Abstract
Accumulating evidence indicates that herpesviruses may be putative pathogens in various types of periodontal diseases. The present study was performed to examine infections with different genotypes of human cytomegalovirus (HCMV) and Epstein-Barr virus (EBV) in subgingival samples from a Chinese population and to analyze the correlation with periodontal status. A nested PCR assay was used to identify the presence of HCMV, EBV type 1 (EBV-1), and EBV-2; and the amplicons were further analyzed by restriction fragment length polymorphism analysis. HCMV was detected in 79.0% of 143 chronic periodontitis (CP) patients, 78.5% of 65 gingivitis patients, and 76.3% of 76 periodontally healthy individuals, while EBV was found in 63.6%, 32.3%, and 30.3% of the three groups of subjects, respectively. The HCMV-positive PCR products from all the samples were identified as corresponding to gB genotype I (gB-I) or gB-II. HCMV gB-II (62.9%), EBV-1 (43.4%), and EBV-2 (18.2%) were associated with CP at higher frequencies (P < 0.05), whereas HCMV gB-I was more often observed in gingivitis patients (40.0%) and healthy individuals (40.8%) (P < 0.05). Furthermore, a higher rate of coinfection with HCMV and EBV was shown in CP patients (52.4%), especially dual infections with HCMV gB-II and EBV-1 (30.8%) or HCMV gB-II and EBV-2 (12.6%), compared with the rates of single infections with HCMV or EBV (P < 0.05). Infection with HCMV gB-II, EBV-1, or EBV-2 was correlated with higher rates of bleeding on probing (P < 0.05). In patients infected with HCMV gB-II or both HCMV and EBV, including HCMV gB-II and EBV-1, a deeper probing depth or more serious attachment loss was found (P < 0.05). These findings clearly indicate that HCMV gB-II is the dominant genotype detected in subgingival samples in CP. HCMV gB-II infection and HCMV gB-II coinfection with EBV-1 are closely associated with periodontal tissue inflammation and destruction.
Periodontal diseases are multifactorial infections elicited by a complex community of bacterial species that interact with host tissues and cells, causing the release of a broad array of inflammatory cytokines, chemokines, and mediators, some of which lead to the destruction of the periodontal structures, including the alveolar bone, periodontal ligament, and cementum around a tooth (34). Even though specific infectious agents are of key importance in the development of periodontitis, it is unlikely that a single agent or even a small group of pathogens are the sole cause or modulator of this heterogeneous disease.
Since the mid-1990s, herpesviruses have emerged as putative pathogens in various types of periodontal diseases (13, 15, 18). In particular, human cytomegalovirus (HCMV) and Epstein-Barr virus (EBV) seem to play important roles in the etiopathogenesis of severe types of periodontitis. The genomes of the two herpesviruses are present at a high frequency in adults with progressive periodontitis and patients with localized and generalized aggressive (juvenile) periodontitis, human immunodeficiency virus (HIV)-associated periodontitis, acute necrotizing ulcerative gingivitis, periodontal abscesses, and some rare types of advanced periodontitis associated with medical disorders (27, 31). HCMV infects periodontal monocytes/macrophages and T lymphocytes, and EBV infects periodontal B lymphocytes (11). Herpesvirus-infected inflammatory cells elicit tissue-destroying cytokines and may exert a diminished ability to defend against bacterial challenge (29). Herpesvirus-associated periodontal sites tend to harbor elevated levels of periodontopathic bacteria, including Porphyromonas gingivalis, Tannerella forsythia, Dialister pneumosintes/Dialister invisus, Prevotella intermedia, Prevotella nigrescens, Treponema denticola, Campylobacter rectus, and Actinobacillus actinomycetemcomitans (28, 30, 32). A novel infectious disease model for human periodontitis was described in which periodontal herpesvirus activation results in the suppression of periodontal immune defenses, the overgrowth of periodontal bacterial pathogens, the release of proinflammatory cytokines and chemokines, the initiation of cytotoxic or immunopathological events, and subsequently, periodontal tissue breakdown (29).
Previous studies have demonstrated that different genotypes of HCMV and EBV might exhibit different pathogenic abilities in other nonoral diseases (6, 17, 22); and the prevalence of these genotypes varies by geography and ethnicity groups. According to the sequence of the gB gene, which encodes a glycoprotein in the outer membrane of HCMV, the virus could arbitrarily be divided into four genotypes (gB genotype I [gB-I] to gB-IV) (3). HCMV gB-I does not infect T lymphocytes, whereas types II and III do (22). Infection with gB-II correlates with retinitis in AIDS patients, but infection with type I does not (5). In Japanese bone marrow transplant recipients, the predominant infecting gB genotypes were I, II, and III; but no cases of type IV infection were found (38). HCMV gB-I was common in an Italian population (4), while gB-II was more prevalent in India (21).
EBV, a double-stranded DNA virus, is one of the eight human herpesviruses (human herpesvirus 4). It is transmitted by salivary contact and establishes a lifelong latent infection that is usually asymptomatic (26). Two types of EBV exist, based on the allelic polymorphisms in the latent gene sequences encoding EBV nuclear antigen 2 (EBNA2) (24). It is frequently detected in blood samples from healthy individuals, and most EBV serum-positive healthy Caucasians are infected with EBV type 1 (EBV-1), while immunosuppressed individuals (HIV-infected and transplant patients) have a high rate of infection with EBV-2 (8, 14). An in vitro study showed that EBV-2 causes more lysis of B lymphocytes than EBV-1 (6). Therefore, it is critical to identify the predominant genotypes of HCMV and EBV present in subgingival samples to further elucidate their roles in the etiology of periodontitis.
The purpose of this study was to investigate the frequency of subgingival infection with different genotypes of HCMV and EBV in Chinese patients with periodontal disease. A nested PCR method was established to detect the gB gene of HCMV and the EBNA2 gene of EBV in subgingival plaque samples from 143 chronic periodontitis (CP) patients and in sulcular samples from 65 gingivitis patients and 76 individuals with healthy periodontia. The amplicons were further characterized by restriction endonuclease digestion, and the relationship between infection with different genotypes of HCMV and EBV in subgingival samples and clinical parameters was analyzed.
MATERIALS AND METHODS
Subjects.
The subjects in this study were 143 Chinese patients with CP (64 males aged 20 to 65 years [mean age, 41.4 ± 7.9 years] and 79 females aged 21 to 68 years [mean age, 42.5 ± 6.9 years]), 65 gingivitis patients (30 males aged 17 to 58 years [mean age, 38.2 ± 7.9 years] and 35 females aged 18 to 60 years [mean age, 37.1 ± 5.4 years]), and 76 individuals with healthy periodontia (35 males aged 22 to 64 years [mean age, 38.8 ± 7.1 years] and 41 females aged 21 to 63 years [mean age, 37.5 ± 6.9 years]) who were referred to the dental clinic in the Second Affiliated Hospital of the School of Medicine of Zhejiang University for dental or periodontal treatment or health monitoring. All the subjects were nonsmokers without any systemic disease and with at least 14 teeth remaining. Those who had received a professional cleaning or who had a history of antibiotic therapy during the preceding 3 months were excluded. All of the patients and the healthy individuals underwent a full-mouth examination. The criteria for the diagnosis of CP and gingivitis were based on the Classification of the Periodontal Diseases issued by the American Academy of Periodontology in 1999 (2). Briefly, the 143 generalized CP patients had >30% of sites showing periodontal probing depths ≥3 mm, clinical attachment loss of >1 mm, and radiographic evidence of alveolar bone loss. The 65 plaque-induced gingivitis patients manifested inflammation of the gingiva, with periodontal probing depths of no more than 3 mm and without any clinical attachment loss or bone loss on X-ray examination. Individuals with periodontal probing depths of less than 3 mm without any clinical attachment loss or radiographic evidence of bone loss and without inflammation of the gingivae were considered periodontally healthy. All subjects and patients were enrolled with informed consent, according to a protocol approved by the Ethics Committee of Zhejiang University.
Sample collection.
Each CP patient contributed four subgingival samples from the four deepest periodontal pockets of the dentition, preferably one pocket from each quadrant. Each of the gingivitis patients and periodontally healthy individuals provided four sulcular samples from the mesial or distal buccal site of a tooth in each quadrant, and usually these four teeth included at least the two first molars. Prior to subgingival sampling, the supragingival plaque was removed with sterile cotton rolls. For all the subjects, three sterile endodontic paper points were placed into the depth of each site for 30 s and were then transferred to a microcentrifuge tube containing 200 μl TE buffer (10 mM Tris-HCl, 1.0 mM EDTA, pH 8.0) and stored at −70°C. The full-mouth clinical parameters recorded included bleeding on probing (BOP), probing pocket depth (PD), and attachment loss (AL) at six sites per tooth.
Viral nucleic acid extraction.
The specimens resuspended in 200 μl TE buffer were vigorously mixed in a vortex mixer. The nucleic acid extraction technique was based on preferential binding to silica particles in the presence of a high concentration of guanidium thiocyanate (GuSCN) (25). Briefly, 200 μl of sample was mixed with 50 μl of silica particles (Sigma Chemical Co., St. Louis, MO) in 800 μl of lysis buffer (120 g of GuSCN; 100 ml of 0.1 mM Tris-HCl, pH 6.4; 22 ml of 0.2 M EDTA, pH 8.0; 2.6 g of Triton X-100), mixed in a vortex mixer for 10 s, and kept at room temperature for 10 min. The nucleic acid-silica complexes were recovered by centrifugation at 12,000 × g for 1 min and washed twice in buffer (GuSCN-Tris-HCl), twice in 70% ethanol, and once in acetone. The sample was then dried at 37°C. The nucleic acid pellet was resuspended in 100 μl of TE buffer containing 0.5 U/ml RNasin (Promega), and the DNA/RNA was separated from the silica particles by incubation at 56°C for 10 min. After centrifugation at 12,000 × g for 2 min, the supernatant was stored at −70°C.
PCR procedures.
A nested PCR method was used to detect the DNA of HCMV, EBV-1, and EBV-2. The primers were demonstrated to have high specificities and no cross-reactivity with the human genome, other viruses, or various microorganisms by other researchers (7, 13, 23, 25, 38). The sequences of the HCMV-specific outer primers were 5′GGC ATC AAG CAA AAA TCT-3′ (forward primer) and 5′-CAG TTG ACG GTA CTG CAC-3′ (reverse primer). The inner primers for HCMV were 5′-TGG AAC TGG AAC GTT TGG G-3′ (forward primer) and 5′-GAA ACG CGC GGC AAT CGG-3′ (reverse primer). The sequences of the EBV-specific outer primers were 5′-AGG GATG CCT GGA CAC AAG A-3′ (forward primer) and 5′-TGG TGC TGC TGG TGG TGG CAA-3′ (reverse primer). The inner primers for EBV-1 were 5′-TCT TGA TAG GGA TCC GCT AGG ATA-3′ (forward primer) and 5′-ACC GTG GTT CTG GAC TAT CTG GAT C-3′ (reverse primer). The inner primers for EBV-2 were 5′-CAT GGT AGC CTT AGG ACA TA-3′ (forward primer) and 5′-AGA CTT AGT TGA TGC TGC CCT AG-3′ (reverse primer).
The first round of the PCR was carried out in a mixture with a total volume of 50 μl that included 5 μl of the template, 1× PCR buffer (pH 8.3) and 2.5 U Ex-Taq DNA polymerase (TaKaRa), 0.2 mM each deoxynucleoside triphosphate, 1.5 mM MgCl2, and 1 μM outer primers. PCR amplification, which was performed in a Perkin-Elmer 2400 DNA cycler, included an initial denaturation step at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, primer annealing at 60°C for 30 s, and extension at 72°C for 1 min, with a final extension step at 72°C for 5 min. A second round of amplification was performed in a new tube with 2 μl of the first-round PCR product; 25 pmol of inner primers; and PCR buffer, Ex-Taq DNA polymerase, deoxynucleoside triphosphates, and MgCl2 at the concentrations described above. The program for the second PCR was 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1.5 min, with a final extension step at 72°C for 7 min.
Positive controls included purified DNA from HCMV (Sigma) and Raji cells containing EBV-1 (American Type Culture Collection, Manassas, VA). For tests for sensitivity, 20 ng/μl of the HCMV or EBV-1 DNA was serially diluted 10-fold, and 5 μl of each dilution containing 10−6 ng to 100 ng DNA was used as the template in the PCR assay. Amplicons were detected by electrophoresis of 10 μl of sample in a 1.5% agarose gel containing 1 μg/ml of ethidium bromide. The expected sizes of the target fragments from HCMV, EBV-1 and EBV-2 were ∼299 to 305 bp, 497 bp, and 165 bp, respectively. The PCR assay was repeated once for each clinical sample.
Restriction endonuclease digestion analysis.
The amplicons were further identified by restriction fragment length polymorphism analysis (RFLP). The HCMV gB gene fragments were digested with endonucleases RsaI and Hinf I (TaKaRa). After RsaI digestion, the HCMV products were cleaved into two fragments (gB-I, 239 and 66 bp; gB-II, 239 and 63 bp) or three fragments (gB-III, 195, 63, and 41 bp; gB-IV 195, 66, and 44 bp). The HCMV products were separated into another two fragments (gB-II, 202 and 100 bp; gB-III, 202 and 97 bp) or three fragments (gB-I and gB-IV, 202, 67, and 36 bp) with endonuclease HinfI. For further identification of EBV, AfaI was used to digest the 497-bp amplicon of EBV-1 into 355- and 142-bp fragments, while StuI was used to turn the 165-bp amplicon of EBV-2 into 118- and 47-bp fragments. The restriction enzyme digests were resolved in an 2.5% agarose gel containing 1 μg/ml of ethidium bromide.
Statistical analysis.
The chi-square test and Fisher's exact test were used to compare the rates of HCMV and EBV positivity in patients and periodontally healthy individuals. Odds ratios (OR) and 95% confidence intervals were calculated to determine whether there was a significant association between the occurrence of HCMV and EBV with periodontal status. An association between the frequencies of infection with HCMV or EBV and clinical parameters was examined by the Kruskal-Wallis test with Stata 8.0 software. A P value equal to or less than 0.05 was considered statistically significant.
RESULTS
Sensitivity and specificity of nested PCR assay.
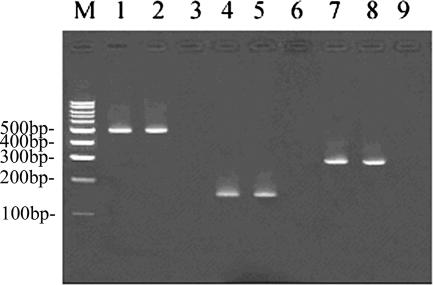
By using 10−6 ng to 100 ng HCMV or EBV-1 DNA as the template in the PCR assay, the nested PCR could detect as little as 0.01 ng of HCMV or EBV-1 DNA (Fig. 1). Repeated PCR results for EBV-1 and EBV-2 were identical for all the clinical samples. All the EBV-1 products could be digested into 355- and 142-bp fragments by AfaI, and StuI could digest all the EBV-2 amplicons into 118- and 47-bp fragments. For 95% of the samples, the results of the two PCR experiments for HCMV were consistent. For all the samples, RFLP analysis indicated that RsaI could cleave the target HCMV amplicons into two 239- and 66-bp fragments by comparison with the 100-bp marker. After HinfI digestion, gB-I was cleaved into three fragments, while gB-II gave two fragments of the expected sizes (Fig. 2). All the HCMV gB gene-positive samples were type I or II. No HCMV gB type III or IV products were found.
FIG. 1.
Results of nested PCR of amplification of the HCMV gB and EBV EBNA2 genes. Lane M, 100-bp DNA marker (Promega); lane 1, HCMV positive control; lane 2, HCMV-positive subgingival sample from a patient with CP; lanes 3, 6, and 9, blank controls; lane 4, EBV-1 positive control; lane 5, EBV-1-positive subgingival sample from a patient with CP; lane 7, EBV-2 positive control; lane 8, EBV-2-positive subgingival sample from a patient with CP.
FIG. 2.
PCR products of EBV-1, EBV-2, and the HCMV gB gene analyzed by RFLP analysis. Lane M, 100-bp DNA marker (Promega); lane 1, 497-bp amplification fragment of EBV-1 from a clinical sample; lane 2, 355-bp and 142-bp fragments of the EBV-1 PCR product digested with AfaI; lane 3, 165-bp amplification fragment of EBV-2 from a clinical sample; lane 4, 118-bp and 47-bp fragments of the EBV-1 PCR product digested with StuI; lane 5, 299- to 305-bp amplification fragment of the HCMV gB gene from a clinical sample; lane 6, 202-, 67-, and 36-bp fragments of the HCMV gB-I PCR product digested with HinfI; lane 7, 202- and 100-bp fragments of the HCMV gB-II PCR product digested with HinfI; lane 8, 239- and 66-bp fragments of the HCMV gB-I PCR product digested with RsaI; lane 9, 239- and 63-bp fragments of the HCMV gB-II PCR product digested with RsaI.
Frequency of infection with HCMV and EBV in subgingival plaque samples from patients and healthy individuals.
HCMV was detected in 79.0% of the CP patients, 78.5% of the gingivitis patients, and 76.3% of the periodontally healthy individuals, while EBV was detected in 63.6%, 32.3%, and 30.3% of the three groups of subjects, respectively (Table 1). The distributions of HCMV in these three groups were similar (χ2 = 0.01 to 0.21; P = 0.091 to 0.922). A majority of HCMV gB-II isolates (62.9%) were detected in CP patients, while the frequency of HCMV gB-I was much lower in CP patients than in gingivitis or periodontally healthy individuals (χ2 = 15.22 to 27.46; P = 0.000). EBV or EBV-1 was more frequently detected in CP patients than in gingivitis or periodontally healthy individuals (χ2 = 5.21 to 22.15; P = 0.000 to 0.022). The rate of EBV-2 detection was also higher in CP patients than in healthy individuals (P = 0.001), but the difference was not statistically significant for the frequency of EBV or of EBV-1 or EBV-2 between gingivitis patients and healthy individuals (P = 0.794, 0.685, 0.248) or for the frequency of EBV-2 between CP and gingivitis patients (P = 0.059).
TABLE 1.
Distribution of the HCMV gB and EBV EBNA2 genotypes in subgingival samples
| Group | Total | No. of cases
|
||||||
|---|---|---|---|---|---|---|---|---|
| HCMV/EBV positive | gB genotype
|
EBNA2 genotype
|
||||||
| Type I | Type II | Type I and II mix | EBV-1 | EBV-2 | EBV-1 and EBV-2 mix | |||
| CP | ||||||||
| Patients | 143 | 113/91a | 15a | 90a | 8 | 62a | 26a | 3 |
| Samples | 520 | 330a/196a | 47a | 283a | 0 | 145b | 51b | 0 |
| Gingivitis | ||||||||
| Patients | 65 | 51/21 | 26 | 22 | 3 | 16 | 5 | 0 |
| Samples | 260 | 128/52 | 69 | 59 | 0 | 44 | 8 | 0 |
| Healthy | ||||||||
| Individuals | 76 | 58/23 | 31 | 24 | 3 | 21 | 2 | 0 |
| Samples | 304 | 129/65 | 71 | 58 | 0 | 63 | 2 | 0 |
P < 0.01 compared with the results for gingivitis or periodontally healthy samples or patients.
P < 0.05 compared with the results for healthy periodontal samples.
As seen in Table 2, the rate of dual infection with HCMV and EBV was higher in CP patients than in gingivitis patients and individuals with healthy periodontia (P = 0.000), while in gingivitis patients and healthy individuals, infection with either HCMV or EBV predominated (P = 0.000). Among the CP patients, infection with HCMV gB-II and EBV-1 was most prevalent, followed by infection with HCMV gB-II and EBV-2 (P = 0.000 to 0.002).
TABLE 2.
Coinfection with different genotypes of HCMV and EBV in subjects
| Virus | No. (%) of patients
|
||
|---|---|---|---|
| CP | Gingivitis | Healthy | |
| HCMV and EBV | 75 (52.4)a | 14 (21.5) | 9 (11.8) |
| HCMV gB-II and EBV-1 | 44 (30.8)a | 3 (4.6) | 2 (2.2) |
| HCMV gB-II and EBV-2 | 18 (12.6)a | 2 (3.1) | 0 |
| HCMV gB-I and EBV-1 | 9 (6.3) | 7 (10.8) | 4 (5.3) |
| HCMV gB-I and EBV-2 | 2 (1.4) | 1 (1.5) | 0 |
| HCMV gB-I, HCMV gB-II, and EBV-1 | 2 (7.0) | 1 (1.5) | 3 (3.9) |
| HCMV or EBV alone | 48 (33.6) | 44 (67.7) | 63 (82.9) |
P < 0.01 compared with the results for gingivitis or periodontally healthy individuals.
A significant association between infection with specific genotypes of HCMV and EBV and the periodontal status was demonstrated (Table 3). HCMV gB-II was associated with CP (OR = 3.679), while HCMV gB-I was closely related to gingivitis (OR = 5.688) and a healthy periodontium (OR = 5.879). EBV-1 and EBV-2 were both found to be associated with CP (ORs = 2.000 and 8.222, respectively). In contrast to a single infection with HCMV or EBV, a higher rate of coinfection with HCMV and EBV was correlated with CP (OR = 10.938), especially dual infection with HCMV gB-II and EBV-1 (OR = 28.875) or HCMV gB-II and EBV-2 (OR = 8.250).
TABLE 3.
Relationship between presence of HCMV and EBV and periodontal status
| Virus | Periodontal status | OR | 95% confidence interval |
|---|---|---|---|
| HCMV gB-II | CPa | 3.679 | 1.96-6.96 |
| HCMV gB-I | Gingivitisb | 5.688 | 2.59-12.67 |
| HCMV gB-I | Healthyb | 5.879 | 2.76-12.76 |
| EBV-1 | CPa | 2.000 | 1.06-3.86 |
| EBV-2 | CPa | 8.222 | 1.95-73.04 |
| HCMV and EBV | CPa | 10.938 | 4.78-27.05 |
| HCMV gB-II and EBV-1 | CPa | 28.875 | 6.82-253.27 |
| HCMV gB-II and EBV-2 | CPc | 8.250 | 1.78-76.22 |
Compared with individuals with healthy periodontia.
Compared with CP patients.
Compared with gingivitis patients.
Association between infection with different genotypes of HCMV and EBV and clinical parameters.
Table 4 describes the correlation between infection or coinfection with different genotypes of HCMV and EBV in CP patients with the mean values for BOP, PD, and AL in the mouth. Compared with CP patients who were negative for both HCMV and EBV, a higher percentage of BOP was observed in patients infected with HCMV gB-II, EBV-1, or EBV-2 or patients coinfected with HCMV gB-II and EBV-1, HCMV gB-II and EBV-2, or HCMV gB-I and EBV-1 (P < 0.05). Moreover, in patients infected with HCMV gB-II or both HCMV and EBV (especially with HCMV gB-II and EBV-1), a deeper probing depth or more serious attachment loss was found compared with the probing depths and attachment loss for patients who were not infected with either of the two viruses (P < 0.05).
TABLE 4.
Correlation between infection or coinfection with different genotypes of HCMV and EBV in CP patients and clinical parameters
| Genotype (no. of patients) | % of patients showing BOP (mean ± SE) | PD (mm [mean ± SE]) | AL (mm [mean ± SE]) |
|---|---|---|---|
| HCMV gB-I (15) | 48.64 ± 9.67 | 3.85 ± 0.55 | 4.22 ± 0.51 |
| HCMV gB-II (80) | 56.78 ± 11.23a | 3.91 ± 0.63b | 4.30 ± 0.65b |
| EBV-1 (62) | 58.34 ± 10.59a | 3.87 ± 0.71 | 4.24 ± 0.70 |
| EBV-2 (26) | 51.29 ± 10.27b | 3.84 ± 0.55 | 4.24 ± 0.62 |
| HCMV and EBV (75) | 54.26 ± 12.25a | 3.89 ± 0.56b | 4.28 ± 0.61b |
| HCMV gB-II and EBV-1 (44) | 58.24 ± 11.17a | 3.91 ± 0.52b | 4.31 ± 0.54b |
| HCMV gB-II and EBV-2 (18) | 53.49 ± 8.49a | 3.86 ± 0.54 | 4.25 ± 0.58 |
| HCMV gB-I and EBV-1 (9) | 51.39 ± 6.79b | 3.85 ± 0.61 | 4.23 ± 0.57 |
| HCMV gB-I and EBV-2 (2) | 53.45 | 3.83 | 4.19 |
| HCMV gB-I, HCMV gB-II, and EBV-1 (2) | 48.75 | 3.88 | 4.27 |
| HCMV and EBV negative (20) | 43.49 ± 8.93 | 3.59 ± 0.41 | 3.96 ± 0.49 |
P < 0.01 compared with the results for patients not infected with either HCMV or EBV.
P < 0.05 compared with the results for patients not infected with either HCMV or EBV.
DISCUSSION
In this study, a nested PCR-RFLP method was designed to detect gB-I to gB-IV of HCMV and EBV-1 and EBV-2 in subgingival samples with a detection limit of 0.01 ng of HCMV and EBV DNA template. The method provided a sensitivity sufficient to allow meaningful data to be obtained from a small volume of specimen removed from periodontal sites. Restriction endonuclease digestion analysis also confirmed the specificity of the method. Moreover, repeating the PCR analysis also showed that the method had a high degree of reproducibility.
Herpesviruses represent some of the most successful viruses in humans, infecting over 90% of humans and persisting for the lifetime of the individuals (8, 26). Of particular interest in the present study with a homogeneous demographic population, we found that the HCMV gB genotypes are distributed differently among subgingival samples from patients with different periodontal status. Four types of HCMV gB genes are known (3, 4), but only two types were detected in subgingival samples from the Chinese individuals. To determine if there are other types of HCMV gB genes in subigingival plaque samples, we expanded our research by recruiting more subjects. A higher frequency of HCMV gB-II was shown in CP patients, whereas HCMV gB-I was more often detected in gingivitis patients or periodontally healthy individuals. Statistical analysis revealed a significant relationship between HCMV gB-II infection and CP. In contrast, HCMV gB-I was found in patients with gingivitis and individuals with healthy periodontia. This study thus represents the first description of the dominant genotypes of HCMV in subgingival samples from Chinese CP patients.
The rate of positivity for EBV-1 was higher in CP patients than gingivitis patients and periodontally healthy individuals, suggesting that EBV-1 infection is associated with the pathogenesis of CP (10, 19). In this study, we demonstrated that infection with either EBV-1 and EBV-2 correlates with CP. The relationship between EBV-2 infection and periodontitis still remains to be established. EBV-2 could be detected in gingival biopsy specimens from HIV-positive periodontitis patients but was not detectable in HIV-negative periodontitis patients (14). Other reports indicate that EBV-2 might be present in subgingival samples from HIV-negative periodontitis patients but be absent from gingivitis patients and periodontally healthy individuals (10, 12). EBV-2 infection correlates with immunosuppression, and EBV-1 and EBV-2 differ in their pathogenicities (6, 14, 17). Taken together, these observations imply that EBV-2 might play a role in the pathogenesis and development of CP, especially in patients with local periodontal immune suppression. Further studies should focus on the immune status of local periodontal tissue to define the correlation between EBV-2 and CP. The results from this study provide strong evidence that specific genotypes of HCMV and EBV infection are associated with the etiology of CP in Chinese populations.
HCMV reactivation could transactivate EBV and other herpesviruses (1, 37). That HCMV-EBV dual infection may give rise to particularly severe pathology has been suggested on the basis of observations for patients with nonoral disease (39), oral ulcers (36), and periodontitis (31). In the present study, coinfection with HCMV and EBV was more frequently detected in CP patients, while the rate of occurrence of a single infection with either HCMV or EBV in patients with gingivitis or periodontally healthy individuals was also high. Furthermore, dual infection with the HCMV gB-II and EBV-1 genotypes was prevalent in CP patients, whereas the most regularly revealed genotypes of coinfection in patients with gingivitis and healthy individuals were HCMV gB-I and EBV-1 (Table 2). These results demonstrate that pathogenesis might be higher during coinfection with HCMV and EBV than infection with only HCMV or EBV and that coinfection with different genotypes of HCMV and EBV might correlate with the different periodontal status of the patients.
It has been proposed that periodontal herpesvirus activation could destroy the normal periodontal defense barrier and impair the local immune system, which would promote subgingival colonization and the proliferation of periodontal bacterial pathogens, finally resulting in periodontal tissue destruction (16, 19, 33, 35). Analysis of the association between infection with different genotypes of HCMV and EBV and clinical parameters indicated that infection with HCMV gB-II, EBV-1, or EBV-2 or coinfection with HCMV gB-II and EBV-1, HCMV gB-II and EBV-2, or HCMV gB-I and EBV-1 correlated with BOP, while HCMV gB-II infection or coinfection with HCMV gB-II and EBV-1 was associated with a deeper probing depth or more serious attachment loss. The results further demonstrated that HCMV gB-II was the disease-contributing genotype in CP patients and that exposure to multiple genotypes of HCMV and EBV acts in concert in the pathogenesis of periodontitis. Previous studies have demonstrated that HCMV and EBV are frequently detected in deep pockets (13, 20, 29), and HCMV infection is predictive of the presence of P. gingivalis (9, 29). EBV-1 may thus promote the development of periodontitis, probably through mechanisms involving periodontopathic species other than P. gingivalis (29). It would be useful to know why single infection with EBV-1 was not related to the risk of periodontal tissue destruction, but statistical significance was shown after the presence of HCMV gB-II was also considered.
In summary, the present study demonstrates that different HCMV genotypes are associated with disease, while others appear to be nonpathogenic. Coinfection with the HCMV gB-II genotype and EBV-1 was dominant in CP patients and was associated with periodontal tissue destruction and inflammation. Although a longitudinal study is still needed, infection with these genotypes of HCMV and EBV are likely to contribute to the onset and development of periodontitis. Future studies on the pathogenic mechanisms of these strains of HCMV and EBV should help to elucidate their role in the etiology of periodontitis.
Acknowledgments
This work was supported by a grant from the Science and Technology Department of Zhejiang Province, China (grant no. N021107286).
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Arcenas, R., and R. H. Widen. 2002. Epstein-Barr virus reactivation after superinfection of the BJAB-B1 and P3HR-1 cell lines with cytomegalovirus. BMC Microbiol. 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage, G. C. 1999. Development of a classification system for periodontal disease and conditions. Ann. Periodontol. 4:1-6. [DOI] [PubMed] [Google Scholar]
- 3.Bale, J. F., L. Miner, and S. J. Petheram. 2002. Congenital cytomagalovirus infection. Curr. Treat. Options Neurol. 4:225-230. [DOI] [PubMed] [Google Scholar]
- 4.Barbi, M., S. Binda, S. Caroppo, V. Primache, P. Dido, P. Guidotti, C. Corbetta, and D. Elotti. 2001. CMV gB genotypes and outcome of vertical transmission: study on dried blood spots of congenitally infected babies. J. Clin. Virol. 21:75-79. [DOI] [PubMed] [Google Scholar]
- 5.Bongarts, A., D. von Laer, C. Vogelberg, K. Ebert, J. van Lunzen, J. Garweg, P. Vaith, F. T. Hufert, O. Haller, and U. Meyer-Konig. 1996. Glycoprotein B genotype of human cytomegalovirus: distribution in HIV-infected patients. Scand. J. Infect. Dis. 28:447-449. [DOI] [PubMed] [Google Scholar]
- 6.Buck, M., S. Cross, K. Krauer, N. Kienzle, and T. B. Sculley. 1999. A-type and B-type Epstein-Barr virus differ in their ability to spontaneously enter the lytic cycle. J. Gen. Virol. 80(Pt 2):441-445. [DOI] [PubMed] [Google Scholar]
- 7.Chou, S., and K. M. Dennison. 1991. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J. Infect. Dis. 163:1229-1234. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, J. I. 2000. Epstein-Barr virus infection. N. Engl. J. Med. 343:481-492. [DOI] [PubMed] [Google Scholar]
- 9.Contreras, A., and J. Slots. 1996. Mammalian viruses in human periodontitis. Oral Microbiol. Immunol. 11:381-386. [DOI] [PubMed] [Google Scholar]
- 10.Contreras, A., M. Umeda, C. Chen, I. Bakker, J. L. Morrison, and J. Slots. 1999. Relationship between herpesviruses and adult periodontitis and periodontopathic bacteria. J. Periodontol. 70:478-484. [DOI] [PubMed] [Google Scholar]
- 11.Contreras, A., H. H. Zadeh, H. Nowzari, and J. Slots. 1999. Herpesvirus infection of inflammatory cells in human periodontitis. Oral Microbiol. Immunol. 14:206-212. [DOI] [PubMed] [Google Scholar]
- 12.Contreras, A., H. Nowzari, and J. Slot. 2000. Herpesviruses in periodontal pocket and gingival tissue specimens. Oral Microbiol. Immunol. 15:15-18. [DOI] [PubMed] [Google Scholar]
- 13.Contreras, A., and J. Slots. 2000. Herpesviruses in human periodontal disease. J. Periodontal Res. 35:3-6. [DOI] [PubMed] [Google Scholar]
- 14.Contreras, A., A. Mardirossian, and J. Slots. 2001. Herpesviruses in HIV-periodontitis. J. Clin. Periodontol. 28:96-102. [DOI] [PubMed] [Google Scholar]
- 15.Idesawa, M., N. Sugano, K. Ikeda, M. Oshikawa, M. Takane, K. Seki, and K. Ito. 2004. Detection of Epstein-Barr virus in saliva by real-time PCR. Oral Microbiol. Immunol. 19:230-232. [DOI] [PubMed] [Google Scholar]
- 16.Kamma, J. J., and J. Slots. 2003. Herpesviral-bacterial interactions in aggressive periodontitis. J. Clin. Periodontol. 30:420-426. [DOI] [PubMed] [Google Scholar]
- 17.Khanna, R., S. R. Burrows, and D. J. Moss. 1995. Immune regulation in Epstein-Barr virus-associated diseases. Microbiol. Rev. 59:387-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konstantinidis, A., D. Sakellari, A. Papa, and A. Antoniadis. 2005. Real-time polymerase chain reaction quantification of Epstein-Barr virus in chronic periodontitis patients. J. Periodontal Res. 40:294-298. [DOI] [PubMed] [Google Scholar]
- 19.Kubar, A., I. Saygun, A. Ozdemir, M. Yapar, and J. Slots. 2005. Real-time polymerase chain reaction quantification of human cytomegalovirus and Epstein-Barr virus in periodontal pockets and the adjacent gingiva of periodontitis lesions. J. Periodontal Res. 40:97-104. [DOI] [PubMed] [Google Scholar]
- 20.Ling, L. J., C. C. Ho, C. Y. Wu, Y. T. Chen, and S. L. Hung. 2004. Association between human herpesviruses and the severity of periodontitis. J. Periodontol. 75:1479-1485. [DOI] [PubMed] [Google Scholar]
- 21.Madhavan, H. N., and K. Priya. 2002. Polymerase chain reaction based restriction fragment length polymorphism for the genotyping of cytomegalovirus (CMV) from patients with CMV disease in Chennai. Indian J. Med. Res. 115:242-247. [PubMed] [Google Scholar]
- 22.Meyer-Konig, U., C. Vogelberg, A. Bongarts, D. Kampa, R. Delbruck, G. Wolff-Vorbeck, G. Kirste, M. Haberland, F. T. Hufert, and D. von Laer. 1998. Glycoprotein B genotype correlates with cell tropism in vivo of human cytomegalovirus infection. J. Med. Virol. 55:75-81. [PubMed] [Google Scholar]
- 23.Meyer-Konig, U., M. Harberland, D. von Laer, O. Haller, and F. T. Hufert. 1998. Intragenic variability of human cytomegalovirus glycoprotein B in clinical strains. J. Infect. Dis. 177:1162-1169. [DOI] [PubMed] [Google Scholar]
- 24.Moss, D. J., S. R. Burrows, S. L. Silin, I. Misko, and R. Khanna. 2001. The immunology of Epstein-Barr virus infection. Philos. Trans. R. Soc. Lond. B 356:475-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parra, B., and J. Slots. 1996. Detection of human viruses in periodontal pockets using polymerase chain reaction. Oral Microbiol. Immunol. 11:289-293. [DOI] [PubMed] [Google Scholar]
- 26.Reeves, M. B., P. A. MacAry, P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 102:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saygun, I., M. Yapar, A. Ozdemir, A. Kubar, and S. Slots. 2004. Human cytomegalovirus and Epstein-Barr virus type 1 in periodontal abscesses. Oral Microbiol. Immunol. 19:83-87. [DOI] [PubMed] [Google Scholar]
- 28.Saygun, I., A. Kubar, A. Ozdemir, M. Yapar, and J. Slots. 2004. Herpesviral-bacterial interrelationships in aggressive periodontitis. J. Periodontal Res. 39:207-212. [DOI] [PubMed] [Google Scholar]
- 29.Slots, J., and A. Contreras. 2000. Herpesviruses: a unifying causative factor in periodontitis? Oral Microbiol. Immunol. 15:277-280. [DOI] [PubMed] [Google Scholar]
- 30.Slots, J., C. Sugar, and J. J. Kamma. 2002. Cytomegalovirus periodontal presence is associated with subgingival Dialister pneumosintes and alveolar bone loss. Oral Microbiol. Immunol. 17:369-374. [DOI] [PubMed] [Google Scholar]
- 31.Slots, J. 2002. Interactions between herpesviruses and bacteria in human periodontal disease, p. 317-331. In K. A. Brogden and J. M. Guthmiller (ed.), Polymicrobial diseases. ASM Press, Washington, DC. [PubMed]
- 32.Slots, J., J. J. Kamma, and C. Sugar. 2003. The herpesvirus-Porphyromonas gingivalis-periodontitis axis. J. Periodontal Res. 38:318-323. [DOI] [PubMed] [Google Scholar]
- 33.Slots, J. 2004. Herpesviruses, the missing link between gingivitis and periodontitis? J. Int. Acad. Periodontol. 6:113-119. [PubMed] [Google Scholar]
- 34.Socransky, S. S., and A. D. Haffajee. 2002. Dental biofilms: difficult therapeutic targets. Periodontol. 2000 28:12-15. [DOI] [PubMed] [Google Scholar]
- 35.Sugano, N., K. Ikeda, M. Oshikawa, M. Idesawa, H. Tanaka, S. Sato, and K. Ito. 2004. Relationship between Porphyromonas gingivalis, Epstein-Barr virus infection and reactivation in periodontitis. J. Oral Sci. 46:203-206. [DOI] [PubMed] [Google Scholar]
- 36.Syrjanen, S., R. Leimola-Virtanen, A. Schmidt-Westhausen, and P. A. Reichart. 1999. Oral ulcers in AIDS patients frequently associated with cytomegalovirus (CMV) and Epstein-Barr virus (EBV) infections. J. Oral Pathol. Med. 28:204-209. [DOI] [PubMed] [Google Scholar]
- 37.Vieira, J., P. O'Hearn, L. Kimball, B. Chandran, and L. Corey. 2001. Activation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) lytic replication by human cytomegalovirus. J. Virol. 75:1378-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wada, K., S. Mizuno, K. Kato, T. Kamiya, and K. Ozawa. 1997. Cytomegalovirus glycoprotein B sequence variation among Japanese bone marrow transplant recipients. Intervirology 40:215-219. [DOI] [PubMed] [Google Scholar]
- 39.Wakiguchi, H., H. Hisakawa, H. Kubota, and T. Kurashige. 1999. Strong response of T cells in infants with dual infection of Epstein-Barr virus and cytomegalovirus. Pediatr. Int. 41:484-489. [DOI] [PubMed] [Google Scholar]