Abstract
For hospital epidemiologists, determining a system of typing that is discriminatory is essential for measuring the effectiveness of infection control measures. In situations in which the incidence of resistant Pseudomonas aeruginosa is increasing, the ability to discern whether it is due to patient-to-patient transmission versus an increase in patient endogenous strains is often made on the basis of molecular typing. The present study compared the discriminatory abilities of pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) for 90 P. aeruginosa isolates obtained from cultures of perirectal surveillance swabs from patients in an intensive care unit. PFGE identified 85 distinct types and 76 distinct groups when similarity cutoffs of 100% and 87%, respectively, were used. By comparison, MLST identified 60 sequence types that could be clustered into 11 clonal complexes and 32 singletons. By using the Simpson index of diversity (D), PFGE had a greater discriminatory ability than MLST for P. aeruginosa isolates (D values, 0.999 versus 0.975, respectively). Thus, while MLST was better for detecting genetic relatedness, we determined that PFGE was more discriminatory than MLST for determining genetic differences in P. aeruginosa.
Pseudomonas aeruginosa is a common pathogen that causes nosocomial infections in intensive care units (ICUs) (1, 28). Resistant P. aeruginosa is an emerging threat to patients (15, 19, 23). Several schemes for the molecular typing of P. aeruginosa have been proposed to determine the relatedness of nosocomial pathogens. These include pulsed-field gel electrophoresis (PFGE), ribotyping (5), and PCR-based fingerprinting (7). For hospital epidemiologists, determining the best type of discriminatory molecular genetic analysis is essential for measuring the effectiveness of infection control measures. Also, in situations in which the incidence of resistant P. aeruginosa is increasing, the ability to discern whether it is due to patient-to-patient transmission versus an increase in patient endogenous strains is often made on the basis of molecular typing. Although other studies have compared numerous molecular typing schemes (2, 13, 26), PFGE is still considered by most to be the “gold standard.”
Recently, multilocus sequence typing (MLST), which is based on the allelic differences among housekeeping genes, has become a popular bacterial typing method (9). MLST typing schemes for determination of the relatedness of numerous microorganisms, including Staphylococcus aureus (8), Salmonella enterica (11), Enterococcus faecalis (20), and Vibrio cholerae (18), have been developed. Recently, Curran and colleagues developed a multilocus sequence typing scheme that discriminates P. aeruginosa isolates by differences in the sequences of the following seven loci: acsA, aroE, guaA, mutL, noD, ppsA, and trpE (4).
In the study described here, we set out to determine the discriminatory abilities and potential utilities of PFGE and MLST for the genetic characterization of P. aeruginosa. In order to compare these two methods, we analyzed 90 P. aeruginosa isolates obtained from perirectal surveillance swab specimens from 90 unique patients among a cohort of ICU patients. We believe that this is the first study to have compared MLST and PFGE by the use of a large cohort of P. aeruginosa isolates.
MATERIALS AND METHODS
Bacterial isolates.
Surveillance perirectal swab specimens taken from a cohort of patients admitted to the medical and surgical ICUs at the University of Maryland Medical Center between 1 September 2001 and 1 September 2004 were used. Perirectal swab specimens for culture were obtained from the patients on admission, weekly, and upon discharge. Non-lactose-fermenting, oxidase-positive isolates were identified as P. aeruginosa if they did not produce an acid reaction in triple sugar iron agar (Becton Dickinson, Franklin Lakes, NJ), grew at 42°C, and produced blue-green pigment on Pseudomonas P agar (Remel, Lenexa, KS). Nonfermenters not matching these characteristics were identified by using API 20NE system (BioMerieux Inc., Durham, NC) test strips. Antimicrobial susceptibility testing was done by disk diffusion according to the CLSI guidelines (3). Forty-six imipenem-resistant isolates (24 isolates susceptible and 22 isolates resistant to piperacillin-tazobactam) and 44 imipenem-susceptible P. aeruginosa isolates (4 isolates susceptible and 40 isolates resistant to piperacillin-tazobactam) from unique patients were randomly chosen to undergo typing by PFGE and MLST.
PFGE.
PFGE was performed as described previously (6). All isolates were digested with SpeI, and the resulting fragments were separated by electrophoresis in 1% agarose gels with a CHEF DR apparatus (Bio-Rad Laboratories, Hercules, CA) for 24 h, with the switch times ranging from 2 s and 40 s in Tris-borate EDTA buffer containing 50 μM thiourea (25). Photographic images of the gels were saved digitally with Geldoc EQ software (Bio-Rad Laboratories) and saved as TIFF files for gel analysis with Fingerprinting II software (Bio-Rad Laboratories). The band patterns were compared by use of the Dice coefficient by using the unweighted pair group method to determine band similarity and the criteria established by Tenover et al. to define the pulsed-field type clusters (27). Isolates with band patterns that were 100% identical were considered to be of identical PFGE types, and isolates that had band patterns with ≥87% similarity were considered to be genetically related.
MLST.
MLST was performed by previously published protocols (4). Briefly, genomic DNA was isolated by using a Prepman Ultura apparatus (Applied Biosystems, Foster City, CA), according to the manufacturer's guidelines. Standard DNA amplification and sequencing of the seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) were performed for all isolates. The nucleotide sequences of both strands were determined by using previously published primers and were compared to existing sequences in the MLST database (www.pubmlst.org/paeruginosa) for assignment of allelic numbers. The isolates were assigned a sequence type (ST) number according to their allelic profiles. The eBURST algorithm (http://pubmlst.org/analysis/) was used for phylogenetic analysis, and isolates that were identical at five or more alleles were considered to be part of a clonal complex (CC).
Data analysis.
Simpson's index of diversity (D) was used to compare the discriminatory powers of PFGE and MLST by use of the following formula (16):
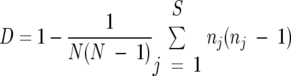 |
where N is the total number of strains in the sample population, S is the total number of types described, and nj is the number of strains belonging to the jth type.
RESULTS
PFGE.
SpeI digestion of the 90 P. aeruginosa isolates from different patients identified 85 unique patterns (designated pattern types [PTs] PT1 to PT85) (Table 1). When the genetic relatedness (defined as ≥87% band identity) of the isolates was compared with that determined by MLST, PFGE revealed 19 isolates that formed into six groups with two or more isolates each, denoted groups A to F (Table 1), and there were 70 unique PFGE patterns with one isolate each. The group with the largest number of isolates of the same PT was group C, which consisted of eight isolates with five related PFGE patterns. All isolates in this group were resistant to piperacillin-tazobactam, six isolates were resistant to imipenem, and two isolates were susceptible to imipenem. The isolates in three of the other five groups were all susceptible to piperacillin-tazobactam but had different susceptibilities to imipenem. However, for the four groups that had more than one isolate with identical PTs, only PT40 contained isolates with different patterns of resistance. Therefore, PFGE did not group the organisms by their susceptibilities. One isolate was not digested by SpeI; however, the isolate was digested with XbaI (data not shown) and was not designated a PFGE type.
TABLE 1.
PFGE types compared to MLST types of 90 clinical P. aeruginosa isolates
| Isolate no. | PFGE no. | PFGE group | MLST ST | Burst group (CC) | Susceptibilitya
|
|
|---|---|---|---|---|---|---|
| Imipenem | Piperacillin- tazobactam | |||||
| 1 | PT01 | 282 | UM08 | R | S | |
| 2 | PT02 | 235 | UM05 | R | R | |
| 3 | PT03 | 444 | UM09 | S | S | |
| 4 | PT04 | 111 | UM02 | R | R | |
| 5 | PT05 | 232 | UM04 | S | R | |
| 6 | PT06 | 471 | UM04 | S | S | |
| 7 | PT07 | 257 | Singleton | S | S | |
| 8 | PT08 | 460 | Singleton | S | S | |
| 9 | PT09 | 469 | Singleton | R | S | |
| 10 | PT10 | 155 | UM03 | R | S | |
| 11 | PT11 | 252 | Singleton | S | S | |
| 12 | PT12 | 17 | UM01 | S | S | |
| 13 | PT13 | 442 | Singleton | R | S | |
| 14 | PT14 | 390 | Singleton | S | S | |
| 15 | PT15 | 244 | UM06 | S | S | |
| 16 | PT16 | 270 | Singleton | S | R | |
| 17 | PT17 | 17 | UM01 | S | S | |
| 18 | PT18 | 439 | Singleton | R | S | |
| 19 | PT19 | 444 | UM08 | R | R | |
| 20 | PT20 | 282 | UM08 | R | S | |
| 21 | PT21 | A | 253 | Singleton | S | S |
| 22 | PT22 | A | 253 | Singleton | R | S |
| 23 | PT23 | 321 | UM01 | R | S | |
| 24 | PT24 | 253 | Singleton | R | S | |
| 25 | PT25 | B | 444 | UM08 | S | S |
| 26 | PT26 | B | 444 | UM08 | R | S |
| 27 | PT27 | 282 | UM08 | S | S | |
| 28 | PT28 | 282 | UM08 | R | S | |
| 29 | PT29 | 282 | UM08 | R | S | |
| 30 | PT30 | 458 | Singleton | R | S | |
| 31 | PT31 | NDb | Singleton | R | S | |
| 32 | PT32 | 466 | UM08 | R | R | |
| 33 | PT33 | 455 | Singleton | S | S | |
| 34 | PT34 | 274 | UM08 | S | S | |
| 35 | PT35 | 453 | Singleton | R | S | |
| 36 | PT36 | C | 111 | UM02 | R | R |
| 37 | PT37 | C | 111 | UM02 | S | R |
| 38 | PT38 | C | 111 | UM02 | R | R |
| 39 | PT38 | C | 111 | UM02 | R | R |
| 40 | PT39 | C | 111 | UM02 | R | R |
| 41 | PT39 | C | 111 | UM02 | R | R |
| 42 | PT40 | C | 111 | UM02 | S | R |
| 43 | PT40 | C | 464 | UM02 | R | R |
| 44 | PT41 | 111 | UM02 | R | R | |
| 45 | PT42 | 445 | UM01 | S | S | |
| 46 | PT43 | 111 | UM02 | R | R | |
| 47 | PT44 | 463 | Singleton | R | S | |
| 48 | PT45 | 282 | UM08 | R | S | |
| 49 | PT46 | 446 | UM10 | S | S | |
| 50 | PT47 | 190 | Singleton | S | S | |
| 51 | PT48 | 385 | UM09 | S | S | |
| 52 | PT49 | 461 | UM03 | R | S | |
| 53 | PT50 | D | 441 | UM06 | S | S |
| 54 | PT51 | D | 462 | UM06 | R | S |
| 55 | PT52 | 244 | UM06 | R | S | |
| 56 | PT53 | ND | UM04 | R | R | |
| 57 | PT54 | 457 | UM05 | R | R | |
| 58 | PT55 | 235 | UM05 | R | R | |
| 59 | PT56 | 459 | Singleton | S | S | |
| 60 | PT57 | 443 | UM09 | S | S | |
| 61 | PT58 | 235 | UM05 | S | S | |
| 62 | PT59 | 27 | UM01 | S | S | |
| 63 | PT60 | 468 | Singleton | S | S | |
| 64 | PT61 | E | 449 | UM01 | R | S |
| 65 | PT62 | E | 27 | UM01 | R | S |
| 66 | PT63 | 452 | Singleton | S | S | |
| 67 | PT64 | 27 | UM01 | S | S | |
| 68 | PT65 | 348 | Singleton | R | R | |
| 69 | PT66 | 164 | Singleton | S | S | |
| 70 | PT67 | 450 | Singleton | S | S | |
| 71 | PT68 | 244 | UM06 | S | S | |
| 72 | PT69 | 440 | UM07 | S | S | |
| 73 | PT70 | 267 | UM07 | S | S | |
| 74 | PT71 | F | 447 | UM11 | R | R |
| 75 | PT71 | F | 447 | UM11 | R | R |
| 76 | PT72 | F | 448 | UM11 | R | R |
| 77 | PT73 | 299 | UM03 | S | S | |
| 78 | PT74 | 179 | Singleton | R | R | |
| 79 | PT75 | 470 | Singleton | S | S | |
| 80 | PT76 | 281 | Singleton | R | S | |
| 81 | PT77 | 111 | UM02 | R | R | |
| 82 | PT78 | 467 | Singleton | S | S | |
| 83 | PT79 | 465 | Singleton | S | S | |
| 84 | PT80 | 456 | Singleton | S | S | |
| 85 | PT81 | 454 | UM10 | S | S | |
| 86 | PT82 | 17 | UM01 | S | S | |
| 87 | PT83 | 275 | Singleton | S | S | |
| 88 | PT84 | 451 | Singleton | S | S | |
| 89 | PT85 | 111 | UM02 | R | R | |
| 90 | ND | ND | Singleton | R | S | |
R, resistant; S, susceptible.
ND, not determined.
MLST.
MLST of the isolates was performed to compare the discriminatory power of MLST to that of PFGE. MLST revealed 60 different STs (Table 2). Among the 60 STs, 36 were not previously submitted to the P. aeruginosa database (17). Fifty-eight STs could be clustered by analysis with the eBURST program into 11 different CCs, while the remaining 32 STs were classified as singletons. The three largest CCs, denoted UM01, UM02, and UM08 (Tables 1 and 2), contained 9, 13, and 12 STs, respectively. In 7 of 12 CCs (CCs UM01, UM02, UM03, UM04, UM05, UM06, and UM08) there were combinations of resistance and susceptibility to piperacillin-tazobactam and imipenem among the isolates. In six of eight cases in which more than one isolate had the same ST, the isolates did not have the same patterns of susceptibility to piperacillin-tazobactam and imipenem. MLST also did not group the isolates by their resistance to these two antimicrobial agents.
TABLE 2.
MLST types and the seven locus STsa
| CC | MLST type | No. of isolates | STa
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| acsA | aroE | guaA | mutL | nuoD | ppsA | trpE | |||
| UM01 | 17 | 3 | 11 | 5 | 1 | 7 | 9 | 4 | 7 |
| UM01 | 27 | 3 | 6 | 5 | 6 | 7 | 4 | 6 | 7 |
| UM01 | 321 | 1 | 6 | 5 | 1 | 7 | 4 | 4 | 7 |
| UM01 | 445 | 1 | 54 | 5 | 1 | 7 | 9 | 4 | 7 |
| UM01 | 449 | 1 | 6 | 5 | 1 | 7 | 4 | 6 | 7 |
| UM02 | 111 | 12 | 17 | 5 | 5 | 4 | 4 | 4 | 3 |
| UM02 | 464 | 1 | 17 | 5 | 5 | 4 | 11 | 4 | 7 |
| UM03 | 155 | 1 | 28 | 5 | 36 | 3 | 3 | 13 | 7 |
| UM03 | 299 | 1 | 17 | 5 | 36 | 3 | 3 | 7 | 3 |
| UM03 | 461 | 1 | 28 | 5 | 36 | 3 | 3 | 13 | 3 |
| UM04 | 232 | 1 | 17 | 5 | 11 | 64 | 4 | 4 | 2 |
| UM04 | 471 | 1 | 17 | 5 | 11 | 5 | 4 | 4 | 2 |
| UM05 | ND | 1 | 38 | 11 | 3 | 13 | 1 | 2 | ND |
| UM05 | 235 | 3 | 38 | 11 | 3 | 13 | 1 | 2 | 4 |
| UM05 | 457 | 1 | 38 | 11 | 10 | 13 | 1 | 2 | 4 |
| UM06 | 441 | 1 | 17 | 5 | 12 | 76 | 14 | 4 | 7 |
| UM06 | 462 | 1 | 17 | 5 | 12 | 76 | 14 | 4 | 2 |
| UM06 | 244 | 3 | 17 | 5 | 12 | 3 | 14 | 4 | 7 |
| UM07 | 440 | 1 | 19 | 5 | 5 | 11 | 11 | 4 | 14 |
| UM07 | 267 | 1 | 19 | 5 | 12 | 11 | 11 | 4 | 14 |
| UM08 | 274 | 1 | 23 | 5 | 11 | 7 | 1 | 12 | 7 |
| UM08 | 282 | 6 | 6 | 5 | 11 | 7 | 3 | 12 | 19 |
| UM08 | 444 | 4 | 6 | 5 | 75 | 7 | 3 | 12 | 19 |
| UM08 | 466 | 1 | 6 | 5 | 11 | 7 | 1 | 12 | 7 |
| UM09 | 443 | 1 | 15 | 5 | 5 | 5 | 50 | 4 | 1 |
| UM09 | 385 | 1 | 15 | 5 | 5 | 5 | 50 | 4 | 14 |
| UM10 | 446 | 1 | 18 | 4 | 5 | 3 | 1 | 17 | 13 |
| UM10 | 454 | 1 | 38 | 4 | 10 | 3 | 1 | 17 | 13 |
| UM11 | 447 | 2 | 40 | 5 | 11 | 3 | 1 | 47 | 60 |
| UM11 | 448 | 1 | 40 | 5 | 11 | 3 | 1 | 47 | 8 |
| Singleton | 164 | 1 | 1 | 5 | 1 | 11 | 4 | 10 | 10 |
| Singleton | 179 | 1 | 36 | 27 | 28 | 3 | 4 | 13 | 7 |
| Singleton | 190 | 1 | 6 | 3 | 1 | 3 | 2 | 14 | 1 |
| Singleton | 252 | 1 | 6 | 28 | 4 | 3 | 3 | 4 | 7 |
| Singleton | 253 | 3 | 4 | 4 | 16 | 12 | 1 | 6 | 3 |
| Singleton | 257 | 1 | 35 | 24 | 36 | 11 | 4 | 15 | 14 |
| Singleton | 270 | 1 | 22 | 3 | 17 | 5 | 2 | 10 | 7 |
| Singleton | 275 | 1 | 11 | 5 | 20 | 5 | 4 | 4 | 52 |
| Singleton | 281 | 1 | 11 | 57 | 1 | 5 | 4 | 4 | 2 |
| Singleton | 348 | 1 | 22 | 20 | 11 | 3 | 3 | 3 | 7 |
| Singleton | 390 | 1 | 39 | 5 | 1 | 3 | 4 | 46 | 56 |
| Singleton | 439 | 1 | 6 | 68 | 20 | 11 | 4 | 4 | 7 |
| Singleton | 442 | 1 | 6 | 5 | 10 | 1 | 1 | 12 | 1 |
| Singleton | 450 | 1 | 28 | 5 | 11 | 77 | 3 | 4 | 1 |
| Singleton | 451 | 1 | 17 | 22 | 11 | 3 | 4 | 4 | 7 |
| Singleton | 452 | 1 | 35 | 5 | 36 | 11 | 4 | 10 | 7 |
| Singleton | 453 | 1 | 16 | 5 | 11 | 7 | 1 | 7 | 61 |
| Singleton | 455 | 1 | 6 | 14 | 12 | 11 | 1 | 4 | 20 |
| Singleton | 456 | 1 | 28 | 68 | 5 | 11 | 3 | 15 | 44 |
| Singleton | 458 | 1 | 13 | 8 | 9 | 3 | 1 | 17 | 3 |
| Singleton | 459 | 1 | 28 | 24 | 10 | 5 | 1 | 6 | 4 |
| Singleton | 460 | 1 | 40 | 57 | 11 | 3 | 4 | 28 | 37 |
| Singleton | 463 | 1 | 6 | 5 | 5 | 3 | 1 | 6 | 3 |
| Singleton | 465 | 1 | 9 | 54 | 78 | 72 | 4 | 20 | 7 |
| Singleton | 467 | 1 | 6 | 5 | 11 | 18 | 4 | 12 | 3 |
| Singleton | 468 | 1 | 5 | 67 | 76 | 3 | 1 | 4 | 62 |
| Singleton | 469 | 1 | 17 | 66 | 5 | 77 | 3 | 4 | 7 |
| Singleton | 470 | 1 | 6 | 6 | 1 | 5 | 27 | 4 | 7 |
| Singleton | ND | 1 | 32 | 8 | 5 | 3 | 5 | 6 | ND |
| Singleton | ND | 1 | 15 | 5 | 77 | 72 | 3 | 6 | ND |
ND, not determined.
Comparison of PFGE and MLST.
The discriminatory abilities of PFGE and MLST were compared by the number of unique STs or patterns determined by each method and by the number of related groups. PFGE determined 85 unique types, with 19 of the isolates grouped into six groups, defined by isolates that were genetically related because they had ≥87% band identity. MLST determined 60 unique STs and grouped the isolates into 11 CCs because of their sequence identity at five or more alleles. Simpson's D value showed that when isolates with identical (or unique) patterns were compared, PFGE had a slightly higher discriminatory power than MLST (D values, 0.999 and 0.975, respectively). The clustering of the isolates by PFGE and MLST was correlated globally, but the details differed. All 32 singleton MLST STs had unique PFGE patterns. The isolates in each PFGE group were found to be members of the same MLST CC and, in most cases, the same ST. Two of the four pairs of isolates with the same PFGE patterns had identical STs. Similarly, for all eight of the STs with multiple isolates, the isolates with the same ST did not have the same PFGE patterns. When PFGE and MLST were compared, although the isolates with similar PTs had different STs, they were grouped in the same CC. Exceptions were PT groups A and B, which contained two isolates with the same MLST type. When multiple isolates with identical MLST types were analyzed, the isolates often had different unrelated PFGE patterns.
DISCUSSION
In this study we compared two different methods, PFGE and MLST, for the molecular typing of P. aeruginosa using 90 isolates obtained from surveillance perirectal swab specimens from patients in ICUs. Using Simpson's D value, we determined that PFGE had a greater discriminatory ability than MLST, with D values of 0.999 and 0.975, respectively. Although both of these methods have high discriminatory abilities, PFGE distinguished more types (85 versus 60 types distinguished by MLST). Neither method grouped the isolates according to their resistance to the antimicrobials imipenem and piperacillin-tazobactam.
Many schemes for the typing of P. aeruginosa have been developed. These include PFGE, ribotyping (5), and PCR-based fingerprinting (5). Although other typing schemes have been developed and show a variety of discriminatory powers, PFGE is known to be the gold standard for the molecular typing of P. aeruginosa. Our study has shown that PFGE is highly discriminatory when it is used for the typing of P. aeruginosa; however, MLST is also highly discriminatory and could be used as a method for the typing of P. aeruginosa. These findings are similar to those of a recent study by Giske et al. that compared multiple methods, including PFGE and MLST, for the molecular typing of 10 multidrug-resistant P. aeruginosa isolates and that determined that PFGE had the greater discriminatory ability (12). MLST is a new typing technique that is becoming popular due to the ease of data analysis, but some studies have shown that MLST had a lower discriminatory ability than PFGE (10, 14, 18, 22, 24). However, other studies have shown that MLST has a greater discriminatory ability than PFGE for Vibrio cholerae and other bacteria (18, 21). Therefore, we believe that our conclusions are specific to P. aeruginosa and not universal for all bacterial species. Comparison studies must be performed with each bacterial species to determine which molecular typing method should be used.
Although our study showed that PFGE had a higher discriminatory ability than MLST, MLST has the advantage that it gives information about the clonal relationships of isolates that PFGE does not. In our study, MLST showed that among the 90 isolates of P. aeruginosa tested, 58 of the isolates fell into 11 CCs. We observed eight STs among the isolates showing different and often unrelated PTs, suggesting that for P. aeruginosa, the PT evolves more rapidly than the ST. In other words, PFGE detects changes in the molecular clock better than MLST does. Giske et al. analyzed 10 VIM-1-like metallo-β-lactamase-producing P. aeruginosa isolates and showed that PFGE was more discriminatory but concluded that the MLST method and the clonal relationship data provided by that method were more advantageous for the typing of P. aeruginosa (12).
In this study we have compared PFGE and MLST to determine their discriminatory abilities and the potential utilities of PFGE and MLST in determining the relatedness of P. aeruginosa isolates. We determined that PFGE is more discriminatory than MLST, and although MLST has the power to establish clonal relationships between the isolates, for eight STs, PFGE discriminated the isolates further than MLST did. Future studies analyzing P. aeruginosa isolates from the same patient or from patients who are known to have epidemiological links are needed to compare PFGE and MLST for their abilities to distinguish isolates that are similar.
Acknowledgments
This study was supported by National Institutes of Health grants K23 AI01752-01A1 and R01 AI60859-01A1 and by the University of Maryland General Clinical Research Center, grant M01RR16500, General Clinical Research Centers Program, National Center for Research Resources (NCRR), NIH.
We acknowledge Gwen Smith for her assistance with the PFGE typing.
Footnotes
Published ahead of print on 19 September 2007.
REFERENCES
- 1.Anonymous. 2000. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992-April 2000, issued June 2000. Am. J. Infect. Control 28:429-448. [DOI] [PubMed] [Google Scholar]
- 2.Botes, J., G. Williamson, V. Sinickas, and V. Gurtler. 2003. Genomic typing of Pseudomonas aeruginosa isolates by comparison of riboprinting and PFGE: correlation of experimental results with those predicted from the complete genome sequence of isolate PAO1. J. Microbiol. Methods 55:231-240. [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically; approved standard, 7th ed. CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Curran, B., D. Jonas, H. Grundmann, T. Pitt, and C. G. Dowson. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denamur, E., B. Picard, P. Goullet, E. Bingen, N. Lambert, and J. Elion. 1991. Complexity of Pseudomonas aeruginosa infection in cystic fibrosis: combined results from esterase electrophoresis and rDNA restriction fragment length polymorphism analysis. Epidemiol. Infect. 106:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deplano, A., O. Denis, L. Poirel, D. Hocquet, C. Nonhoff, B. Byl, P. Nordmann, J. L. Vincent, and M. J. Struelens. 2005. Molecular characterization of an epidemic clone of panantibiotic-resistant Pseudomonas aeruginosa. J. Clin. Microbiol. 43:1198-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elaichouni, A., G. Verschraegen, G. Claeys, M. Devleeschouwer, C. Godard, and M. Vaneechoutte. 1994. Pseudomonas aeruginosa serotype O12 outbreak studied by arbitrary primer PCR. J. Clin. Microbiol. 32:666-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 10.Fakhr, M. K., L. K. Nolan, and C. M. Logue. 2005. Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar Typhimurium. J. Clin. Microbiol. 43:2215-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foley, S. L., D. G. White, P. F. McDermott, R. D. Walker, B. Rhodes, P. J. Fedorka-Cray, S. Simjee, and S. Zhao. 2006. Comparison of subtyping methods for differentiating Salmonella enterica serovar Typhimurium isolates obtained from food animal sources. J. Clin. Microbiol. 44:3569-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giske, C. G., B. Libisch, C. Colinon, E. Scoulica, L. Pagani, M. Fuzi, G. Kronvall, and G. M. Rossolini. 2006. Establishing clonal relationships between VIM-1-like metallo-β-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J. Clin. Microbiol. 44:4309-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundmann, H., C. Schneider, D. Hartung, F. D. Daschner, and T. L. Pitt. 1995. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J. Clin. Microbiol. 33:528-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallin, M., A. Deplano, O. Denis, R. De Mendonca, R. De Ryck, and M. J. Struelens. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45:127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, A., C. Torres-Viera, L. Venkataraman, P. DeGirolami, M. Samore, and Y. Carmeli. 1999. Epidemiology and clinical outcomes of patients with multiresistant Pseudomonas aeruginosa. Clin. Infect. Dis. 28:1128-1133. [DOI] [PubMed] [Google Scholar]
- 16.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbnet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotetishvili, M., O. C. Stine, Y. Chen, A. Kreger, A. Sulakvelidze, S. Sozhamannan, and J. G. Morris, Jr. 2003. Multilocus sequence typing has better discriminatory ability for typing Vibrio cholerae than does pulsed-field gel electrophoresis and provides a measure of phylogenetic relatedness. J. Clin. Microbiol. 41:2191-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landman, D., J. M. Quale, D. Mayorga, A. Adedeji, K. Vangala, J. Ravishankar, C. Flores, and S. Brooks. 2002. Citywide clonal outbreak of multiresistant Acinetobacter baumannii and Pseudomonas aeruginosa in Brooklyn, N.Y.: the preantibiotic era has returned. Arch. Intern. Med. 162:1515-1520. [DOI] [PubMed] [Google Scholar]
- 20.Nallapareddy, S. R., R. W. Duh, K. V. Singh, and B. E. Murray. 2002. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemoy, L. L., M. Kotetishvili, J. Tigno, A. Keefer-Norris, A. D. Harris, E. N. Perencevich, J. A. Johnson, D. Torpey, A. Sulakvelidze, J. G. Morris, Jr., and O. C. Stine. 2005. Multilocus sequence typing versus pulsed-field gel electrophoresis for characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates. J. Clin. Microbiol. 43:1776-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. G. Morris, Jr., D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paterson, D. L. 2006. The epidemiological profile of infections with multidrug-resistant Pseudomonas aeruginosa and Acinetobacter species. Clin. Infect. Dis. 43(Suppl. 2):S43-S48. [DOI] [PubMed] [Google Scholar]
- 24.Revazishvili, T., M. Kotetishvili, O. C. Stine, A. S. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romling, U., and B. Tummler. 2000. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:464-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speijer, H., P. H. Savelkoul, M. J. Bonten, E. E. Stobberingh, and J. H. Tjhie. 1999. Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J. Clin. Microbiol. 37:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]


