution of the CIC stock solution is explained below in Results. RNA purification was carried out from 100 μl of the diluted CIC using the modified in-house extraction method described above (22). A 20-μl aliquot of CIC RNA was tested using the CIC-HCV RT-PCR assay. To study CIC stability at −20°C, 37°C, and 45°C, 10 μl of CIC stock solution was incubated for 5 days at the assigned temperature, and 10−6 dilutions were prepared for each parameter, followed by RNA extraction and the CIC-HCV RT-PCR assay as described above.
To analyze the effect of lyophilization on CIC stability, aliquots of 10 μl of the CIC stock solution were prepared, frozen at −70°C, and then lyophilized for 1 h (FreeZone 12-liter freeze-dry system; Labconco Corporation, Kansas City, MO). After lyophilization was completed, each aliquot was incubated for 5 days at 4°C, 25°C, or 37°C. Lyophilates were reconstituted to the original volume (10 μl) with SM medium, and 10−6 dilutions for each condition were prepared. RNA was extracted and tested using the CIC-HCV RT-PCR assay.
RESULTS
Construction of the CIC rQβ.
To determine the appropriate Qβ genomic region for the insertion of primers KY78 and KY80, we performed an exhaustive analysis of the Qβ sequence to identify the coding and regulatory regions that are essential for phage viability (GenBank accession numbers M99039, X14764, M25167, and AF059242). Based on this analysis, we chose the region coding for the A1 capsid protein, which was reported previously to be capable of being modified without detriment in phage particle production (16, 31). For the inclusion of HCV primer binding sequences into the phage genome, we designed two oligonucleotides, QβBst98I and QβNsiI (Fig. 1A), that contain (i) Bst98I or NsiI restriction sites useful for cloning, (ii) KY78 or KY80 HCV primer binding sequences, and (iii) phage sequences from the A1 Qβ region to support annealing to the wild-type phage genome. These oligonucleotides were used in a PCR assay using pBRT7Qβ as a template (Fig. 1B). The amplicon was digested with Bst98I and NsiI and cloned into pBRT7Qβ cut with the same pair of enzymes that had unique recognition sites within Qβ (Fig. 1C). The correct construction of the new derivative, pBRT7rQβ, was confirmed by sequence analysis demonstrating the replacement of sequences within wild-type A1 Qβ by sequences of the 5′ noncoding region of HCV corresponding to primers KY78 and KY80 (Fig. 1D). It is important that the HCV primer binding sequences inserted into the phage genome provide a 200-bp amplicon. Thus, amplicons produced from rQβ by the CIC-HCV RT-PCR assay would be similar in size to the 244-bp products generated from the HCV target and should therefore share similar amplification efficiencies.
Analysis of rQβ phage particle production and infectivity.
BL21 bacterial cells bearing the pBRT7rQβ or pBRT7Qβ expression plasmid were induced with IPTG to promote viral particle production (rQβ or Qβ, respectively), as described in Materials and Methods. Phage lysates were tested for viral viability and infectivity in phage plaque assays using XL1 Blue bacteria as the receptor strain. In several independent experiments, and using the same methodology, the rQβ lysate consistently gave lower titers than wild-type Qβ. To corroborate the presence of the rQβ derivative in the lysate, we extracted total RNA and performed the CIC-HCV RT-PCR assay using primers KY78 and KY80 and RNA from an HCV-positive plasma as a control for the assay. For rQβ colorimetric detection, we synthesized a 20-bp fluoresceinated probe (VQβ02) from internal phage sequences flanked by KY78/KY80 (Fig. 1D). VQβ02 was specific for Qβ sequences, hybridized to the internal phage region amplified by KY78 and KY80, had a GC content similar to that of the HCV-specific probe (KY88), and was complementary to the biotinylated amplicon strand generated in the CIC-HCV RT-PCR assay. The colorimetric detection was specific for rQβ and HCV since both amplicons could be detected only by their corresponding probes.
The finding of specific PCR products from the phage lysate sample suggested the presence of rQβ in the original lysate. However, to confirm that amplicons were actually derived from rQβ RNA sequences and not from pBRT7rQβ plasmid DNA carrying the rQβ genome, the assay was performed with and without the addition of the RT enzyme to the RT reaction mixture. The results demonstrated that in the absence of RT and despite including a DNase treatment step during particle purification, there were still detectable amounts of DNA (data not shown). To eliminate remnant DNA, an appropriate dilution of the lysate was incubated with additional DNase RQ (Promega, Madison, WI), total RNA was extracted, and a CIC-HCV RT-PCR assay was conducted with and without the RT enzyme. After this treatment, amplification products were detected only in samples incubated with the RT enzyme, confirming the elimination of interfering free DNA and the presence of amplifiable rQβ RNA in the sample (data not shown).
To avoid the need to use extra DNase, which would increase the cost of CIC production, we prepared serial dilutions of the original lysates in order to dilute the interfering pBRT7rQβ plasmid in the rQβ suspension. If this strategy were to be successful, we would be able to obtain an rQβ dilution where the end-point OD values, after colorimetric detection, were positive in the samples treated with RT enzyme and below the cutoff value in its absence. To evaluate this approach, a CIC-HCV RT-PCR assay was carried out with and without the addition of the RT enzyme on RNA extracted from each serial dilution. We found that two dilutions (10−6 and 10−7) of the rQβ stock solution could adequately function as a CIC, as both dilutions had, based on the cutoff value for this particular assay (cutoff value of 0.332), positive OD values in the presence of RT enzyme (OD values of 2.710 and 2.221, respectively), and negative OD values in its absence (OD values of 0.265 and 0.214, respectively). Therefore, the dilution method allowed the selection of an optimal condition where amplification was specific for the RNA target and interfering pBRT7rQβ was eliminated. In this way, it was possible to obtain an alternative to and less expensive procedure than DNase treatment. However, it is worth to noting that the optimal dilution condition has to be determined for each independent phage stock preparation.
To corroborate that amplifiable RNA was indeed packaged in the phage particles and was therefore resistant to RNase, an aliquot of a 10−6 dilution of the stock lysate was incubated with and without RNase A (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The reaction was stopped by the addition of RNase inhibitors to the mixture (Promega, Madison, WI), the samples were then processed for RNA extraction, and the whole protocol was performed as described above. Results from two independent experiments showed that samples treated with and without RNase had similar OD values, indicating that the rQβ RNA genome was indeed packaged into the viral capsids and thus protected from RNase degradation (data not shown).
Phage stability.
As stated above, one of the requirements of an IC for diagnostic RT-PCRs is to be stable over time. The stability of the recombinant phage stock suspension was tested over a period of 452 days. For this purpose, 100 μl of a 10−6 dilution of rQβ was prepared in diethyl pyrocarbonate-H2O from the rQβ stock solution kept at 4°C. RNA was extracted using the in-house method and tested with the CIC-HCV RT-PCR assay at 0, 20, 27, 42, 64, 125, 339, 362, 376, 410, and 452 days. Results are shown in Table 1. At each time point, positive results were obtained after colorimetric detection with the VQB02 probe, indicating that the rQβ stock solution was stable for at least 452 days in SM medium at 4°C.
TABLE 1.
Stability of rQβ at 4°C
| Time (days) | OD value for CIC-HCV RT-PCR assaya | Expt cutoff OD value |
|---|---|---|
| 0 | 1.700 | 0.100 |
| 20 | 2.700 | 0.060 |
| 27 | 2.900 | 0.100 |
| 42 | 2.000 | 0.080 |
| 64 | 2.900 | 0.120 |
| 125 | 2.400 | 0.130 |
| 339 | 1.100 | 0.060 |
| 362 | 0.900 | 0.300 |
| 376 | 1.400 | 0.060 |
| 410 | 1.400 | 0.300 |
| 452 | 2.900 | 0.240 |
Results are expressed as OD values after colorimetric detection with the VQB02 probe.
Subsequently, we investigated rQβ stability at temperatures that could be compatible with different shipping and transportation conditions. Ten microliters of the rQβ stock solution was incubated at −20°C, 37°C, or 45°C for 5 days. Later, 10−6 dilutions were prepared for each temperature condition, followed by RNA extraction and CIC-HCV RT-PCR analysis. Results showed that rQβ was not completely stable at temperatures above or below 4°C (data not shown). In view of these results, we evaluated if lyophilization could improve rQβ stability. Aliquots of 10 μl of rQβ stock solution were lyophilized, kept at 4°C, 25°C, or 37°C for 5 days, resuspended in the original volume, and tested. In order to compare the ODs from the different conditions, the 10−6 dilution of the rQβ stock solution was tested as a reference. As shown in Table 2, again, only aliquots that were lyophilized and kept at 4°C were stable, suggesting that this temperature was required to ensure rQβ stability. Once reconstituted, the lyophilized aliquot was stable for at least 125 days if maintained at 4°C.
TABLE 2.
Effect of lyophilization on the stability of rQβ
| Time (days) | Storage temp (°C) | OD value for CIC-HCV RT-PCR assaya (OD value of 10−6 dilution) |
|---|---|---|
| Postlyophilization | ||
| 5 | 37 | 0.436 (2.187) |
| 5 | 25 | 0.050 (2.187) |
| 5 | 4 | 1.521 (2.187) |
| Postreconstitution | ||
| 12 | 4 | 0.843 (1.157) |
| 35 | 4 | 0.879 (0.920) |
| 82 | 4 | 1.389 (1.276) |
| 125 | 4 | 2.626 (2.729) |
Results are expressed as OD values after colorimetric detection with the VQB02 probe. All experiments showed cutoff values of <0.250. The OD of the 10−6 dilution of the rQβ stock solution tested as a reference is shown in parentheses.
Analytical sensitivity and clinical performance of the CIC-HCV RT-PCR assay.
To analyze the sensitivity for the whole assay, we used a commercially available RNA extraction kit (Nuclisens isolation reagents; bioMérieux) and the WHO International Standard (WHO IS) for HCV RNA (27). Working dilutions of the WHO IS containing 5,000, 500, and 50 IU/ml were prepared in 200 μl of HCV-negative plasma. An aliquot of rQβ stock solution was added to each prepared sample in order to obtain a 10−6 final dilution of the CIC. RNA was extracted and tested with the CIC-HCV RT-PCR assay. Under these conditions, the detection limit of the CIC-HCV RT-PCR assay was 500 IU/ml of HCV RNA, and the addition of the CIC did not affect HCV detection (data not shown). To corroborate these results, two clinical samples with extreme viral loads were analyzed in the presence and absence of the CIC in independent experiments. Results of a representative experiment are shown in Table 3 and demonstrated that the addition of the CIC did not compromise HCV detection independently of the HCV viremia level.
TABLE 3.
CIC-HCV RT-PCR analysis of samples with extreme HCV viral loads
| Sample (viremia level [IU/ml])a | CICb | CIC-HCV RT-PCR assay result (sample OD value/cutoff value)c
|
|
|---|---|---|---|
| HCV (KY88) | rQβ (VQB02) | ||
| A (0) | + | N (0.153/0.288) | P (2.739/0.260) |
| B (≥850,000) | − | P (2.143/0.288) | N (0.086/0.260) |
| C (600) | − | P (2.234/0.288) | N (0.125/0.260) |
| B (≥850,000) | + | P (2.160/0.288) | P (2.775/0.260) |
| C (600) | + | P (0.793/0.288) | P (2.681/0.260) |
Viral loads determined by Amplicor HCV Monitor 2.0 (Roche) (dynamic range, 600 to 850,000 IU/ml).
+, CIC (10−6 dilution of the rQβ stock solution) was added; −, CIC was not added.
Results of the CIC-HCV RT-PCR assay are expressed as positive (P) or negative (N).
Subsequently, to determine the performance of the CIC in a clinical setting, the CIC-HCV RT-PCR assay was used to analyze 16 clinical samples from HCV-infected subjects previously tested with commercially available assays for HCV viral load quantification. A fixed amount of the CIC (see above) was added to each 200-μl aliquot of plasma, and RNAs from both rQβ and HCV were coextracted and, after elution, were tested using the CIC-HCV RT-PCR assay. Results are shown in Table 4. The CIC-HCV RT-PCR assay gave concordant results for all the samples analyzed (100%) compared with commercial kits. The presence of the CIC did not compromise HCV detection independently of the viral load, and the CIC-HCV RT-PCR assay was able to detect the different HCV genotypes prevalent in Argentina as efficiently as commercial kits. However, the advantage of using the CIC was highlighted in sample 331, which was negative by CIC amplification, suggesting a false-negative result for this specimen.
TABLE 4.
Clinical performance of the CIC-HCV RT-PCR assay
| Sample | Viral load (IU/ml)b,c | Genotype | CIC-HCV RT-PCR assay result (sample OD value/cutoff value)a
|
|
|---|---|---|---|---|
| HCV | rQβ | |||
| 169 | >850,000b | 2a/2c | P (2.580/0.280) | P (0.880/0.100) |
| 181 | >850,000b | 1 | P (2.490/0.280) | P (0.700/0.100) |
| 700 | 702,000c | 1a | P (2.960/0.280) | P (0.620/0.100) |
| 701 | >2,000,000c | 1b | P (2.010/0.280) | P (0.330/0.100) |
| 716 | 819,000c | 1a | P (2.900/0.280) | P (0.510/0.100) |
| 171 | >850,000b | 1b | P (1.620/0.160) | P (0.560/0.160) |
| 173 | >850,000b | 1b | P (1.010/0.160) | P (0.850/0.160) |
| 331 | NDb | 2a/2c | N (0.030/0.160) | N (0.150/0.160) |
| 332 | NDb | 1 | N (0.080/0.160) | P (0.800/0.160) |
| 710 | 39c | 3a | P (0.460/0.160) | P (0.660/0.160) |
| 174 | >850,000b | 3a | P (0.860/0.100) | P (0.770/0.080) |
| 179 | >850,000b | 2a | P (0.200/0.100) | P (0.800/0.080) |
| 336 | NDb | 3a | N (0.060/0.100) | P (0.860/0.080) |
| 697 | NDc | 1b | N (0.080/0.100) | P (0.660/0.080) |
| 176 | >850,000b | 1b | P (2.230/0.280) | P (2.770/0.260) |
| 229 | 600b | 1b | P (0.790/0.280) | P (2.680/0.260) |
Results of the CIC-HCV RT-PCR assay are expressed as being positive (P) or negative (N).
Viral load determined by Amplicor HCV Monitor 2.0 (Roche) (dynamic range, 600 to 850,000 IU/ml).
Viral load determined by quantitative RT-PCR (NGI) (dynamic range, 39 to 2,000,000 IU/ml).
DISCUSSION
Since the discovery of PCR and related amplification techniques, it has been known that one of the greatest problems in introducing molecular diagnostics routinely is the false negativity associated in most cases with the great differences in sensitivities of home brew assays (24). This bottleneck becomes more difficult to overcome if RNA is the target molecule to be detected. This fact is reflected by the very few ICs described until now that have fulfilled the ideal requirements. In this paper, we described the construction of a Qβ recombinant derivative to be used as a CIC in RT-PCR detection assays. rQβ was developed to overcome the weaknesses derived from using naked RNA as an IC (13, 17, 21, 30, 33) and the different amplification efficiencies of stable noncompetitive ICs with respect to the target (6, 7) and as an alternative strategy to armored RNA technology (25).
First, we evaluated a system of Qβ phage particle production that consisted of the regulated expression of its genome cloned as cDNA into an expression vector in E. coli, and we demonstrated that this system was an important and useful tool for generating Qβ phage particles. For the introduction of the HCV sequences corresponding to primers KY78 and KY80, we analyzed which phage regions could be modified without detriment in phage viability. The selected region (Fig. 1B) was the Qβ A1 coding region, which was previously reported to support the insertion of heterologous epitopes with no effect on the formation of the icosahedral capsid even though A1 protein may be involved in the infection process (31). Furthermore, by nucleotide sequence comparison analysis, it was demonstrated that this region has a high grade of variability among different members of the family Leviviridae, suggesting that changes in this zone are not important for phage viability (15). However, when rQβ phage particle production and infectivity were analyzed, it was observed that the phage titer for rQβ was consistently lower than that for wild-type phage, indicating some deficiency in the replication of rQβ. It is possible that the recombinant particles were infection defective, because some of the A1 amino acids that had been replaced may be involved in the interaction with the bacterial receptor. However, this loss of infectivity may be an advantage to avoid the excessive replication of a genetically modified agent, a consideration that is important given regulation parameters in various countries related to recombinant agents.
In order to not alter the length and composition of the phage genome, which may affect RNA folding and assembly, the recombinant fragment inserted into the wild-type genome had a size identical to that of the one that replaced it and similar GC content with respect to the HCV amplicon. In this way, both sequences would have similar amplification efficiencies, an essential performance condition, as any factor affecting the amplification would have the same effect on both target templates.
We identified a dilution method to eliminate the remnant pBRT7rQβ DNA carrying the rQβ genome that made our strategy an even less expensive procedure. After that, we demonstrated that rQβ was resistant to RNase degradation and stable in SM medium at 4°C in the test period of 452 days (Table 1). Although rQβ was not completely stable at temperatures other than 4°C, its stability was preserved after lyophilization and reconstitution for 125 days if maintained at 4°C (Table 2). Therefore, shipping of rQβ requires less expense and preparation than shipping of an HCV standard or control, decreasing the cost compared to those associated with dry-ice shipments.
Once rQβ was constructed and its stability was evaluated, we included it in the CIC-HCV RT-PCR assay (Fig. 2). We measured the analytical sensitivity of the whole procedure with a commercial RNA extraction method and the WHO IS, and the detection limit was 500 IU/ml of HCV RNA. Although the sensitivity was lower than that recommended internationally, which is 50 IU/ml (23), it was still within the range accepted by local consensus, equivalent to 50 to 500 IU/ml (1). In our study, we determined the test analytical sensitivity in a 200-μl sample volume because most plasma samples that were available had that maximal volume. Therefore, the sensitivity of CIC-HCV RT-PCR assay could be enhanced further by increasing the sample volume to 1 or 2 ml using the kit format from bioMérieux or with other extraction methods that allow efficient RNA purification.
We compared the clinical performance of the CIC-HCV RT-PCR assay against those of reference tests for viral load measurement, such as Amplicor and quantitative RT-PCR. Although the number of HCV-infected individuals included in our analysis was low, our preliminary data indicated that our assay performed well, with a 100% concordance with the reference test, and, more importantly, the CIC did not lower the detection limit of HCV by the CIC-HCV RT-PCR assay (Tables 3 and 4). The only false-negative result was found in sample 331, which was also negative for CIC detection. It is possible that a defect in the storage and/or handling (i.e., the presence of RNase) of this specimen could be responsible for its lack of amplification in our RT-PCR analysis. Even though sample 331 was previously shown to be HCV negative using the reference test, this result emphasizes the importance of including an IC during viral analysis using in-house molecular methods, which highly improves the accuracy of the results.
Finally, the CIC-HCV RT-PCR assay was able to detect the most prevalent HCV genotypes in South America, where genotypes 1, 2, and 3 are most commonly found (1, 11, 26). Further studies should be undertaken to demonstrate that the CIC-HCV RT-PCR assay exhibits similar HCV sensitivities for all HCV genotypes.
The development of RNase-resistant viral RNA CICs derived from coliphage MS2 using armored RNA technology was previously reported (2, 8). As rQβ, both armored CIC RNAs contain the same primer binding sites as their RNA target but have a different probe region, are packaged into phage particles, and are added to the clinical sample to allow the monitoring of both RNA extraction and RT-PCR efficiencies. The strategy presented here has the same advantages of armored technology for the production of RNase-resistant RNA controls but is cheaper because it is not subjected to patent protection and is easier to synthesize. Moreover, rQβ represents the whole phage genome and mimics the target virus in a more realistic way than armored CIC RNAs.
In summary, we describe an useful and versatile CIC based on the production of a recombinant Qβ phage. The use of this control may guarantee the quality of the results and may contribute to the reduction in false negativity due to inhibitors or improper sample manipulation or amplification reactions. We also assessed its incorporation into an in-house CIC-HCV RT-PCR assay and evaluated the analytical and clinical performance of the whole system. It is recognized that the cost of diagnostic reagents represents a barrier to the implementation of international recommendations for molecular testing in many developing regions around the world (3). Due to limited financial resources and the lack of quality control regulations, it is difficult for most clinical laboratories in those areas to implement standardized in-house or commercial molecular tests. The strategy depicted here has the flexibility of introducing a variety of different RNA sequences and could be adapted to the detection or real-time applications of other RNA pathogens of clinical significance, such as other flaviviruses, HIV, enteroviruses, or any other RNA target.
Acknowledgments
We thank Christof Biebricher from the Max Planck Institute for Biophysical Chemistry (Göttingen, Germany) and Estela Valle from the Instituto de Biología Molecular y Celular de Rosario (Rosario, Argentina) for providing wild-type Qβ phage.
This research has been partially funded by a Fogarty International Center/NIH grant through the AIDS International Training and Research Program of the Mount Sinai School of Medicine Argentina Program (grant 5D43 TW0010137).
Footnotes
Published ahead of print on 15 August 2007.
REFERENCES
- 1.Asociación Argentina para el Estudio de las Enfermedades del Hígado. 2004. Consenso Argentino hepatitis C. Asociación Argentina para el Estudio de las Enfermedades del Hígado, Buenos Aires, Argentina. http://www.aaeeh.org.ar/conce2004.php.
- 2.Beld, M., R. Minnaar, J. Weel, C. Sol, M. Damen, H. van der Avoort, P. Wertheim-van Dillen, A. van Breda, and R. Boom. 2004. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. J. Clin. Microbiol. 42:3059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell Publishing Ltd. 1999. Global surveillance and control of hepatitis C. Report of a WHO consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J. Viral Hepat. 6:35-47. [PubMed] [Google Scholar]
- 4.Chouhy, D., L. Benitez Gil, A. L. Nocito, D. Wojdyla, L. Ornella, J. Cittadini, D. Gardiol, and A. A. Giri. 2006. Development and evaluation of a colorimetric PCR system for the detection and typing of human papillomaviruses. Int. J. Mol. Med. 18:995-1003. [PubMed] [Google Scholar]
- 5.Cleland, A., P. Nettleton, L. Jarvis, and P. Simmonds. 1999. Use of bovine viral diarrhoea virus as an internal control for amplification of hepatitis C virus. Vox Sang. 76:170-174. [DOI] [PubMed] [Google Scholar]
- 6.Dingle, K. E., D. Crook, and K. Jeffery. 2004. Stable and noncompetitive RNA internal control for routine clinical diagnostic reverse transcription-PCR. J. Clin. Microbiol. 42:1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreier, J., M. Störmer, and K. Kleesiek. 2005. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J. Clin. Microbiol. 43:4551-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drosten, C., E. Seifried, and W. K. Roth. 2001. TaqMan 5′-nuclease human immunodeficiency virus type 1 PCR assay with phage-packaged competitive internal control for high-throughput blood donor screening. J. Clin. Microbiol. 39:4302-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisler, D. L., A. McNabb, D. R. Jorgensen, and J. L. Isaac-Renton. 2004. Use of an internal positive control in a multiplex reverse transcription-PCR to detect West Nile virus RNA in mosquito pools. J. Clin. Microbiol. 42:841-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gariglio, R., M. A. Taborda, R. Bortolozzi, J. L. McDermott, I. Martini, M. Borgognone, G. V. Villanova, O. E. Varnier, and A. A. Giri. 2004. Non conventional virological markers in HIV-infected patients: T-HIV DNA, 2LTR-HIV DNA and HIV RNA. Med. Buenos Aires 64:419-428. [PubMed] [Google Scholar]
- 11.Guichón, A., H. Chiparelli, A. Martınez, C. Rodrıguez, A. Trento, J. C. Russi, and G. Carballal. 2004. Evaluation of a new NASBA assay for the qualitative detection of hepatitis C virus based on the NucliSens basic kit reagents. J. Clin. Virol. 29:84-91. [DOI] [PubMed] [Google Scholar]
- 12.Gutfreund, K. S., and V. G. Bain. 2000. Chronic viral hepatitis C: management update. CMAJ 162:827-833. [PMC free article] [PubMed] [Google Scholar]
- 13.Hazari, S., S. K. Acharya, and S. K. Panda. 2004. Development and evaluation of a quantitative competitive reverse transcription polymerase chain reaction (RT-PCR) for hepatitis C virus RNA in serum using transcribed thio-RNA as internal control. J. Virol. Methods 116:45-54. [DOI] [PubMed] [Google Scholar]
- 14.Hietala, S. K., and B. M. Crossley. 2006. Armored RNA as virus surrogate in a real-time reverse transcriptase PCR assay proficiency panel. J. Clin. Microbiol. 44:67-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobson, A. B., R. Arora, M. Zuker, C. Priano, C. H. Lin, and D. R. Mills. 1998. Structural plasticity in RNA and its role in the regulation of protein translation in coliphage Qβ. J. Mol. Biol. 275:589-600. [DOI] [PubMed] [Google Scholar]
- 16.Katanaev, V. L., O. V. Kurnasov, and A. S. Spirin. 1995. Viral Qβ RNA as a high expression vector for mRNA translation in a cell-free system. FEBS Lett. 359:89-92. [DOI] [PubMed] [Google Scholar]
- 17.Kleiboeker, S. B. 2003. Applications of competitor RNA in diagnostic reverse transcription-PCR. J. Clin. Microbiol. 41:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klovins, J., V. Berzins, and J. van Duin. 1998. A long-range interaction in Qβ RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA 4:948-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDermott, J. L., A. A. Giri, I. Martini, M. Bono, M. Giacomini, A. Campelli, L. Tagliaferro, A. Cara, and O. E. Varnier. 1999. Level of human immunodeficiency virus DNA in peripheral blood mononuclear cells correlates with efficacy of antiretroviral therapy. J. Clin. Microbiol. 37:2361-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moradpour, D., A. Cerny, H. H. Markus, and E. H. Blum. 2001. Hepatitis C: an update. Swiss Med. Wkly. 131:291-298. [DOI] [PubMed] [Google Scholar]
- 21.Mueller, J., M. Gessner, A. Remberg, J. Hoch, G. Zerlauth, and P. Hanfland. 2005. Development, validation and evaluation of a homogenous one-step reverse transcriptase-initiated PCR assay with competitive internal control for the detection of hepatitis C virus RNA. Clin. Chem. Lab. Med. 43:827-833. [DOI] [PubMed] [Google Scholar]
- 22.Mulder, J., N. McKinney, C. Chistopherson, J. Sninsky, L. Greenfield, and S. Kwok. 1994. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J. Clin. Microbiol. 32:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Institutes of Health. 2002. NIH consensus statement on management of hepatitis C. NIH Consens. State Sci. Statements 19:1-46. [PubMed] [Google Scholar]
- 24.Niesters, H. G. M. 2004. Molecular and diagnostic clinical virology in real time. Clin. Microbiol. Infect. 10:5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasloske, B. L., C. R. WalkerPeach, R. D. Obermoeller, M. Winkler, and D. B. DuBois. 1998. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J. Clin. Microbiol. 36:3590-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quarleri, J. F., B. H. Robertson, V. L. Mathet, M. Feld, L. Espínola, M. P. Requeijo, O. Mandó, G. Carballal, and J. R. Oubiña. 2000. Genomic and phylogenetic analysis of hepatitis C virus isolates from Argentine patients: a 6-year retrospective study. J. Clin. Microbiol. 38:4560-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saldanha, J., N. Lelie, A. Heath, and the WHO Collaborative Study Group. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Schuppli, D., I. Barrera, and H. Weber. 1994. Identification of recognition elements on bacteriophage Qβ minus strand RNA that are essential for template activity with Qβ replicase. J. Mol. Biol. 243:811-815. [DOI] [PubMed] [Google Scholar]
- 30.Schwab, K. J., M. K. Estes, F. H. Neill, and R. L. Atmar. 1997. Use of heat release and internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 35:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasiljeva, I., T. M. Kozlovska, I. Cielens, A. K. Strelnikova, A. Kazaks, V. Ose, and P. Pumpens. 1998. Mosaic Qβ coats as a new presentation model. FEBS Lett. 431:7-11. [DOI] [PubMed] [Google Scholar]
- 32.WalkerPeach, C. R., M. Winkler, D. B. DuBois, and B. L. Pasloske. 1999. Ribonuclease-resistant RNA controls (armored RNA) for reverse transcription-PCR, branched DNA, and genotyping assays for hepatitis C virus. Clin. Chem. 45:2079-2085. [PubMed] [Google Scholar]
- 33.Wang, Q., K. Chang, M. G Han, S. Sreevatsan, and L. J. Saif. 2006. Development of a new microwell hybridization assay and an internal control RNA for the detection of porcine noroviruses and sapoviruses by reverse transcription-PCR. J. Virol. Methods 132:135-145. [DOI] [PubMed] [Google Scholar]
- 34.Young, K. K. Y., R. M. Resnick, and T. W. Myers. 1993. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J. Clin. Microbiol. 31:882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young, K. K. Y., J. J. Archer, O. Yokosura, M. Omata, and R. M. Resnick. 1995. Detection of hepatitis C virus RNA by a combined reverse transcription PCR assay: comparison with nested amplification and antibody testing. J. Clin. Microbiol. 33:654-657. [DOI] [PMC free article] [PubMed] [Google Scholar]