Abstract
We describe the fourth reported case involving “Mycobacterium ulcerans subsp. shinshuense.” Compared to previous cases, the infection was more invasive with extensive ulceration from the elbow to the forearm. Definitive identification involved IS2404 detection, 16S rRNA gene sequencing, and analysis of the 16S rRNA gene 3′-terminal region and the virulence plasmid pMUM001.
CASE REPORT
On 28 December 2005, a 20-year-old Japanese woman experienced painful swelling, marked by redness, extending from the right elbow to the forearm. As the condition persisted, the patient was seen on 6 January 2006 at the Department of Dermatology, National Hospital Organization, Higashi-Hiroshima Medical Center. On the first examination, marked swelling from the right elbow to the forearm and crusted ulceration, 1 cm in diameter, surrounded by redness and swelling of the right elbow, were observed. On 8 January, the patient was admitted and placed on a parenteral course of ampicillin-sulbactam and clindamycin. The treatment was changed to minocycline on 15 January as a result of increased severity of symptoms. As the magnetic resonance image was suggestive of a potential intramuscle abscess, the patient was transferred to orthopedic surgery where, on 23 January, a 15-cm incision was made and the affected area was cleansed with irrigation. No muscle involvement was evident at the time of the incision. While the patient's condition initially improved, an exacerbation occurred in early February. On 10 February, Ziehl-Neelsen staining of material from the ulcerated area revealed the presence of acid-fast bacilli (AFB) in clumps as well as dispersed. Purulent exudate taken from the ulcer was inoculated into a BBL MGIT (Becton Dickinson, Franklin Lakes, NJ) tube and on two 2% Ogawa egg medium slants for incubation at 27 and 35°C. On the basis of the acid-fast stain result, the patient was placed on isoniazid, rifampin, and ethambutol therapy. The patient's condition continued to worsen with the ulceration growing to the size of the palm with undermined edges and a yellowish-white necrotic base. On 28 March, PCR testing of a biopsy sample targeting a Mycobacterium leprae-specific sequence was negative, while the sample was positive for insertion sequence IS2404, which raised the possibility of Mycobacterium ulcerans or “Mycobacterium ulcerans subsp. shinshuense” as the causative organism.
On 4 April, the ulcer as well as surrounding skin tissue extending 15 cm proximal and distal from the elbow was excised. Following extensive surgical removal of the epidermal, dermal, and subcutaneous skin layers, artificial dermis was placed as a template for dermal regeneration. With the generation of healthy granulation tissue, the freshly formed dermis was covered with ×1.5-meshed thin split-thickness autograft. The patient steadily improved and was discharged on 12 June.
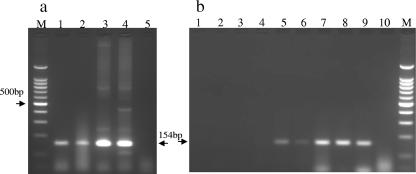
PCR targeting IS2404 specific for M. ulcerans (7) was first performed on a thin section of formalin-fixed paraffin-embedded skin sample taken on 23 January. Briefly, PCR using forward primer PU4F (5′-GCGCAGATCAACTTCGCGGT-3′) and reverse primer PU7R (5′-GCCCGATTGGTGCTCG GTCA-3′) (gene positions 548 to 567 and 702 to 683, respectively) was followed by electrophoresis on a 2% agarose gel and staining with ethidium bromide. A 154-bp PCR product matching M. ulcerans 97-107 (African strain), 5143 (Mexican strain), and 1615 (Malaysian strain) was detected (Fig. 1a). PCR testing of DNA extracted from a fresh skin biopsy specimen taken on 28 March confirmed the presence of IS2404 (Fig. 1b, lane 9). Because of the typical irregular distribution of clumps of AFB in Buruli ulcer, the excised skin specimen taken on 4 April was serially sectioned into eight sections. Each section was screened for IS2404 as well as stained with hematoxylin-eosin and the Fite stain using standard methods. The biopsy section strongly positive for the 154-bp product (Fig. 1b, lanes 7 and 8) corresponded with the presence of AFB on microscopy; however, weakly positive (lanes 5 and 6) and negative (lanes 1 to 4) sections were negative for AFB on microscopy. It is noteworthy that AFB was found within the deeper layers of the dermal tissue despite the normal appearance of the epidermis. AFB from the dermal and subcutaneous layer was purified and extracted. Two strains of “M. ulcerans subsp. shinshuense” and five strains of M. ulcerans (Table 1) were used as controls. Using previously described consensus primers, sequencing of almost the full length of the 16S rRNA gene was performed by direct sequencing of the PCR product with the ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA) (10). PCR primer pairs used were 5′-AGAGTTTGATCCTGGCTCAG-3′ (positions 8 to 27) and 5′-TGCACACAGGCCACAAGGGA-3′ (positions 1047 to 1028) in combination with 5′-GTGTGGGTTTCCTTCCTTGG-3′ (positions 830 to 849) and 5′-AAGGAGGTGATCCAGCCGCA-3′ (positions 1542 to 1523). Nucleotide numbering was based on Escherichia coli 16S rRNA gene sequence as reference. Sequence analysis was performed using DNASIS version 2.1 (Hitachi Software Engineering, Tokyo, Japan). The sequence of the 1,480-bp 16S rRNA gene obtained from the isolate in the present case was completely identical to “M. ulcerans subsp. shinshuense” strain 753 and “M. ulcerans subsp. shinshuense” ATCC 33728. At positions 54 to 510, the sequence of “M. ulcerans subsp. shinshuense” ATCC 33728 in the RIDOM database (15) was identical to that of our isolate and differed from M. ulcerans ATCC 19423 (type strain) and Mycobacterium marinum DSM44344 (type strain) by one base at position 492. As shown in Table 1, our isolate matched perfectly with the 16S rRNA gene 3′-end regions of “M. ulcerans subsp. shinshuense” reported by Portaels et al. (8) to be useful in discriminating “M. ulcerans subsp. shinshuense,” three types of M. ulcerans, and M. marinum.
FIG. 1.
PCR targeting IS2404 specific for M. ulcerans. (a) Lane M, 100-bp ladder marker; lane 1, DNA sample extracted from paraffin-embedded skin (patient); lane 2, M. ulcerans 97-107 (African strain); lane 3, M. ulcerans 5143 (Mexican strain), lane 4; M. ulcerans 1615 (Malaysian strain); lane 5, negative control. (b) Lanes 1 to 8, DNA samples extracted from each of eight serially sectioned skin specimens taken on 4 April; lane 9, DNA sample extracted from a fresh skin biopsy specimen taken on 28 March; lane 10, negative control; lane M, 100-bp ladder marker.
TABLE 1.
16S rRNA gene sequences differentiating M. ulcerans and related species
| Organism (origin) | Differing residue(s) (underlined) at position(s)a:
|
|||
|---|---|---|---|---|
| 492 | 1247 | 1288 | 1449-1451 | |
| Mycobacterium sp. (patient) | TGGGGAA | GGTGCAA | TAAGGCC | ACCC---TTTG |
| “M. ulcerans subsp. shinshuense” 753 (Japan) | TGGGGAA | GGTGCAA | TAAGGCC | ACCC---TTTG |
| “M. ulcerans subsp. shinshuense” ATCC 33728 | TGGGGAA | GGTGCAA | TAAGGCC | ACCC---TTTG |
| M. ulcerans Agy99 (Africa) | TGGAGAA | GGTGCAA | TAACGCC | ACCCTTTTTTG |
| M. ulcerans 1615 (Malaysia) | TGGAGAA | GGTGCAA | TAACGCC | ACCC---TTTG |
| M. ulcerans ATCC 19423Tb | TGGAGAA | GGTGCAA | TAACGCC | ACCC---TTTG |
| M. ulcerans 97-107 (Africa) | TGGAGAA | GGTGCAA | TAACGCC | ACCCTTTTTTG |
| M. ulcerans 5143 (Mexico) | TGGAGAA | GGTGCAA | TAAAGCC | ACCC---TTTG |
| M. marinum ATCC 927Tc | TGGAGAA | GGTACAA | TAAAGCC | ACC----TTTG |
Positions are based on E. coli 16S rRNA genes as the reference.
Type strain of the species.
Sequences are from database accession no. AF456240.
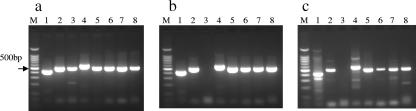
Finally, using primers for eight pMUM001 sequences coding for lipid toxin mycolactone-producing enzymes described by Stinear et al. (12), PCR products for our isolate, M. ulcerans 97-107 (African strain), and “M. ulcerans subsp. shinshuense” 753 were compared. As shown in Fig. 2, all eight bands were detected from the African strain, whereas “M. ulcerans subsp. shinshuense” 753 as well as our isolate lacked the band representing the serine/threonine protein kinase gene MUP011.
FIG. 2.
PCR for the presence of the genes on pMUM001. DNA samples were extracted from M. ulcerans 97-107 (African strain) (a), “M. ulcerans subsp. shinshuense” 753 (b), and purified AFB from a skin specimen taken on 4 April (c). Lanes M, 100-bp ladder markers; lanes 1, repA (413 bp); lanes 2, parA (501 bp); lanes 3, serine/threonine protein kinase gene MUP011 (479 bp); lanes 4, loading domain of mls (560 bp); lanes 5, acyltransferase domain of mls (504 bp); lanes 6, rep type II thioesterase gene (500 bp); lanes 7, rep type III ketosynthase gene (496 bp);lanes 8, rep P450 hydroxylase gene (500 bp).
Of the specimens cultured for mycobacteria, our isolate was recovered from only the specimen taken on 10 February after 11 weeks of incubation at 27°C on a 2% Ogawa egg slant. All characteristics of our isolate with respect to IS2404, 16S rRNA genes, and pMUM001 were identical to data obtained from affected tissue samples that failed to yield mycobacteria on culture. Determination of cultural characteristics as well as identification procedures was performed as previously described (1). The isolate exhibited yellow, rough colonies with pigmentation when grown in the dark. The slowly growing mycobacterium formed visible colonies at 25 and 32°C on a 2% Ogawa egg slant but not at 37 and 42°C. No growth was seen on medium supplemented with 500 μg/ml p-nitrobenzonic acid or 5% NaCl. The isolate was negative for niacin, nitrate reduction, arylsulfatase (3 days), Tween 80 hydrolysis, pyrazinamidase, and iron uptake, while it was positive for catalase and 68°C catalase as well as urease. The in vitro antibiotic susceptibility of this isolate was determined by the microdilution method (16) using the BrothMIC NTM kit (Kyokuto Pharmaceutical Industrial Co. Ltd., Tokyo, Japan), with modification of the incubation temperature to 32°C. MIC testing was performed in triplicate on different days with two of three matching MICs used as the criteria for MIC determination. The MICs of the drugs tested were as follows: streptomycin, 0.25 μg/ml; ethambutol, 1 μg/ml; kanamycin, 0.25 μg/ml; isoniazid, 8 μg/ml; rifampin, ≤0.03 μg/ml; levofloxacin, 0.5 μg/ml; clarithromycin, 0.06 μg/ml; ethionamide, 8 μg/ml; amikacin, ≤0.5 μg/ml.
The taxonomic studies of Tsukamura and Mikoshiba on a mycobacterial strain isolated from the skin lesion of a 19-year-old Japanese woman established the existence of a mycobacterium resembling M. ulcerans which has been subsequently classified as “M. ulcerans subsp. shinshuense” (6, 13, 14). Since the initial report, a case involving this organism from China and another case from Japan have been reported (2, 5).
In the present case, the finding of AFB in the skin lesion raised the suspicion of M. marinum, Mycobacterium haemophilum, M. ulcerans, “M. ulcerans subsp. shinshuense,” and Mycobacterium leprae as etiologic agents. Detection of IS2404 from biopsy specimens taken at different times narrowed the etiology to M. ulcerans and “M. ulcerans subsp. shinshuense.” The detection of IS2404, sequencing of 16S rRNA genes, the presence of genes on pMUM001, and the absence of the serine/threonine protein kinase gene MUP011 identified the organism as “M. ulcerans subsp. shinshuense.” In the case of slow-growing, hard-to-isolate mycobacteria, biopsy followed by molecular diagnostics is the most timely and sensitive diagnostic approach for patient management decisions. As shown in this case report, culturing is not always reliable for some slow-growing mycobacteria and growth may take as long as 11 weeks.
This represents the fourth case of infection involving “M. ulcerans subsp. shinshuense,” including the previous two cases in Japan. Compared to previous reports, this clinical case was more invasive as well as difficult to manage. The severity of the ulcerative lesion resembled Buruli ulcer, a recent M. ulcerans infection case report from Africa (9); however, based on 16S rRNA gene sequencing, our isolate was identical to “M. ulcerans subsp. shinshuense” ATCC 33728 and “M. ulcerans subsp. shinshuense” 753. Furthermore, of the eight pMUM001 gene sequences present in the plasmid responsible for the synthesis of mycolactone in M. ulcerans (11), the organism in this case report exhibited the same seven pMUM001 gene sequences found in the prototype “M. ulcerans subsp. shinshuense” strain (12). Moreover, phenotypic characteristics and in vitro drug susceptibilities were also consistent with those of the other three “M. ulcerans subsp. shinshuense” strains, including the isolate originated from China (2) (data not shown).
Because there were no apparent bacteriological differences to explain the virulence of the present isolate compared to the three other previously reported “M. ulcerans subsp. shinshuense” strains, we cannot explain why the isolate in this case was more invasive than those in previous cases. It is possible that the late administration of an effective drug(s) such as rifampin (MIC, ≤0.03 μg/ml), clarithromycin (MIC, 0.06 μg/ml), and/or amikacin (MIC, ≤0.5 μg/ml) resulted in a bacteriological cure but that the accumulation of the toxic lipid mycolactone led to the worsening of the lesion (3).
While “M. ulcerans subsp. shinshuense” has been rarely reported, there are several phenotypical and molecular differences between “M. ulcerans subsp. shinshuense” and M. ulcerans that should be noted (8, 12, 14). Of particular interest is the novel mycolactone produced by “M. ulcerans subsp. shinshuense” which resembles mycolactone A/B produced by M. ulcerans with only the side chain being structurally different as a result of changes in the coding region for biosynthesis of the side chain (4). Whereas these findings are not conclusive, they lead us to consider whether “M. ulcerans subsp. shinshuense” should be considered a “subspecies” of M. ulcerans. Additional studies may contribute to a better understanding of the evolutionary position as well as the geographical distribution of this organism.
Footnotes
Published ahead of print on 19 September 2007.
REFERENCES
- 1.Della-Latta, P., and I. Weitzman. 1998. Mycobacteriology, p. 169-203. In H. D. Isenberg (ed.), Essential procedures for clinical microbiology, 1st ed. ASM Press, Washington, DC.
- 2.Faber, W. R., L. M. Arias-Bouda, J. E. Zeegelaar, A. H. Kolk, P. A. Fonteyne, J. Toonstra, and F. Portaels. 2000. First reported case of Mycobacterium ulcerans infection in a patient from China. Trans. R. Soc. Trop. Med. Hyg. 94:277-279. [DOI] [PubMed] [Google Scholar]
- 3.George, K. M., D. Chatterjee, G. Gunawardana, D. Welty, J. Hayman, R. Lee, and P. L. Small. 1999. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science 283:854-857. [DOI] [PubMed] [Google Scholar]
- 4.Hong, H., J. B. Spencer, J. L. Porter, P. F. Leadlay, and T. Stinear. 2005. A novel mycolactone from a clinical isolate of Mycobacterium ulcerans provides evidence for additional toxin heterogeneity as a result of specific changes in the modular polyketide synthase. Chem. Biol. Chem. 6:643-648. [DOI] [PubMed] [Google Scholar]
- 5.Kazumi, Y., K. Ohtomo, M. Takahashi, S. Mitarai, I. Sugawara, J. Izumi, A. Andoh, and H. Hasegawa. 2004. Mycobacterium shinshuense isolated from cutaneous ulcer lesion of right lower extremity in a 37-year-old woman. Kekkaku 79:437-441. [PubMed] [Google Scholar]
- 6.Mikoshiba, H., Y. Shindo, H. Matsumoto, M. Mochizuki, and M. Tsukamura. 1982. A case of typical mycobacteriosis due to Mycobacterium ulcerans-like organism. Nippon Hifuka Gakkai Zasshi 92:557-565. [PubMed] [Google Scholar]
- 7.Phillips, R., C. Horsfield, S. Kuijper, A. Lartey, I. Tetteh, S. Etuaful, B. Nyamekye, P. Awuah, K. M. Nyarko, F. Osei-Sarpong, S. Lucas, A. H. Kolk, and M. Wansbrough-Jones. 2005. Sensitivity of PCR targeting the IS2404 insertion sequence of Mycobacterium ulcerans in an assay using punch biopsy specimens for diagnosis of Buruli ulcer. J. Clin. Microbiol. 43:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Portaels, F., P. A. Fonteyene, H. de Beenhouwer, P. de Rijk, A. Guedenon, J. Hayman, and M. W. Meyers. 1996. Variability in 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to geographic origin of isolates. J. Clin. Microbiol. 34:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sizaire, V., F. Nackers, E. Comte, and F. Portaels. 2006. Mycobacterium ulcerans infection: control, diagnosis, and treatment. Lancet Infect. Dis. 6:288-296. [DOI] [PubMed] [Google Scholar]
- 10.Springer, B., W.-K. Wu, T. Bodmer, G. Haase, G. E. Pfyffer, R. M. Kroppenstedt, K.-H. Schröder, S. Emler, J. O. Kilburn, P. Kirschner, A. Telenti, M. B. Coyle, and E. C. Böttger. 1996. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J. Clin. Microbiol. 34:1100-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stinear, T. P., A. Mve-Obiang, P. L. Small, W. Frigui, M. J. Pryor, R. Brosch, G. A. Jenkin, P. D. Johnson, J. K. Davies, R. E. Lee, S. Adusumilli, T. Garnier, S. F. Haydock, P. F. Leadlay, and S. T. Cole. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. USA 101:1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stinear, T. P., H. Hong, W. Frigui, M. J. Pryor, R. Brosch, T. Garnier, P. F. Leadlay, and S. T. Cole. 2005. Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. J. Bacteriol. 187:1668-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukamura, M., and H. Mikoshiba. 1982. A new mycobacterium which caused skin infection. Microbiol. Immunol. 26:951-955. [DOI] [PubMed] [Google Scholar]
- 14.Tsukamura, M., K. Kaneda, T. Imaeda, and H. Mikoshiba. 1989. A taxonomic study on a mycobacterium which caused a skin ulcer in a Japanese girl and resembled Mycobacterium ulcerans. Kekkaku 64:691-697. [PubMed] [Google Scholar]
- 15.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallace, R. J., Jr., D. R. Nash, L. C. Steele, and V. Steingrube. 1986. Susceptibility testing of slowly growing mycobacteria by a microdilution MIC method with 7H9 broth. J. Clin. Microbiol. 24:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]