Abstract
The present investigation focused on genetic diversity and drug resistance of 101 Mycobacterium tuberculosis strains isolated between July 2003 and February 2005 in the Okinawa prefecture, Ryukyu Islands, Japan. A high rate of clustering (87%, eight clusters, 2 to 69 strains/cluster) was observed upon spoligotyping; most of it was due to the lower discriminatory power of this method for the Beijing lineage (n = 72; 71.3% of the isolates). The remaining diversity was limited to seven clusters (two to five isolates/cluster), with the following distribution of major lineages: ill-defined T (n = 13; 12.8%), ancestral East African-Indian (n = 6; 5.9%), Haarlem (n = 4; 4%), Latin American-Mediterranean (n = 2; 2%), X1 (n = 1; 1%), and a total absence of the central Asian clade. Three remaining strains could not be classified on the basis of their spoligotype pattern and were labeled “unknown.” Subtyping with mycobacterial interspersed repetitive units (MIRUs) in association with additional QUB minisatellites was performed to discriminate among the Beijing strains. Based on an “in-house” spoligotyping/MIRU database (n = 694 Beijing strains), eight highly discriminative MIRU loci for Beijing strains were selected (loci numbered 10, 16, 23, 26, 27, 31, 39, and 40). The highest discriminatory power (h) observed in our sample (n = 72; M-26, 0.385; M-10, 0.38; M-31, 0.255; M-16, 0.238) was too low, and 73.6% of the Beijing strains from Okinawa remained clustered. Typing of Beijing strains with additional QUB loci (with the exception of “one-copy” QUB-1451) resulted in higher discriminatory powers: QUB-11b, 0.68; QUB-11a, 0.656; QUB-26, 0.644; QUB-18, 0.553; QUB-4156, 0.5; and QUB-1895, 0.453. A definitive algorithm on the use of QUB markers to subtype Beijing isolates in expanded studies would shed light on their hypervariability, which may sometimes blur recognition between epidemiologically linked Beijing isolates. The total absence of multiple drug resistance among Beijing isolates from Okinawa, as well as the relatively older ages of the patients (majority above 60 years), shows that tuberculosis (TB) is a declining disease in Okinawa, and an adequate TB control program has successfully avoided both the emergence and the spread of multidrug-resistant TB in this insular setting.
Understanding the population structure and transmission dynamics of circulating Mycobacterium tuberculosis clades provides a unique insight into crucial public health issues, such as the appearance and persistence of variants escaping human immune response or the emergence of resistance to antibiotics. PCR-based methods, such as spoligotyping (17), may be a good indicator of strain identity by providing information about epidemiologically important clones, particularly when used in a new setting (17, 30, 40). Although spoligotyping may correctly identify “outbreak” episodes as well as reflect the spread of the disease due to human migratory movements, it is not suitable when used alone for purely epidemiological investigations as it may overestimate clustering of isolates; hence, there is a need for second-line typing methods to know the exact rate of ongoing transmission (29, 32, 37). The PCR-based 12-locus typing of mycobacterial interspersed repetitive units (MIRUs) has provided this new tool which, used in association with spoligotyping, has been adopted as the basis for large-scale, high-throughput genotyping of M. tuberculosis (19, 32). Recent findings favor the use of MIRU-variable-number tandem repeat (VNTR) typing as a more reliable and faster method for transmission analysis than the previous gold standard IS6110-restriction fragment length polymorphism method (37).
In this context, there is a paucity of data on the population structure of tubercle bacilli from Japan, which is composed of four main islands (Kyushu, Shikoku, Honshu, and Hokkaido) and numerous smaller islands, with limited studies from Shikoku (2, 21) and Hokkaido (10). A closer examination of the international spoligotype database SpolDB4 (4), which described 1,939 shared-spoligotype patterns (STs) representative of 39,295 strains from 122 countries, showed that only 0.38% of the isolates were from Japan and none was from the Ryukyus, an island chain at the eastern limit of the East China Sea. Japan is also known to contain high proportions of the M. tuberculosis Beijing genotype (21, 34), which is reportedly more often associated with development of drug resistance (3, 9) and presumably hypervirulence in animal models (7). Consequently, the present investigation focused on tubercle bacilli diversity in Okinawa, situated at the south of the Ryukyus. The study was based on spoligotyping and MIRU-VNTRs, in association to additional minisatellites named QUBs (for Queen University of Belfast [27, 28]) that recently were shown to be useful for further discrimination of the Beijing clade (15, 18), highly prevalent in an initial study from Japan (21, 34).
MATERIALS AND METHODS
Patients, mycobacterial strains, drug susceptibility, and DNA extraction.
M. tuberculosis strains (n = 101) were isolated from patients at the National Okinawa Hospital and University of the Ryukyus over a 20-month period (July 2003 to February 2005). Considering a smear positivity rate of about 9.4/100,000 for Okinawa prefecture (with a total population of 1.26 million), around 118 smear-positive cases are expected per year (tuberculosis [TB] statistics for the year 2004, Ministry of Health, Japan). Thus, an inclusion of 101 isolates in the present study represented roughly one out of two cases in our setting. Drug susceptibility testing for 12 drugs (streptomycin, isoniazid, rifampin, ethambutol, kanamycin, enviomycin, prothionamide, d-cycloserine, p-aminosalicylic acid, levofloxacin, ciprofloxacin, and pyrazinamide) was performed using the proportion method. The genomic bacterial DNA was prepared by the cetyltrimethylammonium bromide method (38) from cultures grown on freshly prepared egg-based Ogawa media at the University of the Ryukyus and sent to the Pasteur Institute of Guadeloupe for spoligotyping, MIRU and QUB typing, and subsequent analysis.
Spoligotyping and database comparison.
Spoligotyping was performed using homemade membranes according to a previously described protocol (17). Spoligotypes in binary format were entered in an Excel spreadsheet and compared to the updated international spoligotyping database of the Pasteur Institute of Guadeloupe. The SpolDB4 database initially contained 39,295 patterns distributed into 1,939 STs and 3,370 orphans (4); ST designates a pattern shared by two or more patient isolates, whereas “orphan” designates a pattern reported for a single isolate. An online version of this database is available (http://www.pasteur-guadeloupe.fr:8081/SITVITDemo). An updated “in-house” SITVIT2 version, not yet available online, contained at the time of this comparison a total of 2,808 STs (now renamed SIT for “spoligotype international type”) corresponding to 63,473 clinical isolates from 122 isolation countries and 160 countries of origin. Major phylogenetic clades were assigned according to signatures provided in SpolDB4 (4), which defines 62 genetic lineages/sublineages (refer to reference 4 for a detailed description). These include specific signatures for various M. tuberculosis subspecies, such as M. bovis, M. microti, M. caprae, M. pinnipedii, and M. africanum, as well as rules defining major lineages/sublineages for M. tuberculosis sensu stricto. The latter include the central Asian clade and its two sublineages, the East African-Indian (EAI) clade with its nine sublineages, the Haarlem clade and its three sublineages, the Latin American-Mediterranean (LAM) clade and its 12 sublineages, the “Manu” family and its three sublineages, the IS6110 low-banding X clade and its three sublineages, and an ill-defined T clade and its five sublineages.
MIRU typing and database comparison.
The PCR-based MIRU typing was performed using the 12-locus system and primers described previously (32). The PCR products were separated by electrophoresis on agarose gel using an in-house protocol, inspired by a high-throughput protocol used by W. Prodinger (Innsbruck Medical University, Austria), with modifications. Briefly, PCR products (8 to 10 μl per amplified sample) were loaded in multiple-slot rows on a 25- by 30-cm agarose gel (1.5%, wt/vol; Invitrogen Corp., Carlsbad, CA) in 1× Tris-borate-EDTA containing three rows of 51 slots (slot width, 2 mm) in a subcell model Fisherbrand HU25 (Fisher-Bioblock, Illkirch, France). A 100-bp molecular size marker (Amersham) was loaded every five to six lanes, and the fragments were separated for 3 h at 150 V. This methodology permits a full 12-locus screening of 10 isolates/apparatus/day (nine test isolates plus an H37Rv parallel control per MIRU locus). Ideally, a single experimenter can simultaneously handle three apparatus per day, which permits an efficient MIRU screening of 27 test isolates/day. The gels were stained with ethidium bromide (Sigma-Aldrich, St. Louis, MO) and photographed using a Gel-Analyst video capture system (Bioprobe, Montreuil, France), followed by the calculation of fragment sizes using Taxotron software (P. A. D. Grimont, Institut Pasteur, Paris, France). The exact MIRU copy number corresponding to the respective band size was calculated according to information provided by Philip Supply (Institut Pasteur, Lille, France) and entered into an Excel spreadsheet. The MIRU data obtained were also entered in the SITVIT2 Pasteur Guadeloupe database (see above), which at the time of this comparison contained 12-locus MIRU patterns for a total of 8,573 isolates and 882 shared types, referred to as MIT (MIRU international type).
Additional typing scheme for Beijing isolates.
Beijing and non-Beijing isolates did not undergo the same MIRU typing scheme, as the 12-locus MIRU typing is known to be less discriminatory for the Beijing genotype than for non-Beijing strains, and additional minisatellite markers, such as QUBs (27, 28), are recommended for further discrimination of Beijing strains (15, 18). Consequently, Beijing and Beijing-like strains in this study were further typed by QUB-11a, -11b, -18, and -26 (28) and QUB-1451, -1895, and -4156c (27). PCR mix for QUB-11a, -11b, -18, and -26 was prepared in a final volume of 60 μl (28) and contained 40 ng of DNA, 1.5 U of recombinant Taq (rTaq) polymerase (Amersham), 6 μl of rTaq buffer mix 10× (Amersham), 2 μl of a mix containing 25 mM of each of the deoxynucleoside triphosphates (Qbiogene), 4.8 μl of 25 mM MgCl2, 6 μl of dimethyl sulfoxide, and 4 μl of each primer diluted at 20 μM. PCRs were run under the following conditions: 94°C for 10 min, followed by 40 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 10 min. PCR mix for QUB-1451, -1895, and -4156c was prepared in a final volume of 30 μl (27) and contained 20 ng of DNA, 0.6 U of rTaq polymerase (Amersham), 3 μl of rTaq buffer mix 10× (Amersham), 1 μl of a mix containing 25 mM of each the deoxynucleoside triphosphates (Qbiogene), 2.1 μl of 25 mM MgCl2, 3 μl of dimethyl sulfoxide, and 2 μl of each primer diluted at 20 μM. PCRs for these three QUBs were run under the same conditions as for routine 12-locus MIRU typing (see above). The PCR products for all seven QUB loci were separated by electrophoresis using 1.5% agar, and the fragment sizes were determined as for MIRUs. The exact copy numbers for QUBs studied were calculated by following tables provided previously (27, 28).
Discriminatory power of MIRU and QUB markers.
The discriminatory power of MIRU and QUB markers was calculated using the Hunter and Gaston discriminatory index (HGDI) (13, 29). This index, h, is based on the probability that two unrelated strains will be placed into different typing groups. An acceptable level for discrimination depends on a number of factors; however, an index of greater than 0.90 is recommended if the typing results are to be interpreted with confidence (13). This index was calculated for each of the 12 MIRU loci for all of the Beijing strains available in our database (n = 694), and the results showed that four loci had HGDI values too low to be used for discrimination among the Beijing clade isolates (Table 1). Consequently, these four loci (MIRU-2, -4, -20, and -24) were excluded for typing Beijing strains. Regarding our samples from Okinawa, the HGDI was calculated for individual MIRU and QUB loci, as well as for associations of markers.
TABLE 1.
Allelic diversity of MIRU markers observed for the Beijing strains from this study (n = 72) and from the updated SITVIT2 database (n = 694)
| MIRU locus | No. of strains of locus type with the following no. of repeats:
|
Total | HGDI | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 14 | |||
| 2 | ||||||||||||||
| Aa | 0 | NDc | ||||||||||||
| Bb | 693 | 1 | 694 | 0.003 | ||||||||||
| 4 | ||||||||||||||
| A | 0 | ND | ||||||||||||
| B | 5 | 2 | 679 | 5 | 3 | 694 | 0.04 | |||||||
| 10 | ||||||||||||||
| A | 18 | 54 | 72 | 0.38 | ||||||||||
| B | 59 | 61 | 567 | 4 | 1 | 2 | 694 | 0.32 | ||||||
| 16 | ||||||||||||||
| A | 7 | 62 | 3 | 72 | 0.25 | |||||||||
| B | 3 | 16 | 634 | 41 | 694 | 0.16 | ||||||||
| 20 | ||||||||||||||
| A | 0 | ND | ||||||||||||
| B | 6 | 688 | 694 | 0.02 | ||||||||||
| 23 | ||||||||||||||
| A | 1 | 1 | 63 | 5 | 2 | 72 | 0.23 | |||||||
| B | 21 | 1 | 3 | 645 | 24 | 694 | 0.134 | |||||||
| 24 | ||||||||||||||
| A | 0 | ND | ||||||||||||
| B | 694 | 694 | 0 | |||||||||||
| 26 | ||||||||||||||
| A | 1 | 2 | 1 | 8 | 56 | 3 | 1 | 72 | 0.385 | |||||
| B | 3 | 1 | 10 | 13 | 236 | 41 | 361 | 17 | 10 | 1 | 1 | 694 | 0.61 | |
| 27 | ||||||||||||||
| A | 1 | 2 | 69 | 72 | 0.08 | |||||||||
| B | 18 | 21 | 646 | 8 | 693 | 0.13 | ||||||||
| 31 | ||||||||||||||
| A | 3 | 2 | 5 | 62 | 72 | 0.255 | ||||||||
| B | 11 | 13 | 29 | 605 | 34 | 1 | 1 | 694 | 0.24 | |||||
| 39 | ||||||||||||||
| A | 4 | 65 | 3 | 72 | 0.18 | |||||||||
| B | 1 | 37 | 561 | 80 | 1 | 13 | 693 | 0.33 | ||||||
| 40 | ||||||||||||||
| A | 2 | 67 | 2 | 71 | 0.11 | |||||||||
| B | 32 | 26 | 605 | 29 | 2 | 694 | 0.235 | |||||||
The rows marked A correspond to results from the study in Okinawa.
The rows marked B correspond to results from the in-house database.
ND, not done.
RESULTS
Patients and drug resistance.
This investigation aimed to describe the population structure and drug resistance patterns of M. tuberculosis clinical strains isolated between July 2003 and February 2005 at the University Hospital, Okinawa prefecture, Ryukyu Islands (n = 101; male-to-female ratio, 3.6). The mean age averaged 63.8 years, with extremes of 21 to 98 years (mean average age for females, 70 years; mean average age for males, 62 years). The age group of 60 to 90 years was the most important and represented more than 55% TB cases, followed by the age group of 30 to 60 years (40%). Interestingly, the remaining 5% of TB cases were more frequent among patients above 90 years of age (four cases) than among those below 30 years (one case). Drug resistance determination showed that 16/101 (15.8%) of the isolates were resistant to at least a single drug tested. However, irrespective of Beijing or non-Beijing genotype status of the strains (see below), none of the isolates were multidrug resistant (MDR), defined as simultaneously resistant to isoniazid and rifampin, with or without additional resistance. Streptomycin resistance concerned five isolates (mean average age of the patients, 60 years; range, 44 to 75 years); in one case it was combined with resistance to isoniazid and in another case to ethambutol. Other cases of monoresistance concerned isoniazid (n = 3) or ethambutol (n = 7), essentially among patients above 60 years of age, and a single case of ciprofloxacin monoresistance occurred in a 38-year-old patient. No monoresistance to rifampin was observed.
Analysis of spoligotyping data.
The spoligotypes obtained and the subsequent clade determinations based on rules provided in SpolDB4 are summarized in Table 2. A high rate of clustering was observed upon spoligotyping (87% of the isolates were clustered in eight clusters containing 2 to 69 strains/cluster). However, most of it was due to the lower discriminatory power of this method for the Beijing genotype, which formed the largest cluster of SIT 1 (Beijing type sensu stricto, n = 69) and a single isolate each for SITs 190, 255, and 1364 (designated pseudo-orphans [see below]), which represented Beijing-like strains in SpolDB4, i.e., 72/101 strains, or 71.3% of the isolates in our setting. Notwithstanding the latter genotype, the diversity observed was limited to seven clusters with 18.8% of the strains, containing two to five isolates per cluster: EAI2-Manilla, Haarlem3, LAM9-var, and ill-defined clades T1, T1-var, T2, and T3-Osaka.
TABLE 2.
Spoligotypes obtained for Beijing and non-Beijing strains, with additional MIRU and epidemiological data for non-Beijing strainsf
| SIT | Binary spoligotype | Clade | No. (%) of strains | 12-Locus MIRU pattern | MIT | Strain | Patient genderc | Patient age (yr) | Patient statusd | Drug resistancee |
|---|---|---|---|---|---|---|---|---|---|---|
| 19 | ▪▪□▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□▪▪▪▪▪▪▪□□□□▪□▪▪▪▪▪▪▪▪▪ | EAI2-Manilla | 5 (4.95) | 22v32vvvv5vva | Pb | RK04 | M | 32 | N | |
| 244326223422 | 729 | RK05 | M | 38 | N | CPFX | ||||
| 254326223432 | 56 | RK114 | M | 55 | N | |||||
| 254326223432 | 56 | RK69 | M | 70 | N | |||||
| 254326223432 | 56 | RK92 | M | 56 | N | |||||
| 256 | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪□▪▪▪▪□▪▪▪▪ | EAI5-var | 1 (0.99) | 264225223533 | 59 | RK95 | M | 81 | N | |
| 50 | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□▪□□□□▪▪▪▪▪▪▪ | Haarlem3 | 3 (2.97) | 325313153324 | Orphan | RK79 | M | 21 | N | |
| 222324153321 | 698 | RK57 | M | 88 | N | |||||
| 222324153321 | 698 | RK58 | M | 91 | N | |||||
| 52 | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪▪▪□▪▪▪ | T2 | 2 (1.98) | 242225152323 | 697 | RK64 | M | 45 | N | |
| 242225152323 | 697 | RK93 | M | 45 | N | |||||
| 117 | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪□▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪▪▪□▪▪▪ | T2-var | 1 (0.99) | 242225152323 | 697 | RK104 | M | 57 | N | |
| Orphan | ▪▪▪▪▪▪▪▪▪▪▪□□□□□□▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪▪▪□▪▪▪ | T2-var | 1 (0.99) | 212325153323 | Orphan | RK66 | M | 61 | N | |
| 53 | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪▪▪▪▪▪▪ | T1 | 3 (2.97) | 223326153311 | 699 | RK83 | F | 69 | N | |
| 222326153311 | 524 | RK22 | M | 78 | N | |||||
| 222326153311 | 524 | RK77 | F | 90 | N | |||||
| 2367 | ▪▪▪▪▪□▪▪□□▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪▪▪▪▪▪▪ | T1-var | 1 (0.99) | 228325163423 | 415 | RK21 | M | 60 | N | |
| 172 | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□□▪▪▪▪▪▪▪ | T1-var | 2 (1.98) | 222326153311 | 524 | RK96 | M | 52 | R | |
| 223326143311 | 723 | RK94 | M | 53 | N | |||||
| 627 | ▪▪▪▪□□□□▪▪▪▪□▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪▪▪▪▪▪▪ | T3-Osaka | 2 (1.98) | 215125113322 | 310 | RK06 | M | 74 | R | |
| 215125113322 | 310 | RK09 | M | 87 | N | |||||
| 2122 | ▪▪▪▪□□□□▪▪▪▪□▪▪▪▪□▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪▪▪▪▪▪▪ | T3-Osaka var | 1 (0.99) | 215125113322 | 310 | RK98 | M | 80 | N | |
| 388 | ▪▪▪□▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪▪▪▪▪▪▪▪□□□□▪▪▪▪▪▪▪ | LAM9-var | 2 (1.98) | 222316153226 | Orphan | RK120 | M | 70 | N | EB |
| 223316153226 | 718 | RK122 | F | 61 | N | |||||
| 742 | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□□□□▪□□□□▪▪▪▪▪▪▪ | Haarlem1-var | 1 (0.99) | 222225153323 | 730 | RK117 | M | 44 | N | EB |
| Orphan | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪□▪□□▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□▪▪▪▪▪▪▪ | X1 | 1 (0.99) | 2423251.2322 | Orphan | RK74 | M | 73 | N | |
| Orphan | ▪▪▪▪▪▪▪▪▪▪□□□▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□□□▪▪▪▪▪▪ | Unknown | 1 (0.99) | 222325153323 | 7 | RK81 | F | 77 | N | |
| 2041 | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□□□□□□□□□□▪▪▪▪▪▪▪ | Unknown | 1 (0.99) | 222325153323 | 7 | RK55 | M | 65 | N | EB |
| 2025 | ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪□□□□□□□▪▪▪▪▪▪ | Unknown | 1 (0.99) | 222325153323 | 7 | RK25 | M | 77 | N | |
| 1 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□▪▪▪▪▪▪▪▪▪ | Beijing | 69 (68) | |||||||
| 190 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□▪▪▪▪▪□▪▪▪ | Beijing-var | 1 (0.99) | |||||||
| 255 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□▪▪▪▪□▪▪▪▪ | Beijing-var | 1 (0.99) | |||||||
| 1364 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□▪▪▪□□▪▪▪▪ | Beijing-var | 1 (0.99) |
v in this MIRU pattern denotes variable locus.
P, polyclonal infection.
M, male; F, female.
N, new patient; R, re-treated patient.
CPFX, ciprofloxacin; EB, ethambutol.
See Table 3 for specific data for Beijing and Beijing-like strains.
Among the non-Beijing strains, a total of 10 isolates were unique in the present study; however, three of these matched shared types were already reported elsewhere (SITs 117, 256, and 742, hereby designated pseudo-orphans), four isolates matched a single orphan in the database, thereby creating new SITs numbered 2025, 2041, 2122, and 2367, and the three remaining isolates (RK66, RK74, and RK81) were not yet reported in the database and may be considered “true orphans” (Table 2). For the three “true orphans” as well as the four newly created SITs (2025, 2041, 2122, and 2367), the exact clade determination was not provided in SpolDB4. Nonetheless, a finer analysis suggested clade attributions for four of them: SITs 2122 and 2367 belong, respectively, to T3-Osaka and T1-var, and the orphan patterns of strains RK66 and RK74 are close to the T2 and the X1 clades, respectively. On the other hand, clades could not be assigned to newly created SITs 2041 and 2025 or to a single orphan strain, RK81.
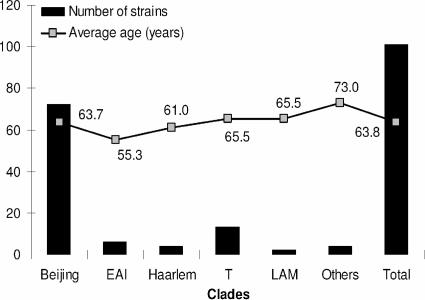
If all of the isolates, including the unique isolates from this study, were taken into account, the distribution of M. tuberculosis clades was as follows (in decreasing order): Beijing, 71.3% (n = 72); the ill-defined T superfamily, 12.8% (n = 13); the ancestral EAI clade, 5.9% (n = 6); the Haarlem clade of presumed European descent, 4% (n = 4); the LAM clade, 2% (n = 2); and the X clade, 1% (n = 1). The major M. tuberculosis clades and corresponding average age of the patients for each of them are summarized in Fig. 1.
FIG. 1.
Major clades of M. tuberculosis and average age distributions of the patients. Compared to the average age of the total sample, the age differences were not statistically significant by use of Student's t test.
MIRU typing of the non-Beijing strains.
The detailed results of MIRU typing of the non-Beijing strains are summarized in Table 2. A total of 16 distinct MIRU profiles, divided into five clusters of three isolates (MITs 7, 56, 310, 524, and 697) and one cluster of two strains (MIT 698), were found in this study. The percentage of clustered strains among the non-Beijing strains by combined spoligotyping and 12-locus MIRUs was 38% (11/29 strains). Ten unique MIRU types were found; among those, three were true orphans while seven matched previous patterns in the SITVIT2 database (Table 2). More than one band for some of the individual MIRU loci was repetitively obtained for a single isolate, RK04, indicating a polyclonal infection, and no PCR product (or one with a very low intensity) was found for one clinical isolate for MIRU-26. When a correlation between spoligotyping and MIRU patterns was made, five clusters remained (SIT19-MIT56, SIT50-MIT698, SIT52-MIT697, SIT53-MIT524, and SIT627-MIT310). Except SIT 52 and SIT 627, which were not split by MIRUs, all other spoligotyping-defined clusters were subdivided into two (SITs 50, 53, 172, and 388) or three (SIT 19) subclusters (Table 2). MITs 310, 524, and 697 were found among spoligotype variants with a single spacer change and are likely to represent true phylogenetic links. Inversely, MIT 7 (pattern 222325153323) was shared by three unrelated spoligotype variants (SITs 2025 and 2041 and an orphan spoligotype).
MIRU typing of the Beijing strains.
As described in Materials and Methods, the 69 Beijing (SIT 1) and three Beijing-like (SITs 190, 255, and 1364) strains were typed by a set of eight highly discriminative MIRU markers. A total of 30 MIRU patterns obtained are summarized in Table 3. These corresponded to 19 unique patterns and 11 clusters containing 2 to 19 strains (a single strain did not provide any result when typed with MIRU-40 and led to an incomplete MIRU profile). A total of 73.6% of the strains of the Beijing genotype were clustered. A comparison with the available data limited to the eight MIRU loci studied with the SITVIT2 database (see above) is summarized in Table 3. The 30 patterns obtained matched with 508 strains (corresponding to 34 MIRU profiles, 23 MIT and 11 orphans) in the database. The higher number of MIRU patterns after comparison (34 instead of 30) arises from the fact that the data were compared to 8 instead of 12 MIRU loci. Indeed, 5/30 patterns from this study matched with two or more 12-locus profiles, 18/30 matched with a unique profile, and 7/30 patterns were unique (Table 3). Parallel spoligotyping data were also available for 304/508 strains, making a detailed comparison possible. Interestingly enough, 296/304 (97.4%) of the isolates with spoligotyping data corresponded to a Beijing or a Beijing-like pattern, underlining that the MIRU patterns obtained based on eight loci might be sufficient to track the Beijing clone.
TABLE 3.
Eight-locus MIRU patterns obtained for Beijing and Beijing-like strains and corresponding 12-locus MIRU patterns in the updated SITVIT2 database and their worldwide distribution
| Match group | Eight-locus MIRU pattern | No. of strains | Corresponding 12-locus pattern (MIT)a | No. of strains in database | Clade (no. of strains with spoligotype) | Geographical distributionb (no. of strains) |
|---|---|---|---|---|---|---|
| A | ..12.5.73533 | 1 | 221225173533 (724) | 2 | Beijing (1) | AUS (1), JPN (1) |
| ..12.5.93533 | 1 | No pattern | None | |||
| ..13.3.83533 | 1 | No pattern | None | |||
| B | ..13.5.43533 | 1 | 221325143533 (orph) | 1 | Beijing (1) | JPN (1) |
| C | ..13.5.73433 | 2 | 221325173433 (711) | 2 | Beijing (2) | JPN (2) |
| ..13.5.73443 | 1 | No pattern | None | |||
| ..13.5.73523 | 1 | No pattern | None | |||
| D | ..13.5.73533 | 8 | 221325173533 (135) | 34 | Beijing (30) | RUS (4), SGP (1), USA (5), JPN (24) |
| E | ..13.8.73533 | 2 | 221328173533 (709) | 5 | Beijing (5) | JPN (5) |
| F | ..32.5.73533 | 4 | 223225173533 (91) | 18 | Beijing (6) | GBR (2), JPN (1), RUS (9), SGP (1), USA (3), ZAF (1) |
| G | ..32.5.83533 | 1 | 223225183533 (orph) | 1 | RUS (1) | |
| H | ..33.1.63533 | 1 | 223321563533 (orph) | 1 | Beijing-var (1) | USA (1) |
| I | ..33.5.33533 | 1 | 223325133533 (300) | 5 | Beijing (2), Beijing-var (1) | AUS (2), USA (3) |
| J | ..33.5.43533 | 1 | 223325143533 (137) | 12 | Beijing (8), Beijing-var (1), T (1) | AUS (2), NLD (1), RUS (4), USA (5) |
| K | ..33.5.53532 | 1 | 223325153532 (438) | 4 | Beijing (1) | JPN (1), RUS (1), GBR (2) |
| ..33.5.63523 | 2 | No pattern | None | |||
| L | ..33.5.63533 | 5 | 223325163533 (83) (n = 45) | 48 | Beijing (24), Beijing-var (1) | AUS (2), BEL (1), CAN (2), ESP (1), GBR (5), JPN (6), RUS (3), SGP (11), USA (14), ZAF (3) |
| 243325163533 (702) (n = 2) | ||||||
| 233325163533 (orph) (n = 1) | ||||||
| M | ..33.5.71533 | 1 | 223325171533 (orph) | 1 | GBR (1) | |
| N | ..33.5.73233 | 3 | 223325173233 (573) | 7 | Beijing (6), Beijing-var (1) | JPN (3), BEL (1), NLD (1), USA (2) |
| O | ..33.5.73333 | 1 | 223325173333 (721) (n = 4) | 5 | Beijing (3) | JPN (2), MTQ (1), GBR (1), SGP (1) |
| 223315173333 (orph) (n = 1) | ||||||
| P | ..33.5.73433 | 2 | 223325173433 (86) | 19 | Beijing (8), Beijing-var (1) | AUS (3), BGD (2), GBR (2), JPN (3), RUS (1), SGP (5), USA (3) |
| Q | ..33.5.73532 | 1 | 223325173532 (97) (n = 15) | 16 | Beijing (9), Beijing-var (1) | JPN (4), NLD (1), SGP (6), USA (5) |
| 233325173532 (orph) (n = 1) | ||||||
| R | ..33.5.73533 | 19 | 223325173533 (17) (n = 268) | 296 | Beijing (143), Beijing-var (16), Haarlem (2), T (1), undefined ST (4) | AUS (21), BEL (12), ESP (1), GBR (38), IND (2), JPN (38), NLD (2), POL (1), RUS (45), SG (40), USA (81), ZAF (15) |
| 203325173533 (214) (n = 10) | ||||||
| 213325173533 (499) (n = 3) | ||||||
| 233325173533 (570) (n = 11) | ||||||
| 223315173533 (858) (n = 2) | ||||||
| 2X3325173533 (orph) (n = 1) | ||||||
| 223325273533 (orph) (n = 1) | ||||||
| S | ..33.5.73534 | 1 | 223325173534 (706) (n = 2) | 3 | Beijing (2) | JPN (2), AUS (1) |
| 203325173534 (orph) (n = 1) | ||||||
| ..33.5.83524 | 1 | No pattern | None | |||
| T | ..33.6.73533 | 3 | 223326173533 (95) | 4 | Beijing-var (2) | USA (2), SGP (2) |
| U | ..33.6.73543 | 2 | 223326173543 (orph) | 1 | Beijing (1) | JPN (1) |
| ..34.5.72333 | 1 | No pattern | None | |||
| V | ..34.5.72533 | 1 | 223425172533 (720) | 5 | Beijing (5) | JPN (5) |
| W | ..34.5.73533 | 1 | 223425173533 (93) | 18 | Beijing (12), Beijing-var (2) | FXX (1), GBR (2), JPN (13), NLD (1), SGP (1) |
| ..33.5.7353? | 1 | No pattern | None |
orph, orphan; n, number of strains.
AUS, Australia; BEL, Belgium; BGD, Bangladesh; CAN, Canada; ESP, Spain; FXX, France; GBR, Great Britain; IND, India; JPN, Japan; MTQ, Martinique; NLA, NLD, The Netherlands; POL, Poland; RUS, Russia; SGP, Singapore; USA, United States; ZAF, South Africa.
Usefulness of QUB typing for further discrimination of Beijing genotype.
All of the Beijing and Beijing-like strains (n = 72) were further typed by QUB-11a, -11b, -18, and -26 (28) and QUB-1451, -1895, and -4156c (27) for further discrimination. Some of the QUB markers used in this investigation could not be amplified uniformly in all cases. QUB-11a appeared to have the highest failure rate, as no PCR product was found in 4 out of 72 strains, and an unusual, very-high-molecular-weight band exceeding 20 copies was obtained for two strains. Uninterpretable results were also obtained for QUB-11b (three strains) and QUB-18 and QUB-1451 (one strain each). For two strains, PCR amplification for QUB-11a and QUB-11b cumulatively provided no result. One strain (isolate RK59) obtained three equal-intensity bands, which could indicate a polyclonal infection. However, no similar result was observed for MIRUs with this strain. Thus, the complete QUB data were available for 63/72 of the isolates. With the exception of QUB-1451, which showed a unique “one-copy” pattern for all Beijing strains, high polymorphism was observed for the remaining six QUB loci (Table 4), and the number of repeats ranged from 2 copies (QUB-11b, -26, -1895, and -4156) to 13 copies (QUB-18). In summary, QUB typing alone resulted in a total of 55 patterns (9 incomplete) and 13 clusters containing 30 isolates (one cluster of four strains, two clusters of three strains, and 10 clusters of two strains [results not shown]). Last, combined typing using eight-locus MIRU and QUBs reduced the number of clustered isolates to 17 (one cluster of three strains and seven clusters of two strains [data shown in the table in the supplemental material]).
TABLE 4.
Parameters of QUB locus variability
| QUB locus | Allelic diversity (HGDI) | Allele no. | Range (no. of repeats) |
|---|---|---|---|
| QUB-11a | 0.656 | 8 | 3-11 |
| QUB-11b | 0.680 | 7 | 2-8 |
| QUB-18 | 0.553 | 8 | 5-13 |
| QUB-26 | 0.644 | 7 | 2-10 |
| QUB-1451 | 0 | 1 | 1 |
| QUB-1895 | 0.453 | 4 | 2-5 |
| QUB-4156 | 0.500 | 4 | 2-5 |
Discriminatory power of MIRU and QUB markers.
As summarized in Table 1, MIRU loci with the highest discriminatory power in our sample from Japan (n = 72) are M-26 (h = 0.385) > M-10 (h = 0.380) > M-31 (h = 0.255) > M-16 (h = 0.238). As none of the MIRU loci displayed an HGDI value greater than 0.385, no MIRU loci could be labeled highly discriminating according to definitions proposed in a previous study (29), and MIRU-10 and -26, despite their highest discriminatory index among the eight MIRU loci, are regarded moderately discriminating. If the same values are assessed on the full sample of Beijing strains in the database (n = 694), the four most discriminative MIRUs were M-26 (h = 0.610) > M-39 (h = 0.331) > M-10 (h = 0.318) > M-31 (h = 0.236). Regarding QUBs, their discriminatory power is significantly higher in our sample than that of MIRUs for Beijing strains and was as high as 0.68 for QUB-11b (Table 4).
DISCUSSION
With 30 new cases per 100,000 inhabitants in 2004, the incidence of TB in Japan remains about six times higher than in the United States and about three times higher than in most of the Western industrialized nations (http://globalatlas.who.int/globalatlas/dataQuery). Furthermore, the proportion of elderly (patients aged 60 or older) among newly registered patients in Japan increased from 18% in 1960 to 55% in 1997 (http://www1.mhlw.go.jp/english/wp_5/vol1/p2c6s2.html), and the incidence of patients over 70 was as high as 90.2/100,000 inhabitants in the year 2001 (10).
We focused on M. tuberculosis clinical isolates collected from Okinawa prefecture in the Ryukyu Islands, which was administrated by the U.S. Civil Administration of Ryukyus (USCAR) from April 1945 to May 1972, following World War II (http://www.jica.go.jp/english/resources/publications/study/topical/okinawa/pdf/okinawa.pdf). After reversion to Japan, a modern system of TB surveillance and control was implemented in 1976, which led to a gradual decrease in the peak of TB incidence for younger age groups. However, TB still remains common among the elderly both in Japan and in Okinawa (http://www.jica.go.jp/english/resources/publications/study/topical/okinawa/pdf/okinawa.pdf). For example, TB morbidity in 1997 was five times more elevated among the elderly (TB morbidity being around 100/100,000 persons for 70 years and above) (http://www1.mhlw.go.jp/english/wp_5/vol1/p2c6s2.html). In the present study, the mean age of the patients was 63.8 years (females, 70 years; males, 62 years), with the age group of 60 and above representing more than 60% of TB cases. The sex ratio at 3.6 was high, suggesting that among the elderly, men more often reactivated their disease in Okinawa, an observation also true for the rest of Japan (22). Indeed, a significant increase in sex ratio of TB patients with increasing age groups in Japan may be deduced through the official TB notification data, e.g., for the year 2000 the sex ratio was equal to 1 for patients aged 20 to 30 years but increased to around 2.7 for those aged 60 to 85 years (22). This is considerably different from the age structure and sex ratios observed for the general population in Japan (https://www.cia.gov/library/publications/the-world-factbook/geos/ja.html#People): 0 to 14 years, 13.8% (sex ratio, 1.06); 15 to 64 years, 65.2% (sex ratio, 1.01); and 65 years and over, 21% (sex ratio, 0.73).
The spoligotyping data and the subsequent M. tuberculosis clade determinations are summarized in Table 2. The high rate of clustering observed (87%) was linked to an elevated proportion of the Beijing genotype in our study (SIT 1, n = 69). Excluding the Beijing genotype and three Beijing-like strains, the diversity observed was limited to seven clusters (18.8% of the strains): EAI2-Manilla, Haarlem3, LAM9-var, T1, T1-var, T2, and T3-Osaka. The high percentage of Beijing strains in Okinawa (71.3%) is in agreement with the overall value of 60% reported for East Asian countries (9) but remains one of the highest in this region (23, 24, 26), with percentages varying from 90% in Beijing and the Hebei province of China (11) to 70% in Korea (24), 68% in Hong Kong (16) and Thailand (25), 54% in Vietnam (1), 44% in Taiwan (14), 33% in peninsular Malaysia (6), and 11% in eastern Malaysia (6). On the other hand, the low proportion of LAM and Haarlem in this study shows the nonexposition of this insular population to any migratory flux from Latin American and European countries. Similarly, the total absence of the central Asian family reconfirms a historical lack of contact with the Indian subcontinent and central Asian countries (4). Last but not least, 6% of the EAI family clade underlines a historical and ongoing relationship with Southeast Asian countries, e.g., Philippines, where it constitutes a predominant clone (4, 8). The observation that the average age of patients carrying EAI family strains (55.3 years) was slightly lower than that of the total sample (63.8 years) and the fact that no retreatment cases were found among the former could indicate that transmission with this genotype is ongoing (Fig. 1). However, this difference was not statistically significant using Student's t test (P = 0.2).
The non-Beijing strains underwent the classical 12-locus MIRU typing (Table 2). The percentage of clustering by combined spoligotyping and 12-locus MIRUs was 38% (11/29 strains in five clusters), suggestive of ongoing transmission with these clones within the Ryukyu Islands. This proportion is close to the 32.6% clustering reported for Osaka by using IS6110-restriction fragment length polymorphism and MIRUs (2). In some cases, different MIRU patterns were found among closely related spoligotype patterns (MITs 310, 524, and 697 among strains showing a single spacer change in their spoligotypes [Table 2]). This translates to the existence of phylogenetically related strains that are not epilinked in our setting. Inversely, a single MIRU profile (MIT 7, pattern 222325153323) was shared by three unrelated spoligotype variants (SITs 2025 and 2041 and an orphan pattern [strain RK81]). Although at first glance this reflects convergence, a stepwise localized evolution of these three spoligotypes from either SIT50 of the Haarlem clade or SIT53 of the T1 sublineage may not be fully excluded.
For some strains that were identical by MIRU, a single spacer change in their spoligotype patterns (MIT 310 shared by SITs 627 and 2122) could indicate the ongoing evolution of certain strains endemic to Japan. Indeed, SIT 627 has been defined previously as the T3-Osaka family (T. Matsumoto, unpublished data) and is characterized by the presence of spacers 1 to 4 and 9 to 12 and the absence of spacers 5 to 8 and 33 to 36. Variants of this pattern exist in our database and are all isolated from Japanese patients. A global comparison of MIRU data also underlined a single very unusual finding. It concerned MIT 310 (pattern 215125113322), for which the most predominant spoligotype is SIT 41, recently shown to be endemic in Turkey and named LAM7-TUR (40). A closer interrogation of the database underlined that MIRU patterns found among T3-Osaka and its variants (MITs 195, 310, and 727) are found among the LAM7-TUR spoligotype (SIT 41) and some (but not all) of its variants (SIT 1937 and 1261). This finding is difficult to explain on the basis of anthropological reasoning, although a probable link between the Japanese and Turkish as two Altaic languages was suspected (5), and a recent paper suggested that the Yayoi culture may be of a central Asian origin (12). Nonetheless, several other reasons, such as casual and/or historical transmission or genetic convergence, could also be involved.
The eight-locus MIRU format resulted in a clustering rate of 73.6% among Beijing isolates (Table 3). A comparison of the results obtained with the SITVIT2 database suggested that 8/12 MIRU loci selected might be sufficient to track Beijing strains. When combined with their worldwide distribution, some MIRU profiles were found to be linked specifically to Japan, e.g., patterns B, C, E, U, and V (≪..13.5.43533≫, ≪..13.5.73433≫, ≪..13.8.73533≫, ≪..33.6.73543≫, and ≪..34.5.72533≫) matched with strains isolated exclusively in Japan. Similarly, profiles D (≪..13.5.73533≫) and W (≪..34.5.73533≫) matched patterns with corresponding strains found predominantly in Japan (70.6% and 72.2% of the isolates). Last, the MIRU pattern ≪..13…[4,6,7]…≫ (the position within brackets represents MIRU-26, with copy numbers that may vary at this position, i.e., 4, 6, or 7 copies) is apparently specific to Japanese Beijing strains, as 48 out of the 59 strains (81.4%) found in the database were from Japan. A cross-examination of available spoligotype information in the database did confirm that this MIRU pattern from Japan is linked strictly to the Beijing genotype. The discriminatory power of the MIRU-QUB markers assessed using HGDI (h) calculations (Tables 1 and 4) showed that MIRU-26 is undoubtedly the most discriminative marker for typing Beijing strains globally (h = 0.385), which is in agreement with published data (20, 33). Furthermore, loci numbered 2, 4, 20, and 24 are apparently not really useful for further discrimination of the Beijing genotype in our data set (data reconfirmed after secondary typing using the four missing MIRUs and subsequent HGDI calculations [results not shown]).
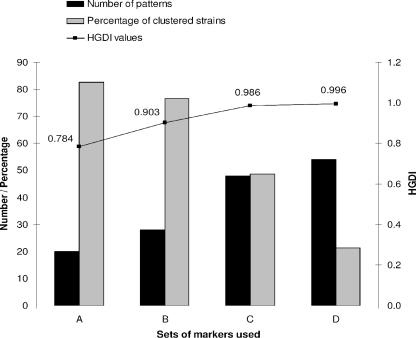
As summarized in Table 4, with the exception of QUB-1451, typing using QUBs showed a higher discriminatory power than MIRUs (e.g., HGDI of 0.680 for QUB-11b), and it reduced the number of clustered Beijing strains from 52 to only 17. A geographically linked variation in HGDI values of QUB has been suspected, e.g., HGDI for QUB-26 ranged from 0.314 in a study realized in Hong Kong (15) to 0.83 in a cosmopolitan study (31). Figure 2 summarizes the data on Beijing genotype discrimination by MIRU typing alone or in combination with QUBs (refer to the supplemental material for detailed results). Patients carrying strains that remained in cluster after combined MIRU and QUB typing are supposed to be epidemiologically linked, a fact that merits to be verified in prospective studies.
FIG. 2.
Discrimination of Beijing strains by MIRU-QUB markers. Column A, a set of four MIRUs (MIRU-10, -26, -31, and -39); column B, set A plus four additional MIRUs (MIRU-16, -23, -27, and -40); column C, set B plus three QUBs (QUB-11a, -11b, and -26); and column D, set C plus three additional QUBs (QUB-18, -1895, and -4156). For each set of markers used, the corresponding values are shown for the number of patterns obtained, the percentage of clustered strains, and the HGDI.
In conclusion, this study corroborates the predominance and historical presence of the Beijing isolates in Japan, including its remote insular entities (71.3% of all isolates in Okinawa), but without any evidence of MDR-TB. Thus, unlike in Russia and the states of the former Soviet Union, where the majority of all MDR isolates belonged to the Beijing genotype as a result of prolonged exposure of the patients to inappropriate antituberculosis treatment (35), the Japanese health system seems well adapted for a better follow-up of the TB patients, with adequate steps to avoid both the emergence and the spread of MDR-TB. Indeed, most of the drug resistance concerned a single drug, and in 16 cases showing any drug resistance, 5 cases concerned streptomycin resistance among the age group of 44 to 75 years (mean average age, 60 years), suggesting cases of reactivation of ancient disease or ongoing circulation of strains from a pool of patients treated previously, as this antibiotic was used extensively in the 1950s. This observation is also corroborated by the fact that most of the TB patients were relatively older (more than 60% of TB cases occurred among the elderly), suggesting that most of the cases probably represented reactivation cases and that TB is a declining disease in Okinawa.
Since the enactment of a new national TB control program in Japan in April 2005 (36), there has been a shift from the current “indiscriminate” screening scheme to a selective one regarding periodic mass health examination. Only subjects aged 65 or older will be eligible for the screening, in addition to selected occupational groups, such as health care providers and school teachers, who are considered to be at a higher risk of TB or who may be a danger to others if they develop TB. The revised law also states the governmental responsibility for medically supervised treatment of TB patients. Last but not least, the new law mandates every prefecture to develop its own TB control plan in order to resolve the problems specific to the respective prefectures in terms of epidemiological parameters or available resources. Our study therefore constitutes a good starting point for TB control strategies to be implemented by the Okinawa prefecture.
Last, the high rate of clustering obtained (74.6%) among Beijing strains after MIRU typing argues for the use of supplemental markers to distinguish epilinked strains from clones that may have evolved locally. In this regard, a recent paper calling for a proposal for standardization of optimized MIRU typing evaluated a total of 29 loci on 824 isolates of tubercle bacilli in a worldwide recruitment and suggested that 5 loci should be excluded due to their lack of robustness and/or stability, as discrepancies among serial or epilinked isolates were observed (31). These included three out of seven QUBs evaluated in the present study, i.e., QUB-1895, QUB-18, and QUB-11a. Hence, before a final algorithm on the potential of QUBs to subtype Beijing isolates can be drawn, a precise correlation of clustered isolates with detailed epidemiological investigations in prospective studies appears to be the ideal way to know if hypervariability of QUBs markers blurs recognition between epidemiologically linked Beijing isolates. This genotype is overrepresented in eastern Asia (3, 4, 39) and Japan (this study), and further studies from this region could provide insight on the usefulness of QUBs for epidemiology and further discrimination of the Beijing lineage.
Acknowledgments
C.M.-S. and N.Y. acknowledge Mutsuo Kuba and Masafumi Matsukawa from the National Okinawa Hospital for providing clinical isolates of M. tuberculosis.
J.M. received a Ph.D. fellowship awarded by the European Union and the Regional Council of Guadeloupe and the International Network of the Pasteur Institutes.
We are grateful to W. Prodinger (Innsbruck Medical University, Austria) for helpful comments regarding the electrophoretic separation of MIRU-VNTR PCR products. We thank Thierry Zozio for helping with experiments with QUB markers and fruitful discussions.
The SITVIT1 database project coordinated by Institut Pasteur de Guadeloupe is an extended version of SpolDB4 (http://www.pasteur-guadeloupe.fr:8081/SITVITDemo), which incorporates MIRU-VNTR data in addition to the spoligotyping data. This project was initially implemented using Java API and MYSQL 5 by Benjamin Liens et al. (18a). It now contains data compiled through 15 June 2006 and a user interface (SUI 1.0) specifically adapted by Christophe Demay. A next-generation SITVIT2 project with online web-based tools and a new user interface, SUI 2.0, is under development and will be released in 2008. Active feedback from more than 100 research groups worldwide towards these databases is greatly acknowledged.
Footnotes
Published ahead of print on 26 September 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Anh, D. D., M. W. Borgdorff, L. N. Van, N. T. Lan, T. van Gorkom, K. Kremer, and D. van Soolingen. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6:302-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ano, H., T. Matsumoto, H. Yoshida, M. Nagai, Y. Tamura, K. Nishimori, K. Kawahara, T. Takashima, and I. Tsuyuguchi. 2006. Molecular epidemiology of tuberculosis by the use of IS6110 restriction fragment length polymorphism: a study from 2001 to 2003. Kekkaku 81:321-328. [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 4.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Gutierrez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, H. M. Ly, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, M. Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. Shemyakin, U. B. Singh, A. Somoskovi, R. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavalli-Sforza, L. L., P. Menozzi, and A. Piazza. 1994. Prehistory and history in East Asia: Japan and Korea, p. 198-202. In L. L. Cavalli-Sforza, P. Menozzi, and A. Piazza (ed.), The history and geography of human genes, 1st ed. Princeton University Press, Princeton, NJ.
- 6.Dale, J. W., R. M. Nor, S. Ramayah, T. H. Tang, and Z. F. Zainuddin. 1999. Molecular epidemiology of tuberculosis in Malaysia. J. Clin. Microbiol. 37:1265-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dormans, J., M. Burger, D. Aguilar, R. Hernandez-Pando, K. Kremer, P. Roholl, S. M. Arend, and D. van Soolingen. 2004. Correlation of virulence, lung pathology, bacterial load and delayed type hypersensitivity responses after infection with different Mycobacterium tuberculosis genotypes in a BALB/c mouse model. Clin. Exp. Immunol. 137:460-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douglas, J. T., L. Qian, J. C. Montoya, J. M. Musser, J. D. Van Embden, D. Van Soolingen, and K. Kremer. 2003. Characterization of the Manila family of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:2723-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. 2006. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg. Infect. Dis. 12:736-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujikane, T., S. Fujiuchi, Y. Yamazaki, H. Matsumoto, M. Takahashi, Y. Fujita, T. Shimizu, and K. Kikuchi. 2004. Molecular epidemiology of tuberculosis in the north Hokkaido district of Japan. Int. J. Tuberc. Lung Dis. 8:39-44. [PubMed] [Google Scholar]
- 11.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer, M. F., T. M. Karafet, H. Park, K. Omoto, S. Harihara, M. Stoneking, and S. Horai. 2006. Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J. Hum. Genet. 51:47-58. [DOI] [PubMed] [Google Scholar]
- 13.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jou, R., C. Y. Chiang, and W. L. Huang. 2005. Distribution of the Beijing family genotypes of Mycobacterium tuberculosis in Taiwan. J. Clin. Microbiol. 43:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kam, K. M., C. W. Yip, L. W. Tse, K. L. Leung, K. L. Wong, W. M. Ko, and W. S. Wong. 2006. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol. Lett. 256:258-265. [DOI] [PubMed] [Google Scholar]
- 16.Kam, K. M., C. W. Yip, L. W. Tse, K. L. Wong, T. K. Lam, K. Kremer, B. K. Y. Au, and D. van Soolingen. 2005. Utility of mycobacterial interspersed repetitive unit typing for differentiating multi-drug resistant Mycobacterium tuberculosis isolates of the Beijing family. J. Clin. Microbiol. 43:306-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovalev, S. Y., E. Y. Kamaev, M. A. Kravchenko, N. E. Kurepina, and S. N. Skorniakov. 2005. Genetic analysis of Mycobacterium tuberculosis strains isolated in Ural region, Russian Federation, by MIRU-VNTR genotyping. Int. J. Tuberc. Lung Dis. 9:746-752. [PubMed] [Google Scholar]
- 18a.Liens, B., C. Sola, K. Brudey, N. Rastogi, and coinvestigators of the SpoIDB4/SITVITI Consortium. 2005. A web site for a global database of M. tuberculosis complex spoligotypes and MIRU-VNTRs-SITVIT1, abstr. O-1, p. 31. Proc. 26th Annu. Cong. Eur. Soc. Mycobacteriol., Istanbul, Turkey, 26 to 29 June 2005.
- 19.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolayevskyy, V., K. Gopaul, Y. Balabanova, T. Brown, I. Fedorin, and F. Drobniewski. 2006. Differentiation of tuberculosis strains in a population with mainly Beijing-family strains. Emerg. Infect. Dis. 12:1406-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohata, R., and A. Tada. 2004. Beijing family and other genotypes of Mycobacterium tuberculosis isolates in Okayama district. Kekkaku 79:47-53. [PubMed] [Google Scholar]
- 22.Ohmori, M., N. Ishikawa, T. Yoshiyama, K. Uchimura, M. Aoki, and T. Mori. 2002. Current epidemiological trend of tuberculosis in Japan. Int. J. Tuberc. Lung Dis. 6:415-423. [PubMed] [Google Scholar]
- 23.Park, Y. K., G. H. Bai, and S. J. Kim. 2000. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated from countries in the Western Pacific Region. J. Clin. Microbiol. 38:191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, Y. K., S. Shin, S. Ryu, S. N. Cho, W. J. Koh, O. J. Kwon, Y. S. Shim, W. J. Lew, and G. H. Bai. 2005. Comparison of drug resistance genotypes between Beijing and non-Beijing family strains of Mycobacterium tuberculosis in Korea. J. Microbiol. Methods 63:165-172. [DOI] [PubMed] [Google Scholar]
- 25.Prodinger, W. M., P. Bunyaratvej, R. Prachaktam, and M. Pavlic. 2001. Mycobacterium tuberculosis isolates of Beijing genotype in Thailand. Emerg. Infect. Dis. 7:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian, L., C. Abe, T. P. Lin, M. C. Yu, S. N. Cho, S. Wang, and J. T. Douglas. 2002. rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from East Asian countries. J. Clin. Microbiol. 40:1091-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 29.Sola, C., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125-133. [DOI] [PubMed] [Google Scholar]
- 30.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53:680-689. [DOI] [PubMed] [Google Scholar]
- 31.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supply, P., R. M. Warren, A. L. Banuls, S. Lesjean, G. D. Van Der Spuy, L. A. Lewis, M. Tibayrenc, P. D. Van Helden, and C. Locht. 2003. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol. Microbiol. 47:529-538. [DOI] [PubMed] [Google Scholar]
- 34.Takashima, T., and T. Iwamoto. 2006. A new era of molecular epidemiology of tuberculosis in Japan. Kekkaku 81:693-707. [PubMed] [Google Scholar]
- 35.Toungoussova, O. S., G. Bjune, and D. A. Caugant. 2006. Epidemic of tuberculosis in the former Soviet Union: social and biological reasons. Tuberculosis (Edinburgh) 86:1-10. [DOI] [PubMed] [Google Scholar]
- 36.Ushio, M. 2005. Amendment of tuberculosis prevention law and prospect of tuberculosis control program. Kekkaku 80:541-546. [PubMed] [Google Scholar]
- 37.van Deutekom, H., P. Supply, P. E. de Haas, E. Willery, S. P. Hoijng, C. Locht, R. A. Coutinho, and D. van Soolingen. 2005. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J. Clin. Microbiol. 43:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Soolingen, D., P. E. de Haas, P. W. Hermans, and J. D. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 39.van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zozio, T., C. Allix, S. Gunal, Z. Saribas, A. Alp, R. Durmaz, M. Fauville-Dufaux, N. Rastogi, and C. Sola. 2005. Genotyping of Mycobacterium tuberculosis clinical isolates in two cities of Turkey: description of a new family of genotypes that is phylogeographically specific for Asia Minor. BMC Microbiol. 5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]