Abstract
The need to evaluate antiviral treatment response and the emergence of resistance have made the human immunodeficiency virus (HIV) viral load assay a major feature of the diagnostic monitoring of HIV-infected individuals. The objective of this study was to evaluate the utility of the recently In Vitro Diagnostic Medical Devices Directive-approved Roche COBAS AmpliPrep/TaqMan96 real-time PCR assay by comparison with the existing Roche COBAS AmpliPrep/AMPLICOR MONITOR conventional PCR assay. EDTA-treated plasma samples from 191 HIV-1-infected individuals were tested for HIV-1 RNA by the AMPLICOR assay and the TaqMan assay. This was a prospective study using 191 pairs of samples from the same bleed per patient. The correlation coefficient of the assays was 98.08%. The mean difference between the assays was 0.05 log10 copy/ml plasma, with a standard deviation (SD) of 0.27 log10 copy/ml plasma. Thirteen samples gave results with variances greater than 0.5 log10 copy/ml plasma, which is our clinical cutoff. Two samples were more than 3 SD different (0.81 log10 copy/ml plasma). The TaqMan assay appeared to be slightly more sensitive at the lower end of the dynamic range. The assays correlated significantly (P > 0.95) with each other, and the regression analysis was also highly significant (R2 > 0.95).
The utility of monitoring human immunodeficiency virus type 1 (HIV-1) RNA viral load in plasma is well documented (1). While the HIV-1 RNA viral load and CD4 lymphocyte count are related to prognosis, the issue now seems more complex than first thought (4, 9). However, with the advent of combination HIV drug therapy in 1996, the need to evaluate antiviral treatment response and the emergence of resistance mean this assay has remained a major feature in monitoring HIV infections (8).
Many patented methodologies have been made commercially available to diagnostic laboratories, and the most widely used in large United Kingdom diagnostic laboratories have been quantitative PCR and quantitative branched-chain DNA assays. The early PCR assays were difficult to perform as high-volume diagnostic tests, and it was not until the introduction of semiautomated systems, such as the Roche COBAS AMPLICOR analyzer, that PCR became a more robust tool for use in this clinical setting. The automation of the nucleic extraction step of the assay using the AmpliPrep extractor further facilitated reliable high-volume testing.
The Roche COBAS TaqMan quantitative HIV 1 RNA assay is more automated than the AMPLICOR assay, requiring no manual intervention between the initial addition of sample to an assay tube and the generation of the quantitative result. Additionally the assay, being a real-time PCR assay, has a wider dynamic range than its predecessor when trying to establish results with samples with high viral loads (>4.87 log10 copies/ml plasma) (COBAS AmpliPrep/COBAS TaqMan HIV-1 test kit insert; Roche Molecular Diagnostics, Basel, Switzerland). The assay uses the TaqMan real-time PCR technology (3). The primer and probes of both of the Roche assays in comparison target conserved regions of the gag region of HIV-1.
Here we present a prospective study, carried out to evaluate the utility of the COBAS AmpliPrep/TaqMan96 (referred to hereafter as TaqMan) assay for HIV-1 RNA in comparison with the existing Roche COBAS AmpliPrep/AMPLICOR Monitor (referred to hereafter as AMPLICOR) assay in use in our laboratory. The quantitative results of the assays are compared.
MATERIALS AND METHODS
EDTA plasma samples from 191 HIV-1-infected individuals were tested for HIV-1 RNA by the AMPLICOR assay and the TaqMan assay. The HIV clinics at the Royal London and St. Bartholomew's Hospitals were asked to provide duplicate samples from the same bleed for a 2-week period to enable testing by both methods.
All samples were centrifuged at 3,000 rpm for 20 min, and the plasma was frozen at −80°C before testing as per the laboratory's standard operating procedure. From each pair, one sample was tested by the AMPLICOR assay and the second by the TaqMan assay. The assays were performed strictly according to the manufacturer's instructions.
When performing the AMPLICOR assay, a decision was made, based upon the available patient information, as to whether the “ultrasensitive” assay (dynamic range, 50 to 75,000 copies/ml) or the “standard” assay (dynamic range, 400 to 750,000 copies/ml) would be performed. Of the 191 samples, 47 were tested with the “standard” assay and 144 with the “ultrasensitive” assay. For the purposes of the analysis, the cutoff for the limit of detection was deemed to be 50 copies/ml for both the TaqMan and AMPLICOR assays.
Three samples initially giving results outside the dynamic range of the AMPLICOR ultrasensitive assay required dilution and retesting. These were diluted using the kit's negative control.
In routine clinical use, a difference in viral load of more than 0.5 log10 copy/ml is deemed clinically significant and this cutoff has been used in the analysis for guidance of significance.
Results were transformed into log10 copies/ml plasma before the statistical analyses, which involved the paired z test of all results and a Student's t test of the clade results. Linear regression and correlation tests were also performed to judge the relationship of the assays to one another.
RESULTS
A total of 189 (98.95%) pairs of results gave differences falling within 3 standard deviations (SD), and those within our clinical cutoff of ±0.5 log10 copy/ml numbered 178 (93.19%). Figure 1 shows the correlation data for the two assays plotted as a scattergram. The correlation coefficient of the assays was 98.08%. The difference between the data sets was not significant (P = 0.73), as determined by the paired z test.
FIG. 1.
HIV-1 RNA by TaqMan and AMPLICOR scatter plot.
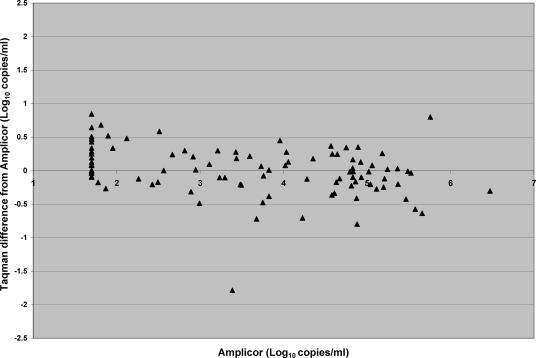
Figure 2 demonstrates the log difference of the TaqMan assay from the AMPLICOR assay plotted against the AMPLICOR log result. The mean log difference was 0.05 log10 copy/ml, and the SD was 0.27 log10 copy/ml, giving a 99% confidence interval at 3 SD of 0.81 log copy/ml. Fifteen samples were ≥0.5 log different from the mean, and 2 of these were ≥3 SD different. Seven (3.67%) samples gave TaqMan results ≥0.5 log copy/ml higher than the AMPLICOR, and 6 (3.14%) samples gave results ≥0.5 log10 copy/ml lower. The mean AMPLICOR viral load of the seven samples ≥0.5 log10 copy/ml higher than the AMPLICOR result was 2.44, and that for six samples ≥0.5 log10 copy/ml lower than the AMPLICOR result was 3.70. One (0.52%) sample was >0.81 log10 copy/ml higher than the AMPLICOR result, having an AMPLICOR value of 1.70 log10 copies/ml, and one sample (0.52%) was >0.81 log10 copy/ml lower, with an AMPLICOR value of 3.39. The data showed a cluster of 12 samples which were detectable by TaqMan at 1.7 to 2.5 log10 copies/ml but were <1.7 log10 copies/ml by AMPLICOR assay. Four of the 13 samples differing by >0.5 log10 copy/ml were tested with the standard AMPLICOR assay and 9 with the ultrasensitive assay.
FIG. 2.
Difference plot of HIV-1 RNA by TaqMan and AMPLICOR assays.
Figure 3 demonstrates the frequency of difference between log values of the two assays.
FIG. 3.
Difference frequencies in HIV-1 RNA by TaqMan and AMPLICOR assays.
Table 1 shows the previous AMPLICOR results and the number of days the sample was taken prior to the TaqMan-AMPLICOR pairs. Some inference may be drawn from previous values of three samples (05M008218, 05M008560, and 05M008665) due to the temporal proximity of the previous sample to the evaluation pair. In all three cases, the previous sample was closer to the TaqMan value and in two cases would make the difference fall within the clinical cutoff.
TABLE 1.
AMPLICOR results from previous samples
| Laboratory no. | Log10 copies/ml plasma
|
No. of days prior to TaqMan samplea | |||
|---|---|---|---|---|---|
| AMPLICOR | TaqMan | Difference | Previous AMPLICOR result | ||
| 05M008665 | 1.70 | 2.54 | 0.85 | 2.05 | 21 |
| 05M008775 | 5.76 | 6.56 | 0.80 | 5.84 | 105 |
| 05M008541 | 1.81 | 2.49 | 0.68 | ||
| 05M008357 | 1.70 | 2.34 | 0.64 | 1.86 | 85 |
| 05M008373 | 2.51 | 3.10 | 0.58 | 5.92 | 125 |
| 05M007996 | 1.90 | 2.42 | 0.52 | 3.73 | 28 |
| 05M008460 | 1.70 | 2.21 | 0.51 | 2.46 | 162 |
| 05M008265 | 5.58 | 5.00 | −0.57 | 4.69 | 63 |
| 05M008218 | 5.66 | 5.03 | −0.63 | 5.55 | 1 |
| 05M008706 | 4.23 | 3.52 | −0.70 | ||
| 05M008087 | 3.68 | 2.96 | −0.72 | ||
| 05M008560 | 4.88 | 4.08 | −0.80 | 4.19 | 0 |
| 05M008220 | 3.39 | 1.60 | 1.78 | 1.92 | 42 |
Number of days previously sample was taken for AMPLICOR prior to TaqMan-AMPLICOR pairs.
Twenty-four patients tested were of known genotype: 11 B clades and 13 non-B clades. Four patients of known genotype varied by more than 0.5, and of these three were non-B clades. The mean of the clade B differences was 0.22 log10 copy/ml with an SD of 0.23. The mean of the difference of non-B clades of 0.37 log10 copy/ml was less than the clinical cutoff; it was still higher than the clade B mean difference and had a higher SD of 0.46 log10 copy/ml. The difference, however, was not statistically significant as determined by one-tailed Student's t test (P = 0.15).
DISCUSSION
The aim of this study was to evaluate the utility of the TaqMan assay by comparing it with the AMPLICOR assay. Our results demonstrate that the assays compare favorably. The AMPLICOR assay is one of the most widely used assays both for clinical monitoring and for clinical trial purposes, and it has been evaluated in many different studies (2). While the technology used to produce the signal has changed from conventional PCR to real-time PCR, the principle of comparing the signal of the unknown viral load in a sample to that of the introduced control of known copy number remains the same. This minimizes interassay variation and interlot variation. Such an approach is essential for high-throughput laboratories requiring tight quality control (6). It is possible that the results having the greatest variance may be due to the manual manipulation stages of the AMPLICOR assay. Full automation should further improve the reproducibility of these results. A recent study (7) compared the AMPLICOR assay with the semiautomated TaqMan 48 assay; however, the extraction system used (High Pure kit) is different from the automated AmpliPrep extraction system used on the fully automated TaqMan96 analyzer.
Previous studies (5, 10, 11) have shown that clade is an important factor in the quantitation of HIV plasma viral load. Our clade results, although limited in number, have shown the variation is within the clinical cutoff and well within statistical significance limits, although non-clade B genotype virus strains gave more variable results than those of clade B.
In conclusion, the assays correlate significantly (P > 0.95), and the regression analysis is also highly significant (R2 > 0.95). With respect to the practical considerations of a diagnostic virology laboratory, we considered that the TaqMan assay was suitable for direct substitution for the AMPLICOR assay. Due to the increased sensitivity at the lower end of the dynamic range, the advice to our requesting clinicians following the introduction of the assay was that patients previously with <1.7 log10 copies/ml may now have detectable low-level viral load. In addition to this, the lower limit of the linear range for the TaqMan assay is 1.6 log10 copies/ml, which will add to the numbers of samples with quantifiable RNA being detected if the manufacturer's reporting guidance is used. This will obviously entail close liaison with the requesting clinicians to ensure that the significance of these results is fully explained in the light of the change of assay.
Acknowledgments
We gratefully acknowledge the staff in Specimen Reception and those of the Molecular Virology Section at the Royal London Hospital for their assistance with the management of the samples. We also thank Roche Diagnostics for supplying the TaqMan reagents for this comparison. Finally, we thank the staff in Infection and Immunity at Barts and the London NHS Trust for assisting with the procurement of the samples.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.BHIVA Executive Committee. 2005. British HIV Association (BHIVA) guidelines for the treatment of HIV-infected adults with antiretroviral therapy. HIV Med. 6(Suppl. 2):1-61. [DOI] [PubMed] [Google Scholar]
- 2.Erali, M., and D. R. Hillyard. 1998. Evaluation of the ultrasensitive Roche Amplicor HIV-1 Monitor assay for quantitation of human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 37:792-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 5.Michael, N. L., S. A. Herman, S. Kwok, K. Dreyer, J. Wang, C. Christopherson, J. P. Spadoro, K. K. Young, V. Polonis, F. E. McCutchan, J. Carr, J. R. Mascola, L. L. Jagodzinski, and M. L. Robb. 1999. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J. Clin. Microbiol. 37:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niesters, H. G. 2004. Molecular and diagnostic clinical virology in real time. Clin. Microbiol. Infect. 10:5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perrin, L., J. M. Pawlotsky, M. Bouvier-Alias, C. Sarrazin, S. Zeuzem, and G. Colucci. 2006. Multicenter performance evaluation of a new TaqMan PCR assay for monitoring human immunodeficiency virus RNA load. J. Clin. Microbiol. 44:4371-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riddler, S. A., and J. W. Mellors. 1997. HIV-1 viral dynamics and viral load measurement: implications for therapy. AIDS Clin. Rev. 1997-1998:47-65. [PubMed]
- 9.Rodriguez, B., A. K. Sethi, V. K. Cheruvu, W. Mackay, R. J. Bosch, M. Kitahata, S. L. Boswell, W. C. Mathews, D. R. Bangsberg, J. Martin, C. C. Whalen, S. Sieg, S. Yadavalli, S. G. Deeks, and M. M. Lederman. 2006. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. JAMA 296:1498-1506. [DOI] [PubMed] [Google Scholar]
- 10.Swanson, P., C. de Mendoza, Y. Joshi, A. Golden, R. L. Hodinka, V. Soriano, S. G. Devare, and J. Hackett, Jr. 2005. Impact of human immunodeficiency virus type 1 (HIV-1) genetic diversity on performance of four commercial viral load assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v1.5, VERSANT HIV-1 RNA 3.0, and NucliSens HIV-1 QT. J. Clin. Microbiol. 43:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Triques, K., J. Coste, J. L. Perret, C. Segarra, E. Mpoudi, J. Reynes, E. Delaporte, A. Butcher, K. Dreyer, S. Herman, J. Spadoro, and M. Peeters. 1999. Efficiencies of four versions of the AMPLICOR HIV-1 MONITOR test for quantification of different subtypes of human immunodeficiency virus type 1. J. Clin. Microbiol. 37:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]