Abstract
Salmonella enterica serotype Typhi clinical isolates (n = 91) resistant to nalidixic acid (Nalr) were collected from sporadic cases and minor outbreaks throughout Vietnam between 1996 and 2004. These isolates were typed and compared by four methods: Vi phage typing, PstI ribotyping, XbaI and SpeI pulsed-field gel electrophoresis (PFGE), and single-nucleotide polymorphism (SNP) analysis. The results indicated that 65% of the isolates were not typeable by Vi phage typing. In contrast, the ribotyping and, with more accuracy, the SNP analysis methods indicated that all Nalr isolates belonged to a single clone (ribotype 3a, haplotype H58) that was found previously and that largely consisted of plasmid-encoded multidrug-resistant serotype Typhi isolates. PFGE demonstrated the occurrence of microevolution within this clone. We identified two major combined PFGE profiles: X1-S1 and X3-S6. X3-S6 predominated between 1996 and 2002 but was replaced by X1-S1 after 2002. Nevertheless, PFGE, with a Simpson's index of 0.78, was not considered an optimal discriminatory method for investigating typhoid fever outbreaks in Vietnam. The rate of quinolone resistance increased and the rate of multidrug resistance decreased during the study period. From 2002 to 2004, 80.6% of the isolates from South Vietnam were resistant only to Nal. The mechanism of Nal resistance in most of the isolates (94%) was a mutation in the quinolone resistance-determining chromosomal region of gyrA that led to the amino acid substitution Ser83Phe. No plasmid-located qnrA, qnrB, or qnrS was detected.
In the last 30 years, high rates of multiple-drug resistance (MDR) to all first-line antimicrobial agents encoded by large conjugative plasmids in Salmonella enterica serotype Typhi have been reported around the world, but the Indian subcontinent and Southeast Asian countries are particularly affected (43).
In Vietnam MDR serotype Typhi isolates with self-transferable plasmids of the H1 incompatibility group, which code for chloramphenicol, streptomycin, sulfonamides, and tetracycline resistance, were first isolated in 1971; the rate of such MDR increased to 85% in 1975 (3, 25, 34). MDR serotype Typhi isolates with additional resistance to ampicillin and sulfamethoxazole-trimethoprim were first reported in southern Vietnam at the beginning of the 1990s (29). The economic reforms that took place in Vietnam in the early 1990s have resulted in a boom in private pharmacies, and all first-line antibiotics for typhoid fever could easily be bought as over-the-counter medicines without a prescription (40). Over 90% of the sporadic or epidemic serotype Typhi isolates in the north, central, and south regions of Vietnam were MDR from 1995 to 2002 (18). Since then, fluoroquinolones have become a first-line treatment for typhoid fever. However, MDR serotype Typhi isolates with additional chromosomally encoded resistance to nalidixic acid (Nalr) and with reduced susceptibilities to fluoroquinolones have been reported more and more in the Indian subcontinent since the early 1990s and have later been reported in various Asian countries (13, 23, 24, 33, 38, 41). MDR serotype Typhi Nalr isolates were first reported in southern Vietnam in 1993 and made up 20% of the isolates in 1996 (30, 41). Data collected by the Laboratoire d'Épidémiologie de la Résistance Bacterienne, Institut National d'Hygiène et d'Épidémiologie, Hanoi, Vietnam, show that the percentage of Nalr isolates did not change between 2000 and 2004. However, the percentages differed by region: about 80% of 243 isolates in the south of the country were Nalr, 50% of 70 isolates in the central part of the country were Nalr, and rare isolates in the north were Nalr. These differences might be explained by two factors. Typhoid fever is more prevalent in densely populated agricultural tropical southern Vietnam, in particular, in the Mekong River Delta. Of the 187,318 cases reported nationally between 1991 and 2001 (approximately 17,000 per annum), 75.8% and 6.9% occurred in the Mekong River Delta and in the southeastern part of the country, respectively (16). In southern Vietnam over-the-counter antimicrobial drugs are readily available in private pharmacies (including new of antibiotics).
The emergence of MDR S. enterica serotype Typhi Nalr isolates is of great concern because these strains are associated with slow clinical responses to fluoroquinolones and treatment failures (5, 39, 41) and require the use of expensive expanded-spectrum cephalosporins. This study investigates the genetic diversity of MDR serotype Typhi Nalr isolates to determine whether one or several clones were involved in the recent small outbreaks and sporadic cases of typhoid fever in Vietnam. These findings should facilitate determination of the mode through which MDR serotype Typhi Nalr isolates spread and should be useful for the implementation of rational strategies and suitable measures in the field of public health to prevent this disease. We used four subtyping methods to analyze a representative collection of 91 Nalr serotype Typhi isolates recovered in Vietnam between 1996 and 2004. We found that only one MDR serotype Typhi Nalr clone was circulating during this period but identified microevolution (generation-to-generation small-scale genetic changes in a population) within this clone.
MATERIALS AND METHODS
Bacterial isolates.
Ninety-one Nalr serotype Typhi isolates (one per patient) were selected from the Laboratoire d'Épidémiologie de la Résistance Bacterienne collection. These were epidemiologically independent, i.e., 11 from the north, 10 from the center, and 70 from the south. The isolates were collected during an 8-year period (1996 to 2004) from blood cultures in hospitals located in the three regions of Vietnam: in the north at Hanoi (Back Mai Hospital, Saint-Paul Hospital, 103 Military Hospital, Institut National d'Hygiène et d'Épidémiologie, and Pediatric Hospital), in the center at Hué (Central Hospital), and in the south at Ho Chi Minh City (Institut Pasteur and Pediatric Hospital No. 1). Unfortunately, no precise data on the prevalence of the Nalr serotype Typhi isolates were available from these hospitals. The isolates were identified with the API 20E system (BioMérieux, Marcy l'Étoile, France) and by serotyping with antisera specific for the O:9, Vi, and H:d antigens (Bio-Rad, Hanoi, Vietnam). Escherichia coli ATCC 25922 and a pansusceptible serotype Typhi isolate were used as controls for susceptibility testing. S. enterica serotype Braenderup H9812 was used as a molecular size marker for the PFGE experiment. E. coli Lo, Klebsiella pneumoniae Kp25, and Enterobacter cloacae AME (gifts from L. Poirel, Hôpital de Bicêtre, France) were used as qnrA-, qnrB-, and qnrS-positive controls, respectively.
Antimicrobial susceptibility testing.
The susceptibilities of the 91 Nalr serotype Typhi isolates to 32 antimicrobials were determined by disk diffusion (Bio-Rad, Marnes la Coquette, France) on Mueller-Hinton agar (Bio-Rad), according to the guidelines of the Comité de l'Antibiogramme de la Societé Française de Microbiologie, as described previously (46). The MICs of nalidixic acid (Sigma, St Louis, MO) were determined by standard agar doubling dilution, and the MICs of ciprofloxacin (Cip) were determined by Etest (AB Biodisk, Solna, Sweden).
PCR amplification and DNA sequencing.
Total DNA was extracted with an InstaGene matrix kit (Bio-Rad), in accordance with the manufacturer's recommendations. Oligonucleotide primers (Table 1) were synthesized by MWG-Biotech (Ebersberg, Germany) for amplification of the quinolone resistance-determining chromosomal regions (QRDCRs) of gyrA, gyrB, and parC (6); for amplification of the plasmid-mediated qnrA, qnrB, and qnrS genes (35, 44); and to test for the presence of the locus coding for the Vi antigen (42). All amplifications were done in 50-μl volumes containing DNA (2.5 μl), primers (50 pmol each), deoxynucleoside triphosphates (200 μM), Taq DNA polymerase (1.25 U; Ampli Taq Gold; Roche) and its buffer, MgCl2 (2 mM), and dimethyl sulfoxide (10%). The cycling conditions included 10 min for denaturation at 94°C (1 cycle), followed by 35 cycles of 30 s for denaturation at 94°C; 30 s for annealing at 52°C for qnrA, 53°C for qnrS and parC, 55°C for tviB and qnrB, 60°C for gyrA, 62°C for SPI-7, and 63°C for gyrB; and 1 min for polymerization at 72°C, followed by 10 min for extension at 72°C.
TABLE 1.
Oligonucleotide primers used in the study
| PCR target | Primer name | Oligonucleotide sequence (5′→3′) | Reference or source | PCR product size (bp) |
|---|---|---|---|---|
| gyrA | GyrA-F | CTGAAGCCGGTACACCGTCG | 6 | 290 |
| GyrA-R | TCGGCCATCAGTTCGTGGGC | 6 | ||
| gyrB | GyrBTyphi-Fa | TTATCGATGCTGCGCGTGC | This study | 1,280 |
| GyrB-R | TCGCCGCTTTCAGGGCGTTC | 6 | ||
| parC | ParC-F | CGCCTACTTAAACTACTCCA | 6 | 540 |
| ParC-R | ATCAGCGTAATCGCCGCTTT | 6 | ||
| qnrA | QnrA-F | TCAGCAAGAGGATTTCTCA | 44 | 625 |
| QnrA-R | GGCAGCACTATTACTCCCA | 44 | ||
| qnrB | QnrB-F | GATCGTGAAAGCCAGAAAGG | 35 | 469 |
| QnrB-R | ACGATGCCTGGTAGTTGTCC | 35 | ||
| qnrS | QnrS-F | ACGACATTCGTCAACTGCAA | 35 | 417 |
| QnrS-R | TAAATTGGCACCCTGTAGGC | 35 | ||
| viaB locus | TviB-F | CGAGTGAAACCGTTGGTACA | 42 | 846 |
| TviB-R | CAATGATCGCATCGTAGTGG | 42 | ||
| SPI-7 | DE0032-F | GCTCAGTCGGTAGAGCAGGGGATT | 42 | 1,275b |
| DE0083-R | TCATCTTCAGGACGGCAGGTAGAATG | 42 |
Primer GyrBTyphi-F is 1 bp shorter than the forward primer, described elsewhere (6), which is designed to anneal to gyrB of serotype Typhimurium.
If the isolate is SPI-7 negative.
Both strands of the purified amplicons were sequenced by the Public Health Platform PF8 (Institut Pasteur), and nucleotide sequences were obtained by the use of BigDye (version 3.1) chemistry (Applied Biosystems, Foster City, CA) on an ABI 3700 apparatus (Applied Biosystems).
Lasergene software (DNAstar, Madison, WI) was used to analyze the nucleotidic sequences. The BLASTN program of NCBI (http://www.ncbi.nlm.nih.gov) was used for database searches.
Vi phage typing.
The 91 Nalr serotype Typhi isolates in this study were analyzed by the method described by Craigie and Yen (8, 9). Phage suspensions were kindly provided by the Health Protection Agency, Colindale, United Kingdom.
Ribotyping.
A subset of 71 Nalr S. enterica serotype Typhi isolates, selected on the basis of their diversity (geographic area of isolation, year of isolation, phenotype of resistance, and pulsed-field gel electrophoresis [PFGE] profile), were analyzed by manual PstI ribotyping, as described previously (18). Nine isolates exhibiting all representative ribotypes were analyzed by use of the RiboPrinter microbial characterization system (Qualicon, Wilmington, DE), a fully automated and standardized ribotyping method that creates a database. Ribotype numbering was generated by this system. The ribotype profiles were compared to those in the RiboPrinter database of the Unité de Biodiversité des Bactéries Pathogènes Émergentes, Institut Pasteur, Paris, France (1997 to 2004, which contains 339 PstI ribotypes of serotype Typhi isolates).
PFGE.
PFGE of XbaI (Roche)- and SpeI (Roche)-digested genomic DNA was carried out with all 91 Nalr serotype Typhi isolates in this study, as described previously (45). The running conditions and the molecular size marker (XbaI-digested DNA from S. enterica serotype Braenderup H9812) were the same as those described in the standardized PulseNet protocol (15). BioNumerics software (version 4.0) was used for image normalization and the construction of similarity matrices. Bands were assigned manually. Clustering was carried out by the unweighted pair-group method with arithmetic averages (UPGMA), based on the Dice similarity index. A 0.5% optimization parameter and a 1% band position tolerance were used. Each profile that differed by one or more bands of >100 kb was assigned a type (e.g., type X1). Profiles that differed by a band(s) of less than 100 kb were assigned to subtypes (e.g., subtypes X1a and X1b).
SNP analysis.
A subset of 40 Nalr S. enterica serotype Typhi isolates, selected on the basis of their diversity (geographic area of isolation, year of isolation, PFGE profile, and ribotype), were examined by single-nucleotide polymorphism (SNP) analysis of 55 polymorphic coding gene fragments by denaturing high-performance liquid chromatography at the Max Planck Institute for Infectious Biology, Berlin, Germany, as described previously (36). The haplotypes (combination of the 55 alleles) of these 40 Nalr serotype Typhi isolates were compared to those of 527 isolates in the Max Planck Institute for Infectious Biology database.
Calculation of discrimination indices.
Simpson's index of diversity was used to calculate the discriminatory abilities of the methods used in this study, as described previously (14).
Statistical analysis.
We analyzed our results to assess whether the increase in the rates of resistance to fluoroquinolones from 1996 to 2004 was significant. Statistical testing of the differences in proportions was conducted by the chi-square test with Epi Info (version 6.04) software (CDC, Atlanta, GA); P values <0.05 were considered significant.
RESULTS
Resistance phenotypes: MICs and mechanisms of resistance to quinolones.
Of the 91 Nalr isolates, 69 (76%) were MDR; 66 had the same patterns of resistance to ampicillin, streptomycin, chloramphenicol, tetracycline, and co-trimoxazole; and 3 had other patterns due to the loss of one or several markers (Table 2). Nalr isolates lost more and more of their plasmid-encoded MDR during the period of this study: whereas 97.4% (38/39) of the Nalr isolates in the south were MDR before 2002, only 42% (13/31) were MDR between 2002 and 2004 (Table 2).
TABLE 2.
Phenotypic and molecular characteristics of Nalr S. enterica serotype Typhi isolates in Vietnam from 1996 to 2004a
| Region | Yr | No. of isolates | Antimicrobial resistance pattern | MICs (mg/liter)
|
Phage type | PFGE combined type (XbaI-SpeI) | Ribotype | gyrA mutation(s) | |
|---|---|---|---|---|---|---|---|---|---|
| Nal | Cip | ||||||||
| South | 1996-1998 | 22 | ASCTeSulSxtNal (22) | 64 (1), 128 (4), 256 (15), 1,024 (2) | 0.125 (1), 0.25 (13), 0.5 (8) | UT (13), UVS (6), Evar (2), E1 (1) | X3a-S6a (8), X1a-S1a (4), X3a-S9 (1), X3b-S6b (2), X5c-S4 (1), X20-S27 (1), X21-S19a (1), X23-S22 (1), X11a-S5a (1), X11b-S5b (1), X14-S19b (1) | 3a (17), 341 (1), S007 (1), ND (3) | Phe83 (16), Tyr83 (1), ND (5) |
| 1999-2001 | 17 | ASCTeSulSxtNal (16), Nal (1) | 128 (3), 256 (3), 512 (1), 1,024 (1), >1,024 (9) | 0.25 (7), 0.5 (10) | UVS (6), E1 (5), Evar (4), UT (2) | X3a-S6a (4), X1a-S1a (3), X3b-S6b (3), X5a-S4 (1), X5a-S1b (1), X1a-S2 (1), X1b-S1b (1), X17-S17 (1), X18-S18 (1), X19-S1b (1) | 3a (11), S007 (1), 142 (1), ND (4) | Phe83 (11), ND (6) | |
| 2002-2004 | 31 | Nal (18), ASCTeSulSxtNal (11), ASTeSulSxtNal (1), CTeTmpNal (1), | 256 (11), 512 (5), >1,024 (15) | 0.125 (1), 0.25 (5), 0.5 (25) | UT (20), E1 (7), Evar (2), UVS (2), | X1b-S1b (16), X1a-S1a (3), X8-S12 (4), X5b-S11 (2), X1b-S5b (1), X3a-S6a (1), X3b-S6b (1), X5c-S15 (1), X6-S1a (1), X9-S13 (1) | 3a (14), 73/34/8 (4), 177 (2), 23a (1), ND (10) | Phe83 (22), ND (9) | |
| Center | 1996-1998 | 0 | |||||||
| 1999-2001 | 5 | ASCTeSulSxtNal (4), ANal (1) | 256 (1), >1,024 (4) | 0.5 (5) | UT (3), UVS (2) | X3a-S6a (3), X3b-S6b (2) | 3a (4), ND (1) | Phe83 (4), ND (1) | |
| 2002-2004 | 5 | ASCTeSulSxtNal (3), Nal (2) | 256 (1), >1,024 (4) | 0.25 (1), 0.5 (4) | UVS (3), UT (2) | X3b-S6b (2), X3b-S7 (1), X1a-S1a (1), X26-S14 (1) | 3a (4), ND (1) | Phe83 (4), ND (1) | |
| North | 1996-1998 | 8 | ASCTeSulSxtNal (8) | 1,024 (8) | 0.06 (1), 0.25 (4), 0.5 (3) | E1 (4), Evar (4) | X1a-S1a (3), X1b-S1b (1), X24-S24 (1), X5c-S4 (1), X11b-S16 (1), X16-S20 (1), | 3a (4), S007 (1), 350 (1), 351 (1), ND (1) | Phe83 (4), Tyr83 (2), ND (2) |
| 1999-2001 | 1 | ASCTeSulSxtNal | 1,024 (1) | 0.25 (1) | E1 (1) | X1a-S1a (1) | 3a (1) | Phe83 (1) | |
| 2002-2004 | 2 | ASCTeSulSxtNal (1), Nal (1) | 128 (1), >1,024 (1) | 0.125 (1), 0.5 (1) | E1 (2) | X1a-S1a (1), X13-S1b (1) | 3a (2) | Phe83 (1), Gly87 (1) | |
Numbers in parentheses refer to the number of isolates. Abbreviations: UVS, untypeable Vi isolate; UT, not typeable (Vi negative); var, variant; A, amoxicillin; S, streptomycin; C, chloramphenicol; Te, tetracycline; Sul, sulfonamides; Tmp, trimethoprim; Sxt, sulfamethoxazole-trimethoprim; Nal, nalidixic acid; ND, not determined.
Analysis of the MICs of Nal and Cip (Table 2) shows that susceptibility to Cip decreased in all but one of the Nalr isolates. The MICs of Cip were between 0.06 and 0.5 mg/liter (Table 2). There was a significant increase in the rates of resistance to Cip over the period of the study (P = 0.0047): from 1996 to 1998, 36.4% (8/22) of the isolates in the south had a Cip MIC of 0.5 mg/liter; from 1999 to 2001, 58.8% (10/17) had this level of resistance; and from 2002 to 2004, 80.6% (25/31) had this level of resistance.
DNA sequencing of the QRDCR of gyrA revealed one frequent mutation that led to the replacement of serine with phenylalanine at position 83 of GyrA in 63/67 isolates analyzed (94%). Other point mutations led to the replacement of serine with tyrosine at position 83 in three isolates and the replacement of asparagine with glycine at position 87 in one isolate. We screened a subset of 12 isolates that had various Cip MICs for QRDCRs of gyrB and parC. All of these isolates had the same sequence as the quinolone-susceptible isolates sequenced to date (isolates CT18 and Ty2), indicating that no additional mutation within the sequences of gyrB and parC was amplified by the primers used in this study. We did not detect plasmid-located qnrA, qnrB, or qnrS.
Molecular and phage typing.
The 51 Vi-positive isolates were predominantly (63%) of Vi phage types E1 (n = 20) and E variant (Evar; n = 12). Evar isolates had lysis profiles (lysis with typing phages E3 and E9) between those of E3 (lysis with typing phages E3, E4, and E9) and E9 (lysis with typing phage E9). The 19 other Vi-positive isolates were insensitive to the phages used in this study. Forty isolates were not typeable (Vi negative). All but four isolates had an internal fragment of the tviB gene (42), encoded within the viaB operon, as detected by PCR. This fragment is essential for the production of the Vi antigen. We found that Salmonella pathogenicity island 7 (SPI-7) (31), which includes the viaB operon, was absent from its normal location (between the pheU and the phoN genes) in these four isolates.
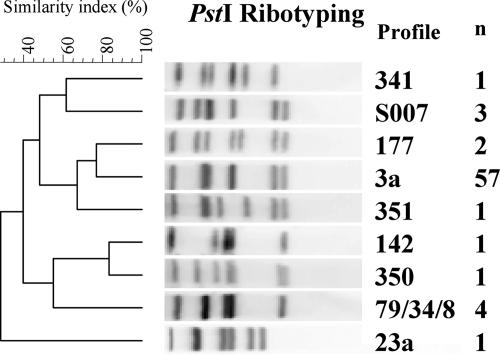
We found nine PstI ribotypes among the 71 isolates tested (Fig. 1). We most frequently observed ribotype 3a (57/71, 80.3%) (Table 2). Simpson's discrimination index for ribotyping was 0.35.
FIG. 1.
Representative PstI ribotypes obtained from a subset of 71 S. enterica serotype Typhi isolates under study. Ribotype numbering is indicated. The dendrograms generated by the BioNumerics software show the results of cluster analysis on the basis of PstI ribotyping. Similarity analysis was performed by using the Dice coefficient, and clustering was done by UPGMA. n, the number of isolates for each ribotype is indicated.
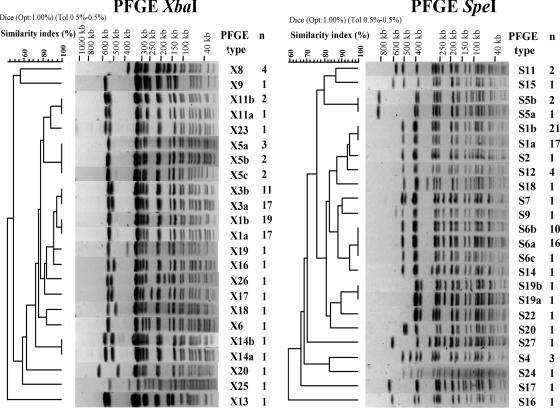
We found 17 distinct profiles by XbaI PFGE (Table 3; Fig. 2). Simpson's discrimination index for XbaI PFGE was 0.739. The most prevalent profiles were X1 (36/91; 39.6%) and X3 (28/91; 30.8%), which differed by a single additional band of approximately 500 kb. We subdivided five profiles (profiles X1, X3, X5, X11, and X19) into two to three subtypes (profile numbers followed by a, b, and c) on the basis of the absence or the presence of bands of less than 100 kb in size. These bands possibly corresponded to a plasmid(s) and, in particular, to the large MDR plasmid: the fully sequenced MDR plasmid pHCM1 (218 kb) of the serotype Typhi strain CT18 (GenBank accession number AL513383) has nine XbaI restriction sites, yielding fragments of from 4 kb to 78.3 kb; also, subtype a correlated with MDR S. enterica serotype Typhi Nalr isolates, and subtype b correlated with Nalr isolates. Both subtype X1a and subtype X3a were displayed by 33 MDR serotype Typhi Nalr isolates and only 1 Nalr isolate. There were discrepancies for both subtype X1b and subtype X3b displayed by 13 MDR serotype Typhi Nalr isolates and 17 Nalr isolates. When they were retested by antibiotic susceptibility tests, the 13 initially MDR serotype Typhi Nalr isolates displaying either subtype X1b or subtype X3b were only Nalr. This has been attributed to the loss of the MDR plasmid during subcultivation. With these new results there was a strict correlation between subtype a and MDR serotype Typhi Nalr isolates and between subtype b and Nalr isolates.
TABLE 3.
Correlation between haplotypes and ribotypes and PFGE profiles
| Haplotype | Combined type (XbaI PFGE type-SpeI PFGE type-ribotype)a |
|---|---|
| H58 | X1-S1-3a (8), X1-S2-3a (1), X1-S5-3a (1), X3-S6-3a (6), X3-S7-3a (1), X3-S9-3a (1), X5-S4-S007 (2), X5-S1-3a (1), X5-S11-177 (1), X6-S1-3a (1), X11-S5-3a (1), X11-S16-350 (1), X13-S1-3a (1), X16-S20-351 (1), X18-S18-3a (1), X19-S1-3a (1), X20-S27-341 (1), X21-S19-3a (1), X25-S24-3a (1), X26-S14-3a (1) |
| H60 | X5-S15-3a (1) |
| H61 | X23-S22-3a (1) |
| H63 | X14-S19-3a (1), X17-S17-142 (1), X1-S1-3a (1), X3-S6-3a (2) |
Numbers in parentheses refer to the number of isolates.
FIG. 2.
Representative XbaI and SpeI PFGE profiles obtained for the 91 S. enterica serotype Typhi isolates under study. The PFGE profile numbering is indicated. The dendrograms generated by the BioNumerics software show the results of cluster analysis on the basis of PFGE fingerprinting. Similarity analysis was performed by using the Dice coefficient, and clustering was done by UPGMA. n, the number of isolates for each PFGE profile is indicated.
There was a shift in the prevalence of profiles X1 and X3 during the study period. X3 represented 50% (11/22) of the isolates in the south from 1996 to 1998, and X1 represented 18.2% (4/22). However, 6.5% (2/31) of the isolates recovered from 2002 to 2004 had the X3 profile and 64.5% (20/31) had the X1 profile. There was also an approximately 150-kb fragment missing in the four Vi-negative isolates (profiles X14a, X14b, X20, and X23) (Fig. 2).
We found 19 distinct profiles by SpeI PFGE (Table 3; Fig. 2). The most prevalent profiles were S1 (38/91; 41.2%) and S6 (27/91; 29.7%), which differed by two bands of 300 and 230 kb. There was also a shift in the prevalence of these two profiles during the study period. We found profile S6 in 45.5% (10/22) of the isolates collected from the south in 1996 to 1998 and profile S1 in 18.2% (4/22). However, 9.7% (3/31) of the isolates recovered from 2002 to 2004 had the S6 profile and 64.5% (20/31) had the S1 profile. Simpson's discrimination index for SpeI PFGE was 0.746. We subdivided four profiles (profiles S1, S5, S6, and S19) into two to three subtypes on the basis of the absence or the presence of bands of less than 100 kb in size that was attributed to a plasmid(s) (plasmid pHCM1 has six SpeI restriction sites, yielding fragments from 0.1 kb to 99.7 kb).
The two PFGE fingerprints were consistent, enabling the discrimination of 25 combined types (Table 2) with a Simpson's discrimination index of 0.780. We observed two major combined types, types X1-S1 and X3-S6, which accounted for 37.4% (34/91) and 28.6% (26/91) of the isolates, respectively. Two isolates with the X1 profile had profiles other than S1. Also, two isolates with the X3 profile had profiles other than S6.
SNP analysis indicated that haplotype H58 was highly frequent in Vietnam (33 of 40 isolates; 82.5%) (Table 3). Derivative haplotypes from H58 were also identified (H60, 2.5%; H61, 2.5%; and H63, 12.5%). These three variants differed from H58 by only one SNP. Simpson's discrimination index was 0.31.
DISCUSSION
We assessed the genetic diversity of MDR S. enterica serotype Typhi Nalr isolates collected throughout the three regions of Vietnam by four subtyping methods and by exploring determinants with various evolution rates.
The SNP method showed that all the strains belonged to haplotype H58 and its closest variants, indicating that the same MDR serotype Typhi Nalr clone had spread throughout Vietnam. Additionally, 94% of these isolates carried the same gyrA mutation, leading to the nonsynonymous substitution Ser83Phe, which was identical to that described in Vietnam in 1997 (41). These results are also consistent with those published by Roumagnac et al. (36), which showed that Nalr H58 has recently spread throughout southern Asia. Although SNP analysis revealed that the Vietnamese isolates belong to this recent Asian strain, it is not suitable for subtyping the detailed populations within this strain.
Vi phage typing, the oldest method for the subtyping of S. enterica serotype Typhi (described in 1938), can discriminate among more than 100 phage types. However, it has limitations: (i) the method is nondiscriminatory if there is a predominant common phage type or a few predominant phage types in an area; (ii) the acquisition or loss of lysogenic phages might change the phage type; (iii) some Vi isolates are not typeable, due to insensitivity to phage suspensions or to loss of Vi antigen; and (iv) this method is often restricted to reference laboratories. E1 and E3 (retrospectively reassigned to Evar) were shown previously (18) to be the most frequent phage types observed among epidemiologically independent MDR serotype Typhi isolates collected throughout Vietnam between 1995 and 2002. As E1 and Evar belong to the same E group, it is very probable that Evar (lysis with typing phages E3 and E9) was derived from E1 (lysis with typing phages E1 to E10) by acquisition of a new lysogenic phage, abolishing various reactions with typing phage suspensions. Our results are consistent with this and are in accord with clonal expansion. However, the percentage of untypeable (Vi-negative) isolates was much higher in the present study than in the previous one: 40/91 (44%) and 18/81 (11%), respectively. Consequently, the phage typing method does not seem to be suitable for the subtyping of serotype Typhi isolates in Vietnam. We have no simple hypothesis to explain the increase and the present high rate of Vi-negative isolates. The absence of Vi agglutination was not due to the loss of the Vi locus in 36/40 isolates. This finding is reassuring, as Vi vaccination started in 1997 and is now carried out in half of the Vietnam provinces (data from the Vietnamese Ministry of Health). The Vi-negative isolates did not have a particular PFGE type (data not shown), and Vi negativity was not considered a phenotypic marker for a particular group of strains. Vi production in serotype Typhi is downregulated by various environmental conditions, including osmolarity. Thus, the most probable hypothesis is that Vi production in our strains was low and not detected by slide agglutination but could have been detected by a more sensitive phenotypic method, e.g., immunofluorescence, as demonstrated by Wain et al. (42). We do not know if the deletion in the four isolates missing SPI-7 occurred in vivo in the peripheral blood of patients or during storage of the bacterial cultures (2, 4).
PstI ribotyping is a stable, reproducible, and sensitive method that has been used since 1989 (1, 12, 18, 20, 27, 28); but it has several limitations: (i) manual ribotyping is technically demanding and automated ribotyping is expensive; (ii) due to the high degree of plasticity of the S. enterica serotype Typhi genome, genetic rearrangements produced by homologous recombinations between rrn operons can occur during the emergence of an outbreak, leading to various ribotypes among outbreak isolates; and (iii) a single genetic rearrangement can substantially modify the banding pattern, precluding an accurate cluster analysis of epidemiologically related isolates (10, 21). PstI ribotyping indicated that most of the Vietnamese isolates belonged to the same clonal expansion, confirming the results obtained with the SNP method. However, ribotyping indicated that 19.7% of the isolates tested had ribotypes other than ribotype 3a. Other methods confirmed that these isolates belonged to the clone. Nevertheless, difficulty estimating the genetic relationship between isolates by ribotyping, even after a single genetic rearrangement within rrn operons, precluded their assignment to the clone. This is a clear limitation of the ribotyping method. The predominant ribotype found in this study (ribotype 3a) is the same as that found in a previous study of molecular typing of MDR Nals serotype Typhi isolates in Vietnam (18). This suggests that the Nalr serotype Typhi isolates are derived from the previous MDR serotype Typhi isolates.
PFGE has successfully been used since 1994 and is currently the method for the subtyping of sporadic or epidemic serotype Typhi isolates (7, 11, 17, 28, 37). Until recently, differences in PFGE running conditions precluded accurate comparisons with the profiles of serotype Typhi isolates typed by other laboratories. Now, the interlaboratory exchange of PFGE profiles at the national and international levels is facilitated by the use of a standardized PFGE protocol, the PulseNet protocol (15). The fingerprints that we obtained for the Nalr Typhi isolates by XbaI and SpeI PFGE pointed out the limited rearrangements that occurred within the clone during the study period. This is possibly due to the influence of more types of DNA rearrangements (insertions, deletions, mutations that modify a restriction site, inversions) on PFGE profiles than on those obtained by the SNP method (mainly punctual mutations within targeted genes). Given that two major combined PFGE types, X1-S1 and X3-S6, accounted for 66% of the Nalr serotype Typhi isolates, PFGE was not a very discriminatory method for the investigation of typhoid fever outbreaks in Vietnam. Simpson's index of 0.78 was far below the 0.90 value considered the lower limit for a discriminatory typing method (14). Therefore, new reproducible methods with a better discriminatory index (DI) must be implemented to reveal the Asian dynamics of the H58 strain and to detect relationships among Vietnamese H58 isolates collected locally over short time intervals. Nair et al. (26) have reported that amplified fragment length polymorphism analysis of serotype Typhi isolates from Papua New Guinea had a DI of 0.88, which is more discriminatory than PFGE (DI, 0.74). That study did not show the antimicrobial resistance status of the strains. Another method, multiple-locus variable-number tandem-repeat analysis, which was successfully used to subtype S. enterica serotype Typhimurium DT104 clone (19), has been developed for serotype Typhi. Two schemes have been published (22, 32). They use three to six polymorphic loci. However, only 20 French and 61 Asian serotype Typhi isolates, without antibiotic resistance data, have been tested. We will soon examine our Nalr isolates by amplified fragment length polymorphism analysis and multiple-locus variable-number tandem-repeat analysis.
This study also showed that since 2002, more and more Nalr isolates belonging to the 3a/H58 clone, with an untypeable Vi phage and with the X1-S1 PFGE profile, were no longer MDR, probably due to the loss of the MDR plasmid. This trend apparently occurred after the replacement of classic antimicrobial treatments by fluoroquinolones. This might lead to the cautious reuse of classical first-line antibiotics such as phenicols, co-trimoxazole, and aminopenicillins.
These results suggest that this serotype Typhi clone has followed a particular course of evolution. First, the strain received an MDR plasmid and profited by clonal expansion under pressure of the classical first-line antibiotics. Then it maintained a chromosomal point mutation under fluoroquinolone pressure and extended clonal expansion. It may be of interest to explore the characteristics of this strain that govern these changes.
The main conclusions of this study are that (i) SNP analysis and, to a lesser extent, ribotyping are more suitable methods for detecting the current Vietnamese Nalr serotype Typhi strain, (ii) PFGE is useful for detecting very recent microevolution within this clone, and (iii) suitable measures in the field of antibiotic administration are needed to prevent increasing levels of resistance to fluoroquinolones, in particular regarding the reuse of classical antibiotics.
Continued monitoring of clonal expansion and antimicrobial resistance among serotype Typhi isolates and communication between physicians and reliable medical biology laboratories will facilitate determination of prevention (vaccination and individual hygiene practices) and treatment policies.
Acknowledgments
We thank Le Quoc Thinh, Pediatric Hospital No. 1, Ho Chi Minh City; Nguyen Thi Nam Lien, Hospital for Infectious Diseases, Hue; Doan Mai Phuong, Hospital for Infectious Diseases, Hanoi; Nguyen Thi Mai Hoa, Saint Paul Hospital, Hanoi; and Ngo Thi Thi, Pediatric Hospital, Hanoi, for providing strains; Nguyen Thi Nga, Tran Van Phuong, and Nguyen Hiep Le Yen, the National Institute for Hygiene and Epidemiology, Hanoi, for technical assistance; and Nguyen Binh Minh and Mark Achtman for their support.
This work was supported by the Délégation Générale au Réseau International des Instituts Pasteur et Instituts Associés and the Université Paris Nord, Bureau des Relations Internationales.
We have no potential conflicts of interest to report.
Footnotes
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Altwegg, M., F. W. Hickman-Brenner, and J. J. Farmer III. 1989. Ribosomal RNA gene restriction patterns provide increased sensitivity for typing Salmonella typhi isolates. J. Infect. Dis. 160:145-149. [DOI] [PubMed] [Google Scholar]
- 2.Baker, S., Y. Sarwar, H. Aziz, A. Haque, A. Ali, G. Dougan, J. Wain, and A. Haque. 2005. Detection of Vi-negative Salmonella enterica serovar Typhi in the peripheral blood of patients with typhoid fever in the Faisalabad region of Pakistan. J. Clin. Microbiol. 43:4418-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, J. D., M. Duong Hong, and E. R. Rhoades. 1975. Chloramphenicol-resistant Salmonella typhi in Saigon. JAMA 231:162-166. [PubMed] [Google Scholar]
- 4.Bueno, S. M., C. A. Santiviago, A. A. Murillo, J. A. Fuentes, A. N. Trombert, P. I. Rodas, P. Youderian, and G. C. Mora. 2004. Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar Typhi. J. Bacteriol. 186:3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butt, T., R. N. Ahmad, A. Mahmood, and S. Zaidi. 2003. Ciprofloxacin treatment failure in typhoid fever case, Pakistan. Emerg. Infect. Dis. 9:1621-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casin, I., J. Breuil, J. P. Darchis, C. Guelpa, and E. Collatz. 2003. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica Typhimurium isolates in humans. Emerg. Infect. Dis. 9:1455-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connerton, P., J. Wain, T. T. Hien, T. Ali, C. Parry, N. T. Chinh, H. Vinh, V. A. Ho, T. S. Diep, N. P. Day, N. J. White, G. Dougan, and J. J. Farrar. 2000. Epidemic typhoid in Vietnam: molecular typing of multiple-antibiotic-resistant Salmonella enterica serotype Typhi from four outbreaks. J. Clin. Microbiol. 38:895-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craigie, J., and C. H. Yen. 1938. The demonstration of types of B. typhosus by means of preparation of type II Vi phage. I. Principles and technique. Can. Public Health J. 29:448-483. [Google Scholar]
- 9.Craigie, J., and C. H. Yen. 1938. The demonstration of types of B. typhosus by means of preparation of type II Vi phage. II. The stability and epidemiological significance of V form types of B. typhosus. Can. Public Health J. 29:484-496. [Google Scholar]
- 10.Echeita, M. A., and M. A. Usera. 1998. Chromosomal rearrangements in Salmonella enterica serotype Typhi affecting molecular typing in outbreak investigations. J. Clin. Microbiol. 36:2123-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampton, M. D., L. R. Ward, B. Rowe, and E. J. Threlfall. 1998. Molecular fingerprinting of multidrug-resistant Salmonella enterica serotype Typhi. Emerg. Infect. Dis. 4:317-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermans, P. W., S. K. Saha, W. J. van Leeuwen, H. A. Verbrugh, A. van Belkum, and W. H. Goessens. 1996. Molecular typing of Salmonella typhi isolates from Dhaka (Bangladesh) and development of DNA probes identifying plasmid-encoded multidrug-resistant isolates. J. Clin. Microbiol. 34:1373-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirose, K., A. Hashimoto, K. Tamura, Y. Kawamura, T. Ezaki, H. Sagara, and H. Watanabe. 2002. DNA sequence analysis of DNA gyrase and DNA topoisomerase IV quinolone resistance-determining regions of Salmonella enterica serovar Typhi and serovar Paratyphi A. Antimicrob. Agents Chemother. 46:3249-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size standard isolate for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly-Hope, L. A., W. J. Alonso, V. D. Thiem, D. D. Anh, D. G. Canh, H. Lee, D. L. Smith, and M. A. Miller. 2007. Geographical distribution and risk factors associated with enteric diseases in Vietnam. Am. J. Trop. Med. Hyg. 76:706-712. [PubMed] [Google Scholar]
- 17.Kubota, K., T. J. Barrett, M. L. Ackers, P. S. Brachman, and E. D. Mintz. 2005. Analysis of Salmonella enterica serotype Typhi pulsed-field gel electrophoresis patterns associated with international travel. J. Clin. Microbiol. 43:1205-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le, T. A. H., M. Lejay-Collin, P. A. D. Grimont, T. L. Hoang, T. V. Nguyen, F. Grimont, and M. R. Scavizzi. 2004. Endemic, epidemic clone of Salmonella enterica serovar Typhi harboring a single multidrug-resistant plasmid in Vietnam between 1995 and 2002. J. Clin. Microbiol. 42:3094-3099. (Erratum, 43:3594, 2005.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindstedt, B. A., E. Heir, E. Gjernes, and G. Kapperud. 2003. DNA fingerprinting of Salmonella enterica subsp. enterica serovar Typhimurium with emphasis on phage type DT104 based on variable number of tandem repeat loci. J. Clin. Microbiol. 41:1469-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling, J. M., N. W. Lo, Y. M. Ho, K. M. Kam, N. T. Hoa, L. T. Phi, and A. F. Cheng. 2000. Molecular methods for the epidemiological typing of Salmonella enterica serotype Typhi from Hong Kong and Vietnam. J. Clin. Microbiol. 38:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, S. L., and K. E. Sanderson. 1996. Highly plastic chromosomal organization in Salmonella typhi. Proc. Natl. Acad. Sci. USA 93:10303-10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y., M. A. Lee, E. E. Ooi, Y. Mavis, A. L. Tan, and H. H. Quek. 2003. Molecular typing of Salmonella enterica serovar Typhi isolates from various countries in Asia by a multiplex PCR assay on variable-number tandem repeats. J. Clin. Microbiol. 41:4388-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madhulika, U., B. N. Harish, and S. C. Parija. 2004. Current pattern in antimicrobial susceptibility of Salmonella Typhi isolates in Pondicherry. Indian J. Med. Res. 120:111-114. [PubMed] [Google Scholar]
- 24.Mermin, J. H., R. Villar, J. Carpenter, L. Roberts, A. Samaridden, L. Gasanova, S. Lomakina, C. Bopp, L. Hutwagner, P. Mead, B. Ross, and E. D. Mintz. 1999. A massive epidemic of multidrug-resistant typhoid fever in Tajikistan associated with consumption of municipal water. J. Infect. Dis. 179:1416-1422. [DOI] [PubMed] [Google Scholar]
- 25.Meyruey, M. H., J. A. Goudineau, P. Berger, H. Pelloux, and J. Queinnec. 1975. Actualité de la fièvre typhoïde au Sud Vietnam. Rev. Epidemiol. Med. Soc. Santé Publique 23:345-358. [PubMed] [Google Scholar]
- 26.Nair, S., E. Schreiber, K. L. Thong, T. Pang, and M. Altwegg. 2000. Genotypic characterization of Salmonella typhi by amplified fragment length polymorphism fingerprinting provides increased discrimination as compared to pulsed-field gel electrophoresis and ribotyping. J. Microbiol. Methods 41:35-43. [DOI] [PubMed] [Google Scholar]
- 27.Nastasi, A., C. Mammina, and M. R. Villafrate. 1991. rDNA fingerprinting as a tool in epidemiological analysis of Salmonella typhi infections. Epidemiol. Infect. 107:565-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro, F., T. Llovet, M. A. Echeita, P. Coll, A. Aladuena, M. A. Usera, and G. Prats. 1996. Molecular typing of Salmonella enterica serovar Typhi. J. Clin. Microbiol. 34:2831-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen, T. A., K. Ha Ba, and T. D. Nguyen. 1993. La fièvre typhoïde au Sud Vietnam, 1990-1993. Bull. Soc. Pathol. Exot. 86:476-478. [PubMed] [Google Scholar]
- 30.Parry, C., J. Wain, N. T. Chinh, H. Vinh, and J. J. Farrar. 1998. Quinolone-resistant Salmonella typhi in Vietnam. Lancet 351:1289. [DOI] [PubMed] [Google Scholar]
- 31.Pickard, D., J. Wain, S. Baker, A. Line, S. Chohan, M. Fookes, A. Barron, P. O. Gaora, J. A. Chabalgoity, N. Thanky, C. Scholes, N. Thomson, M. Quail, J. Parkhill, and G. Dougan. 2003. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 185:5055-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramisse, V., P. Houssu, E. Hernandez, F. Denoeud, V. Hilaire, O. Lisanti, F. Ramisse, J. D. Cavallo, and G. Vergnaud. 2004. Variable number of tandem repeats in Salmonella enterica subsp. enterica for typing purposes. J. Clin. Microbiol. 42:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renuka, K., A. Kapil, S. K. Kabra, N. Wig, B. K. Das, V. V. Prasad, R. Chaudhry, and P. Seth. 2004. Reduced susceptibility to ciprofloxacin and gyrA gene mutation in North Indian isolates of Salmonella enterica serotype Typhi and serotype Paratyphi A. Microb. Drug Resist. 10:146-153. [DOI] [PubMed] [Google Scholar]
- 34.Ricosse, J. H., J. A. Goudineau, J. C. Doury, J. F. Vieu, M. H. Meyruey, and H. Pelloux. 1979. La fièvre typhoïde au Vietnam. Etude bactériologique: 607 souches de salmonelles isolées de 1961 à 1975 à l'hôpital Grall de Saïgon. Med. Trop. 39:415-424. [PubMed] [Google Scholar]
- 35.Robicsek, A., J. Strahilevitz, D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50:2872-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roumagnac, P., F. X. Weill, C. Dolecek, S. Baker, S. Brisse, N. T. Chinh, T. A. H. Le, C. J. Acosta, J. Farrar, G. Dougan, and M. Achtman. 2006. Evolutionary history of Salmonella Typhi. Science 31:1301-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thong, K. L., Y. M. Cheong, S. Puthucheary, C. L. Koh, and T. Pang. 1994. Epidemiologic analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed-field gel electrophoresis. J. Clin. Microbiol. 32:1135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Threlfall, E. J., and L. R. Ward. 2001. Decreased susceptibility to ciprofloxacin in Salmonella enterica serotype Typhi, United Kingdom. Emerg. Infect. Dis. 7:448-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Threlfall, E. J., L. R. Ward, J. A. Skinner, H. R. Smith, and S. Lacey. 1999. Ciprofloxacin-resistant S. typhi and treatment failure. Lancet 353:1590-1591. [DOI] [PubMed] [Google Scholar]
- 40.Van Duong, D., C. W. Binns, and T. Van Le. 1997. Availability of antibiotics as over-the-counter drugs in pharmacies: a threat to public health in Vietnam. Trop. Med. Int. Health 2:1133-1139. [DOI] [PubMed] [Google Scholar]
- 41.Wain, J., N. T. Hoa, N. T. Chinh, H. Vinh, M. J. Everett, T. S. Diep, N. P. Day, T. Solomon, NJ White, L. J. Piddock, and C. M. Parry. 1997. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin. Infect. Dis. 25:1404-1410. [DOI] [PubMed] [Google Scholar]
- 42.Wain, J., D. House, A. Zafar, S. Baker, S. Nair, C. Kidgell, Z. Bhutta, G. Dougan, and R. Hasan. 2005. Vi antigen expression in Salmonella enterica serovar Typhi clinical isolates from Pakistan. J. Clin. Microbiol. 43:1158-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wain, J., and C. Kidgell. 2004. The emergence of multidrug resistance to antimicrobial agents for the treatment of typhoid fever. Trans. R. Soc. Trop. Med. Hyg. 98:423-430. [DOI] [PubMed] [Google Scholar]
- 44.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weill, F. X., M. Demartin, D. Tandé, E. Espié, I. Rakotoarivony, and P. A. D. Grimont. 2004. Extended-spectrum-β-lactamase (SHV-12 like)-producing strains of Salmonella enterica serotypes Babelsberg and Enteritidis isolated in France among infants adopted from Mali. J. Clin. Microbiol. 42:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weill, F. X., F. Guesnier, V. Guibert, M. Timinouni, M. Demartin, L. Polomack, and P. A. D. Grimont. 2006. Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003). J. Clin. Microbiol. 44:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]