Abstract
Wound botulism is a growing problem among injecting drug users. The condition is often difficult to diagnose, with laboratory confirmation in only 50% of the cases. Here we present a real-time PCR-based method for the diagnosis of wound botulism caused by Clostridium botulinum. The assay includes an internal amplification control which is amplified simultaneously with the genes encoding neurotoxin types A, B, and E. This method was used to detect the first case of wound botulism in an injecting drug user in Sweden. In addition, to the best of our knowledge, this is the first reported case of wound botulism caused by C. botulinum type E.
Wound botulism has been, except in times of war, a very rare form of botulism, occurring mainly in patients with traumatic and surgical wounds that were contaminated with dirt containing Clostridium botulinum spores. The spores germinate and botulinum neurotoxin is produced in vivo. The symptoms of botulism are caused by the irreversible binding of the neurotoxin to presynaptic motor-neuron terminals, which blocks acetylcholine transmission across the neuromuscular junction. Botulism typically presents as a progressive, symmetrical, flaccid, descending paralysis without sensory involvement; it creates a pathognomonic clinical picture, but due to the rarity of the condition, the diagnosis of botulism is frequently delayed or even missed. Between 1943 and 1990, only 47 cases were reported to the Centers for Disease Control and Prevention, Atlanta, GA, and in the literature (9). Since then, however, there has been an upsurge of cases (1, 3, 5, 6, 9, 20, 21, 26, 34). The new risk group is injecting drug users (IDUs), especially those that use skin or muscle “popping,” in which the drug is injected subcutaneously or intramuscularly (26). The first case of botulism in an IDU was identified in New York in 1982 and involved cocaine (8), but most of the recent cases have involved heroin (1, 3, 5, 20, 21, 25, 26, 28, 30, 34). Soft-tissue infections resulting in abscesses at injection sites or associated with cellulitis can be caused by several different Clostridium spp., as well as by Bacillus cereus (11, 25). These infections may be due to spore-contaminated injecting paraphernalia or drugs. To make them suitable for injection, drugs are often dissolved in acids, which enhance tissue damage and thus create a favorable milieu for bacterial spores to germinate and initiate a wound infection. So far, all confirmed cases of wound botulism have been caused by C. botulinum types A and B (31).
To confirm a clinical diagnosis of botulism, samples should be sent to a specialized reference laboratory. The diagnostic procedure consists of the detection of botulinum neurotoxin by the mouse lethality assay and/or the isolation of C. botulinum. If wound botulism is suspected, a serum sample is examined for botulinum toxin and pus or tissue samples are examined for the presence of C. botulinum and, if the samples are of sufficient size, for the presence of toxin.
However, microbiological confirmation of wound botulism has been possible in only around 50% of the cases (31). The reason may be incorrectly taken samples, insufficient sample sizes, low levels of circulating toxin, or the start of antibiotic treatment before sampling. If the latter is the cause, dead cells may still be present in the wound, thus permitting PCR detection. Several different PCR methods have been developed for the detection of C. botulinum in food and clinical samples (4, 7, 10, 15, 22, 23, 33). However, most of these methods have been developed for conventional PCR and require gel electrophoresis and/or probes to confirm the identity of the target. In addition, they do not usually contain an internal amplification control (IAC), which has been suggested to be mandatory by both the European Committee for Standardization and the International Organization for Standardization (18).
We have previously developed a real-time PCR method for the detection of C. botulinum type B (10, 24). In this report, we describe assays for the detection of C. botulinum types A and E. The assays include an IAC that is amplified simultaneously with the target genes encoding botulinum neurotoxin types A, B, and E. This real-time PCR approach was used to detect the first case of wound botulism in an IDU in Sweden, and this case is, to our knowledge, the first reported case of wound botulism caused by C. botulinum type E.
CASE REPORT
The patient was a 24-year-old homeless woman who had been injecting heroin and amphetamine for over 2 years. Previously, she had been treated for recurrent skin and soft-tissue abscesses following intramuscular and subcutaneous injections. Ten days before hospital admission, an abdominal wall abscess had been incised by a general practitioner, and antibiotic therapy with flucloxacillin had been prescribed. The culture of wound secretions yielded growth of Staphylococcus aureus, beta-hemolytic group G streptococci, and unspecified anaerobic bacteria.
Eight days later, the patient started to experience dysphagia and throat pain and sought medical care in an emergency room 1 day after the onset of these symptoms. At that time, she complained of generalized weakness and difficulty in speaking, swallowing, and holding her head upright. She was afebrile and manifested a normal level of consciousness. A psychogenic cause for her symptoms was suspected, and she was referred for psychiatric assessment. Antibiotic therapy was changed to clindamycin due to persistent purulent drainage from multiple gluteal and abdominal wall abscesses.
On the following day, the patient's condition worsened with progressive cranial nerve symptoms and weakness of all extremities. She was admitted for observation in a neurological ward. One day after this admission, she developed respiratory arrest, after which she was intubated and transferred to an intensive care unit (ICU).
Upon ICU admission, the patient was tetraplegic and had bilateral ptosis and facial paralysis but was fully conscious with intact sensorium. Lumbar puncture and brain-imaging studies did not reveal any pathological changes. An electromyogram showed a nonspecific pattern. A clinical diagnosis of wound botulism was made, but antitoxin was not administered due to the extended duration of symptoms before the diagnosis was considered. A tracheostomy was performed 2 days after ICU admission. A chest X ray obtained after tracheal intubation showed right-lower-lobe infiltrates compatible with aspiration pneumonia, and antibiotic therapy with cefuroxime was started.
After 2 months, the patient began to gradually recover muscle power in the extremities, but she still required ventilatory support for 134 days before she could be completely weaned from mechanical ventilation. Subsequently, she made a full recovery of muscular function.
MATERIALS AND METHODS
Culture conditions and DNA extraction.
All strains used for specificity studies are listed in Table 1. All anaerobic strains were grown anaerobically in tryptone-peptone-glucose-yeast extract broth at 37°C, as described previously (10). All aerobic strains were grown in tryptone broth (Oxoid Ltd., Basingstoke, United Kingdom) at 37°C.
TABLE 1.
Bacterial strains used in the specificity test of the PCR assays
| Species | Strain(s)a |
|---|---|
| C. botulinum | |
| Type A | ATCC 3502, ATCC 19397, Atlanta 2204-4, Johannesson 263, ATCC 25763, SMRICC 87, SMRICC 89, Madison 109A |
| Type B | ATCC 7949, ATCC 17841, Eklund 2b, SMRICC 91, SMRICC 92, SMRICC 93, Johannesson 105-66, Atlanta 3025, BL 105/66 |
| Type E | CB-S3E, CB-S4E, CB-S6E, CB-S21E, CB-S25E, CB-K27E, CB-K44E, CB-K58E, CB-K83E, CB-K114E, CB-124E, ATCC 9564, BL E64 S20 St15, BL E823, BL 1177, BL 1890 |
| Type F | ATCC 35415, ATCC 23387, Craig 610B8-6F, Atlanta 2821, Johannesson, ATCC 25765, BL F1 |
| Clostridium butyricum | ATCC 19398 |
| Clostridium perfringens | ATCC 14810, ATCC 3629, ATCC 12925, ATCC 3624, ATCC 3626, ATCC 13124 |
| Clostridium septicum | SVA G2, BL 8/65 |
| Clostridium sporogenes | ATCC 19404, ATCC 11437, K54 SBL, CCUG 15941 |
| Clostridium novyi | BL SVA 89 |
| Clostridium difficile | ATCC 9689 |
| Clostridium ramosum | BL 174 |
| Escherichia coli | ATCC 25922, O157 |
| Staphylococcus aureus | NCTC 7428, ATCC 8181 |
| Streptococcus agalactiae | CCUG 4208, NCTC 8181 |
| Proteus mirabilis | ATCC 43071 |
| Proteus vulgaris | CCUG 6327 |
| Pseudomonas aeruginosa | ATCC 27853 |
| Enterococcus faecalis | ATCC 29212 |
| Serratia marcescens | ATCC 13880 |
| Stenotrophomonas maltophilia | CCUG 5866 |
| Klebsiella pneumoniae | ATCC 10031 |
| Bacteroides thetaiotaomicron | ATCC 29741 |
| Bacteroides fragilis | ATCC 25285 |
| Listeria monocytogenes | NCTC 1194 |
| Campylobacter jejuni | CCUG 10937, CCUG 11284 |
| Yersinia ruckeri | CCUG 21537 |
| Yersinia pseudotuberculosis | CCUG 5855 |
| Yersinia enterocolitica | JH 19, JH 23, Box 27/3 |
| Yersinia frederiksenii | 176-36/80 |
| Yersinia kristensenii | Y891 |
| Salmonella enterica serovar Typhimurium | 51K61 |
Strains with designations including the following abbreviations are from the indicated sources: ATCC, American Type Culture Collection; SMRICC, Swedish Meat Research Institute Culture Collection; SVA, Swedish National Veterinary Institute; CCUG, Culture Collection, University of Göteborg, Göteborg, Sweden; NCTC, National Collection of Type Cultures, Central Public Health Laboratory, London, England.
One milliliter of an overnight culture was harvested at 10,000 × g for 5 min. After the discarding of the supernatant, the cells were resuspended in 200 μl of Tris-EDTA buffer. For gram-positive strains, 32 μl of lysosyme (50 mg/ml) and 1.6 μl of mutanolysin (5,000 U/ml; Sigma Chemicals Co., St Louis, MO) were added and the sample was incubated at 37°C for 30 min before extraction using the Easy-DNA kit according to the instructions of the manufacturer (protocol no. 3; Invitrogen BV, Groningen, The Netherlands). The DNA concentration was measured on a TD-700 fluorometer (Turner Designs, Sunnyvale, CA) using the PicoGreen double-stranded DNA quantitation kit (Molecular Probes, Inc., Leiden, The Netherlands).
Primer and probe design.
The primers and probes used in this study are listed in Table 2. The assay for the type B neurotoxin gene has previously been described by Lövenklev et al. (24). An alignment of sequences was made using the BioEdit sequence alignment editor (16). Published nucleotide sequences for the neurotoxin genes of C. botulinum type E (accession numbers X62683, AB039264, AB040123, AB040125, AB040126, AB040128, AB082519, AB088207, X62088, X62089, EF028403, EF028404, and AB037704) and type A (accession numbers M30196, AF461540, AF488749, EF033126, EF028393, EF028392, EF028391, AY953275, X52066, X52088, and X73423) were collected from the GenBank Sequence Database (http://www.ncbi.nih.gov/Genbank/). Several different primers and probes were tested, some from the literature and some designed using the LightCycler probe design software, version 1.0 (Idaho Technology Inc., Salt Lake City, UT). The specificity was checked against published sequences by using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and tested using PCR. Several probes were designed and compared. The hybridization probes consisted of two parts: a donor probe labeled with fluorescein at the 3′ end and an acceptor probe with the 5′ labeled with LC Red 640 and the 3′ hydroxyl blocked with a phosphate molecule. The acceptor probe for the IAC was labeled with LC Red 705.
TABLE 2.
Sequences and fluorescent dye of primers and hybridization probes used for real-time PCR
| Target gene | GenBank accession no. | Primer or probe | Nucleotide positions | Nucleotide sequence (5′→3′) | Source or reference |
|---|---|---|---|---|---|
| cntA | M30196 | AS-11d | 435-455 | TG(C/T)AGGACAAATGCAACCAGT | Modified from reference 33 |
| AS-22 | 697-717 | TCCACCCCAAAATGGTATTCC | 33 | ||
| ToxA-Fluo | 545-570 | TGAAACTGGAACTTGTTTTGCTTCTG-FLa,b | This study | ||
| ToxA-Red | 517-542 | LC640-GGTGGATTTAAATCTCCTTCTTCAGG-pa,c | This study | ||
| cntE | X62683 | fToxE | 1658-1673 | TGAATCAGCACCTGGA | This study |
| rToxE | 1974-1982 | GGTTAGCTTCAGTAGTAAA | This study | ||
| ToxE-Fluo | 1913-1936 | TGTCAATAAACCTGTGCAAGCAGC-FLb | This study | ||
| ToxE-Red | 1939-1973 | LC640-TATTTGTA(A/G)GCTGGATACAACAAGT (A/G)TTAGTAGAT-pc | This study | ||
| IAC | LC143-3′Fluo | GCGGTGGGTTTTGTTGTCTTCTCTATT-FLb | Modified from reference 27 | ||
| LC150-5′Red705 | LC705-CTATCCTTGAGCCGTAGGCCACTATCG-pd | 27 |
The probe is located on the antisense strand.
The donor probe is labeled with fluorescein (FL) at the 3′ end.
The acceptor probe is labeled with LC Red 640 (LC640) at the 5′ end, and the 3′ hydroxyl is blocked with a phosphate molecule (p).
The acceptor probe is labeled with LC Red 705 (LC705) at the 5′ end, and the 3′ hydroxyl is blocked with a phosphate molecule (p).
Construction of the IAC.
The method of Stöcher et al. (32) was used to generate multiple internal controls that were amplified by the same primers used for the detection of the neurotoxin genes of C. botulinum types A, B, and E. A DNA sequence (IC T69) already used as an internal control for Salmonella spp. and containing a binding site for a pair of hybridization probes (27) was used as starting material, and additional primer-specific sequences were added in a conventional preparative PCR with composite primers (Table 3). By using the previous PCR product as a template in the next preparative PCR, after a purification step with the QIAquick purification kit (QIAGEN, Hilden, Germany), the primer-specific sequences were added, one by one, to the original DNA fragment. Preparative PCRs were performed in a GeneAmp 9700 thermal cycler (Perkin-Elmer Cetus, Norwalk, CT) using a mixture of 0.2 μM (each) deoxynucleoside triphosphates (dNTPs; Roche Diagnostics GmbH, Mannheim, Germany), 1× Taq reaction buffer (Roche), 1 U of Taq DNA polymerase (Roche), 0.5 μM (each) composite primers (Sigma-Genosys), and PCR-grade water added to reach a final mixture volume of 20 μl, to which 5 μl of the template (1 μg/ml) was added. The following cycle parameters were applied for the first PCR: 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 60°C, and 40 s at 72°C; and 7 min at 72°C. In the following PCRs, the times for each step of the cycles were increased to 40 and 60 s, respectively.
TABLE 3.
Composite primers used for preparative PCR for the generation of the multiple IAC
| PCR | Primer | Nucleotide sequence (5′→3′) |
|---|---|---|
| 1 | Forward: fBn-P139 | AAAGTAGATGATTTACCAATTGTAGTGAAATTATCGCCACGTTCGGGCAA |
| Reverse: rBn-P141 | GTTAGGATCTGATATGCAAACTATCATCGCACCGTCAAAGGAACCGTAA | |
| 2 | Forward: As11-fBn | TGCAGGACAAATGCAACCAGTAAAGTAGATGATTTACCAATTGTA |
| Reverse: As22-rBn | TCCACCCCAAAATGGTATTCCGTTAGGATCTGATATGCAAACTA | |
| 3 | Forward: fToxE-As11 | TGAATCAGCACCTGGATGCAGGACAAATGCAACCAGT |
| Reverse: rToxE-As22 | GGTTAGCTTCAGTAGTAAATCCACCCCAAAATGGTATTCC |
After each preparative PCR, the product was examined by agarose gel electrophoresis. For final purification, DNA was cut out from the gel and purified using the QIAquick gel extraction kit (QIAGEN). The final product was examined in a conventional PCR with the respective C. botulinum-specific primers (Table 2). To confirm the expected nucleotide sequences, the generated internal control was subjected to sequence analysis. Competitive real-time PCR was performed with C. botulinum DNA, toxin types A and E, reference strains ATCC 3502 and ATCC 9564, and different amounts of the internal control to determine the optimum amount.
PCR conditions.
Real-time PCR amplification was carried out in a LightCycler instrument (Roche). For type B, the protocol from Lövenklev et al. (24), which has previously been used to detect a case of infant botulism (M. Lövenklev, personal communication), was followed. The PCR mixture specific for the cntE gene contained 1× Tth PCR buffer (Roche), 4 mM MgCl2 (Roche), 0.2 mM (each) dNTPs (Roche), 0.3 μM (each) primers (Sigma-Genosys), 50 copies of the IAC, 0.15 μM probes for the IAC, 0.3 μM probes for cntE (TIB Molbiol, Berlin, Germany), 1.25 U of Tth DNA polymerase (Roche), and 4 μl of template solution. The PCR mixture specific for the cntA gene contained 1× Tth PCR buffer (Roche), 3 mM MgCl2 (Roche), 0.2 mM (each) dNTPs (Roche), 0.5 μM (each) primers, 50 copies of the IAC, 0.15 μM probes for the IAC, 0.225 μM probes for cntA (TIB Molbiol), 1.25 U of Tth DNA polymerase (Roche), and 4 μl of template solution. The total volume added to each capillary was 20 μl. The LightCycler amplification protocol used started with an initial denaturation at 95°C for 60 s, followed by 45 cycles of 95°C for 0 s, annealing and fluorescence acquisition at 56°C for 5 s, and elongation at 72°C for 25 s. The specificity was confirmed using a melting-curve analysis. It consisted of 95°C for 0 s and 50°C for 15 s, followed by an increase in temperature of 0.2°C/s up to 90°C. The cnt gene was detected in channel F2, and the IAC was detected in channel F3. A color-compensating file was created to avoid spectral overlap of multiple fluorophores.
Sensitivity and amplification efficiency (AE) were tested using series of DNA dilutions. The AE was calculated using the following equation: AE = 10(−1/s) − 1, where s is the slope of the line formed by plotting the crossing-point value against the logarithm of the DNA concentration (22a). For type A, DNA from strain ATCC 3502 was used, and for type E, the DNA was from strain ATCC 9564.
Sequencing.
Conventional PCR was used for amplification, and the amplicon was purified using the QIAquick PCR purification kit (QIAGEN). The fragment was sequenced using the BigDye kit (Applied Biosystems) according to the chain termination method (29). The PCR mixture used contained 40 to 100 ng of a PCR fragment, 2 μl of BigDye premix, 0.5 μM primer (Sigma-Genosys), and water added to reach a final mixture volume of 10 μl. The PCR program consisted of 25 cycles of 95°C for 30 s, 50°C for 15 s, and 60°C for 4 min. The DNA was precipitated with a mixture of 1 μl of 3 M sodium acetate (pH 5.2) and 29 μl of 96% ethanol, and after centrifugation, washing with 70% ethanol, and drying, the DNA was sequenced on an ABI PRISM 3100 genetic analyzer (Applied Biosystems) at the BM Unit, Lund University, Lund, Sweden. The sequencing was done three times in each direction, and the results were compared using the BioEdit sequence alignment editor (16).
Laboratory tests.
Pus specimens from two abdominal wall abscesses were cultured anaerobically on selective agar (C. botulinum isolation) medium and fastidious-anaerobe agar (LabM) and examined for the presence of C. botulinum (12). Both samples were also inoculated into enrichment chopped-meat-carbohydrate (CMC) medium (17). After incubation for 2 days, the CMC cultures were used for subculturing onto fastidious-anaerobe agar and fresh CMC medium. Samples for DNA extraction and PCR were taken both from the original culture and from plates. Unfortunately, the original sample material was insufficient to allow direct extraction. Cell-free CMC culture supernatants were tested for the presence of botulinum neurotoxins by the mouse bioassay (17). In addition, a serum sample was examined for the presence of botulinum toxin. The pus samples (swab samples) could not be tested for toxin directly, due to the insufficient sample size.
One milliliter of culture was harvested and DNA was extracted as described above. Samples taken from a plate with a sterile loop were dissolved in 200 μl of Tris-EDTA before proceeding as described above. The samples were analyzed as described above using 4 and 8 μl of template, undiluted, diluted 10-fold, or diluted 100-fold. When an increase in fluorescence could be seen in the late cycles, an additional 10 cycles were added to get clearer detection.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in GenBank under accession numbers EF517320 to EF517333.
RESULTS
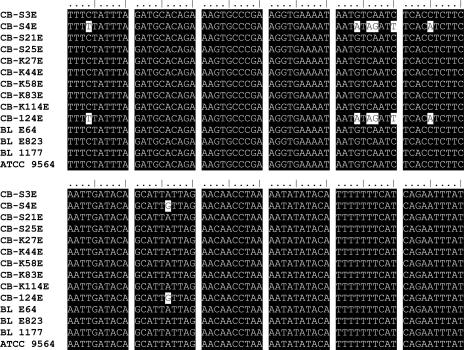
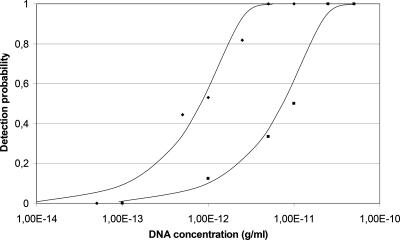
Oligonucleotides specific for part of the type A and type E neurotoxin genes were constructed (Table 2). During the design of the type E hybridization probes, it was found that two strains, CB-S4E and CB-124E, displayed punctual mutations compared to the other strains sequenced in this study and the sequences reported in GenBank (Fig. 1). The differences are in the same places in both strains. The ToxE-Red probe was therefore redesigned with two degenerated positions. The selectivity of both the type A and type E real-time PCR assays was evaluated using 40 strains of C. botulinum, 16 strains of other Clostridium spp., and 26 non-Clostridium strains (Table 1). The PCR products of the primers selected were shown on an agarose gel to be the predicted 335 bp for type E strains, 283 bp for type A strains, and 216 bp for type B strains. None of the other strains gave amplified PCR products when tested with conventional PCR or any fluorescent signal when tested with real-time PCR, and there was no cross-reaction between strains. The IAC gave the expected sizes of 0.33 kb with type B primers, 0.37 kb with type A primers, and 0.41 kb with type E primers. The amount of the IAC added did not change the detection limit of the assay (data not shown). The detection limit was at least 20 fg per reaction for type A and 100 fg for type E (Fig. 2). The lowest amount detected was 2 fg for type A, equivalent to 0.5 genomes, and 4 fg for type E, equivalent to 1 genome, assuming an approximate genome size of 3.89 Mbp. The AE with the IAC present was 1.01 for type A and 0.92 for type E, calculated in the linear range between 1 μg/ml and 0.1 ng/ml.
FIG. 1.
Alignment of part of the cntE nucleotide sequences determined in this study with the corresponding region, positions 1787 to 1906, of the sequence from NCTC 11219 (accession number X62683).
FIG. 2.
Detection probability for purified DNA in water. Diamonds represent C. botulinum type A, and squares represent C. botulinum type E.
The real-time PCR approach to detect C. botulinum types A, B, and E was used on the clinical abscess material from a patient with suspected botulism (see Case Report). Abscess samples were cultivated in CMC medium for 48 h prior to DNA extraction. The PCR results were positive for C. botulinum type E. The laboratory confirmation was obtained by the isolation of C. botulinum type E and the detection of botulinum toxin type E by the mouse bioassay. A toxin assay of serum failed to demonstrate the presence of botulinum toxin.
DISCUSSION
In this study, we report a sensitive real-time PCR approach to detect C. botulinum types A, B, and E. It is proposed to be used to complement the conventional mouse bioassay and to reduce the number of animals needed for the detection and typing of C. botulinum. The hope is that eventually the mouse bioassay can be phased out and completely replaced by other techniques, as is recommended by international legislation (Directive 86/609/EEC). Even though there are several published real-time PCR methods for C. botulinum, few include an IAC that is amplified by the same primers. In addition, our approach appears to be more sensitive, with a detection limit of 20 fg for type A and 100 fg for type E. In comparison, Yoon et al. (35) report a detection limit of 0.1 ng for type A, Akbulut et al. (2) report a detection limit of 70 fg for type A and 195 fg for type E, and Kimura et al. (22) report a detection limit of 0.5 pg for type E. Fenicia et al. (14), who include an IAC, do not state a detection limit for pure DNA but claim that it is less than 60 genomic copies for type A, i.e., equivalent to approximately 230 fg. Akbulut et al. (2) also used an IAC; however, it is an exogenous control and uses different primers, which may be affected in different ways by inhibitors (19). In addition, they do not seem to have added it to all samples, since it uses the same dye as one of their probes. Several of our samples were shown to be highly PCR inhibitory (data not shown), which shows the need for an IAC to avoid getting false-negative results.
It appears that delayed diagnosis of wound botulism is not rare (28), and it is even possible that the diagnosis may have been missed in some cases. In the present case, in which antibiotics had been administered before botulism was suspected and samples were taken, it took several days of culturing and repeated subculturing to isolate the bacteria, while PCR could be performed on material from an early stage in the process. The clinical presentation of this patient, i.e., symmetrical, descending, flaccid paralysis in a female IDU, is highly suggestive of wound botulism, but due to the rarity of the disorder, the patient's symptoms and signs were initially misinterpreted. Several other diagnoses, including hysteria, were considered before botulism was considered. Early diagnosis might have allowed for treatment with antitoxin, which is effective in reducing the severity of the symptoms of botulism and the length of hospitalization if administered early in the course of the disease. In a study from California, a lower proportion of patients who had received antitoxin within 12 h of presentation required mechanical ventilation (28) than those who received it later. In addition, the duration of mechanical ventilation was shorter among such patients (median, 45 versus 60 days). Our patient required full ventilatory support for more than 4 months after the disease onset.
Although wound botulism cases in various geographical regions have been described previously, no cases in Sweden have hitherto been reported. Reported risk factors for wound botulism are subcutaneous or intramuscular injection of heroin (skin popping) and female gender (26). We have not been able to identify the source of this patient's infection, but it is likely to be heroin contaminated with spores of C. botulinum. In California, black tar heroin is imported from Mexico, but in southern Sweden, most black heroin originates from Afghanistan. We were not able to get access to heroin to test for C. botulinum, and the contamination may be restricted to certain batches. The most probable source of contamination is the “cutting” process, when the drug is diluted before sale (26). Since type E spores are the most common in Swedish soil (13), the cutting was probably done locally. It is also possible that the syringe was contaminated.
Outbreaks of wound botulism have occurred in many geographical locations, but so far we are not aware of other cases in IUDs in Sweden. However, this case illustrates that wound botulism is yet another complication of injecting drug use that has to be considered by clinicians and demonstrates the utility of real-time PCR as a complement to traditional methods of diagnosis.
Acknowledgments
This work was supported by the Swedish Governmental Agency for Innovation Systems (VINNOVA) and the Swedish Research Council for Environment, Agricultural Sciences, and Special Planning (Formas).
Footnotes
Published ahead of print on 19 September 2007.
REFERENCES
- 1.Akbulut, D., J. Dennis, M. Gent, K. Grant, V. Hope, C. Ohai, J. McLauchlin, V. Mithani, O. Mpamugo, F. Ncube, and L. De Souza-Thomas. 2005. Wound botulism in injectors of drugs: upsurge in cases in England during 2004. Euro Surveill. 10:172-174. [PubMed] [Google Scholar]
- 2.Akbulut, D., K. A. Grant, and J. McLauchlin. 2004. Development and application of real-time PCR assays to detect fragments of the Clostridium botulinum types A, B, and E neurotoxin genes for investigation of human foodborne and infant botulism. Foodborne Pathog. Dis. 1:247-257. [DOI] [PubMed] [Google Scholar]
- 3.Alpers, K., U. van Treeck, and C. Frank. 2005. Outbreak of wound botulism in injecting drug users in Germany, October-December 2005. Euro Surveill. 10:E051215.4. [DOI] [PubMed] [Google Scholar]
- 4.Alsallami, A. A., and R. Kotlowski. 2001. Selection of primers for specific detection of Clostridium botulinum types B and E neurotoxin genes using PCR method. Int. J. Food Microbiol. 69:247-253. [DOI] [PubMed] [Google Scholar]
- 5.Brett, M. M., G. Hallas, and O. Mpamugo. 2004. Wound botulism in the UK and Ireland. J. Med. Microbiol. 53:555-561. [DOI] [PubMed] [Google Scholar]
- 6.Burnens, A. 3 February 2000, posting date. Cases of wound botulism in Switzerland. Eurosurveill. Wkly. 2:000203. http://www.eurosurveillance.org/ew/2000/000203.asp#1. [Google Scholar]
- 7.Campbell, K. D., M. D. Collins, and A. K. East. 1993. Gene probes for identification of the botulinal neurotoxin gene and specific identification of neurotoxin types B, E, and F. J. Clin. Microbiol. 31:2255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CDC. 1982. Wound botulism associated with parenteral cocaine abuse, New York City. Morb. Mortal. Wkly. Rep. 31:87-88. [PubMed] [Google Scholar]
- 9.Cherington, M. 1998. Clinical spectrum of botulism. Muscle Nerve 21:701-710. [DOI] [PubMed] [Google Scholar]
- 10.Dahlenborg, M., E. Borch, and P. Rådström. 2001. Development of a combined selection and enrichment PCR procedure for Clostridium botulinum types B, E, and F and its use to determine prevalence in fecal samples from slaughtered pigs. Appl. Environ. Microbiol. 67:4781-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dancer, S. J., D. McNair, P. Finn, and A. B. Kolsto. 2002. Bacillus cereus cellulitis from contaminated heroin. J. Med. Microbiol. 51:278-281. [DOI] [PubMed] [Google Scholar]
- 12.Dezfulian, M., L. M. McCroskey, C. L. Hatheway, and V. R. Dowell. 1981. Selective medium for isolation of Clostridium botulinum from human feces. J. Clin. Microbiol. 13:526-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodds, K. L. 1993. Clostridium botulinum in the environment., p. 21-51. In A. H. W. Hauschild and K. L. Dodds (ed.), Food science and technology series, vol. 54. Clostridium botulinum: ecology and control in foods. Marcel Dekker, Inc., New York, NY. [Google Scholar]
- 14.Fenicia, L., F. Anniballi, D. De Medici, E. Delibato, and P. Aureli. 2007. SYBR green real-time PCR method to detect Clostridium botulinum type A. Appl. Environ. Microbiol. 73:2891-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franciosa, G., J. L. Ferreira, and C. L. Hatheway. 1994. Detection of type A, B, and E botulism neurotoxin genes in Clostridium botulinum and other Clostridium species by PCR: evidence of unexpressed type B toxin genes in type A toxigenic organisms. J. Clin. Microbiol. 32:1911-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 17.Holdeman, L. V., E. P. Cato, and W. E. C. Moore. 1977. VPI anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg, VA.
- 18.Hoorfar, J., N. Cook, B. Malorny, M. Wagner, D. De Medici, A. Abdulmawjood, and P. Fach. 2003. Making internal amplification control mandatory for diagnostic PCR. J. Clin. Microbiol. 41:5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, T., D. Jacobsen, E. von der Lippe, M. S. Heier, and B. Selseth. 1998. Clinical wound botulism in injecting drug addicts. Tidsskr. Nor. Laegeforen. 118:4363-4365. (In Norwegian.) [PubMed] [Google Scholar]
- 21.Jensenius, M., R. Z. Lövstad, G. Dhaenens, and L. M. Rörvik. 2000. A heroin user with a wobbly head. Lancet 356:1160. [DOI] [PubMed] [Google Scholar]
- 22.Kimura, B., S. Kawasaki, H. Nakano, and T. Fujii. 2001. Rapid, quantitative PCR monitoring of growth of Clostridium botulinum type E in modified-atmosphere-packaged fish. Appl. Environ. Microbiol. 67:206-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Klein, D., P. Janda, R. Steinborn, M. Muller, B. Salmons, and W. H. Gunzburg. 1999. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis 20:291-299. [DOI] [PubMed] [Google Scholar]
- 23.Lindström, M., R. Keto, A. Markkula, M. Nevas, S. Hielm, and H. Korkeala. 2001. Multiplex PCR assay for detection and identification of Clostridium botulinum types A, B, E, and F in food and fecal material. Appl. Environ. Microbiol. 67:5694-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lövenklev, M., E. Holst, E. Borch, and P. Rådström. 2004. Relative neurotoxin gene expression in Clostridium botulinum type B, determined using quantitative reverse transcription-PCR. Appl. Environ. Microbiol. 70:2919-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell, P. A., and P. T. Pons. 2001. Wound botulism associated with black tar heroin and lower extremity cellulitis. J. Emerg. Med. 20:371-375. [DOI] [PubMed] [Google Scholar]
- 26.Passaro, D. J., S. B. Werner, J. McGee, W. R. Mac Kenzie, and D. J. Vugia. 1998. Wound botulism associated with black tar heroin among injecting drug users. JAMA 279:859-863. [DOI] [PubMed] [Google Scholar]
- 27.Perelle, S., F. Dilasser, B. Malorny, J. Grout, J. Hoorfar, and P. Fach. 2004. Comparison of PCR-ELISA and LightCycler real-time PCR assays for detecting Salmonella spp. in milk and meat samples. Mol. Cell. Probes 18:409-420. [DOI] [PubMed] [Google Scholar]
- 28.Sandrock, C. E., and S. Murin. 2001. Clinical predictors of respiratory failure and long-term outcome in black tar heroin-associated wound botulism. Chest 120:562-566. [DOI] [PubMed] [Google Scholar]
- 29.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro, R. L., C. Hatheway, and D. L. Swerdlow. 1998. Botulism in the United States: a clinical and epidemiologic review. Ann. Intern. Med. 129:221-228. [DOI] [PubMed] [Google Scholar]
- 31.Sieradzan, K. A. 2005. Wound botulism. Pract. Neurol. 5:46-52. [Google Scholar]
- 32.Stöcher, M., V. Leb, G. Hölzl, and J. Berg. 2002. A simple approach to the generation of heterologous competitive internal controls for real-time PCR assays on the LightCycler. J. Clin. Virol. 25(Suppl. 3):S47-S53. [DOI] [PubMed] [Google Scholar]
- 33.Takeshi, K., Y. Fujinaga, K. Inoue, H. Nakajima, K. Oguma, T. Ueno, H. Sunagawa, and T. Ohyama. 1996. Simple method for detection of Clostridium botulinum type A to F neurotoxin genes by polymerase chain reaction. Microbiol. Immunol. 40:5-11. [DOI] [PubMed] [Google Scholar]
- 34.Werner, S. B., D. Passaro, J. McGee, R. Schechter, and D. J. Vugia. 2000. Wound botulism in California, 1951-1998: recent epidemic in heroin injectors. Clin. Infect. Dis. 31:1018-1024. [DOI] [PubMed] [Google Scholar]
- 35.Yoon, S. Y., G. T. Chung, D. H. Kang, C. Ryu, C. K. Yoo, and W. K. Seong. 2005. Application of real-time PCR for quantitative detection of Clostridium botulinum type A toxin gene in food. Microbiol. Immunol. 49:505-511. [DOI] [PubMed] [Google Scholar]