Abstract
Spoligotyping was performed to study the population structure of Mycobacterium tuberculosis complex strains (n = 224) from Bangladesh. Strains were split into principal genetic group 1 (PGG 1 [75.0%]) and PGG 2 and 3 (25%). Forty-nine strains with a new spoligotype signature and considered as south or southeast Asian-linked emerging clones were designated as “Matlab type.”
Spoligotyping is a powerful and simple fingerprinting technique for simultaneous detection and differentiation of Mycobacterium tuberculosis complex (MTC) (12, 15). The discriminatory power of spoligotyping is less than that of IS6110 restriction fragment length polymorphism typing (16). However, spoligotyping is more efficient for differentiating MTC strains with a small number of IS6110 copies, including Mycobacterium bovis (2). Limited genotypic information about clinical isolates of MTC is available from Bangladesh (1, 6). Therefore, this study was planned to search for the existence of internationally-recognized phylogenetic clades of MTC in Bangladeshi tuberculosis (TB) patients and to identify any new spoligotyping signature(s) phylogeographically specific for Bangladesh.
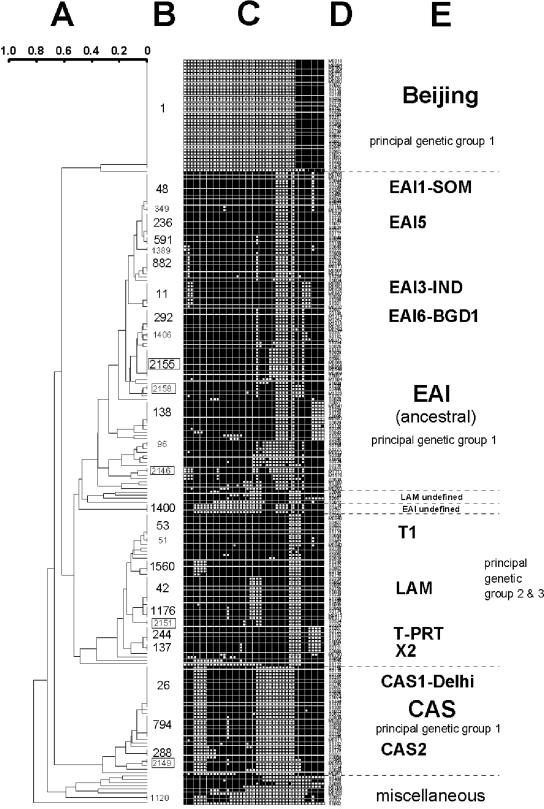
During a period from July 2001 through November 2003, 224 MTC strains were isolated from nonhospitalized TB patients of selected urban (Dhaka) and rural (Matlab) areas of Bangladesh. These strains were genotyped using the standard spoligotyping technique (20). Spoligotype patterns were recorded as octal and binary formats in Excel spreadsheets and compared with the SpolDB4 database (5). A spoligotype-based unrooted dendrogram of all the isolates (n = 224) was built (Fig. 1) (14) using the Taxotron software (11).
FIG. 1.
Comparison of 224 MTC strain spoligotypes from this study with those in the Spo1DB4 database. (A) Dendrogram built using Taxotron (P. A. D. Grimont, Taxolab, Institut Pasteur, 1994-2000 [Jaccard index and unweighted-pair group method using average linkages]). (B) ST designation number of spoligotype according to SpolDB4 nomenclature. Two-isolate clusters appear with smaller letters. (C) Binary description of spoligotypes. (D) Isolate number designation. M, Matlab, rural area; S, urban area. (E) Clade/subclade designation. Except for T1, LAM, T-PRT, and X2, which cluster with PGG 2 and 3, all other branches cluster with PGG 1 isolates.
Age and gender information was available from 211 of 224 TB patients (47 rural and 177 urban). The male (n = 63)/female (n = 148) ratio was 1.42, and mean age was 36.5 years (range, 15 to 83 years; standard deviation, 16.1 years). Thirty percent of patients were within the 15- to 24-year age group.
Using the standard spoligotyping format, 193 (86.0%) of 224 isolates were clustered into 31 shared types (ST) containing from 2 to 34 strains, whereas 31 strains (14.0%) were unique. After matching with the SpolDB4 database (available online from http://www.pasteur-guadeloupe.fr:8081/SITVITDemo), these strains (n = 224) were further clustered into four major clades or lineages (Fig. 1). Seventy-five percent of all strains belonged to principal genetic group 1 (PGG 1) (18), which includes Beijing, East African-Indian (EAI), and Central Asian (CAS), whereas 25.0% belonged to PGG 2 and 3.
EAI is the most prevalent clade and constitutes about 44.20% of all isolates (n = 99; 30 from rural and 69 from urban TB patients). Based on the absence of specific spacers of the direct repeat region, most strains of the EAI clade could be classified into EAI1 to EAI5 (Fig. 1). Within the EAI clade, 49 strains had a new spoligotype signature and were designated EAI6-Bangladesh 1 (EAI6-BGD1). Other known EAI subclades, such as EAI2 (ST 19; Manila) (7), EAI4 (ST 139; Vietnam), and EAI8 (ST 109; Madagascar) (8), were not found in this study. Other EAI variants are likely to be present in Bangladesh.
The sublineage EAI6-BGD1 was designated “Matlab type” after the name of the rural field site Matlab, where these strains were first isolated and subsequently isolated from both rural and urban areas. Known as an “ancient” TB clade (4), strains of the EAI clade are predominant in Bangladesh, arguing in favor of the historical presence of endemic strains in this country. It is also evident from the comparison of this study with results from a previous one (17; Rigouts et al., unpublished data) (n = 510) obtained in another setting in Bangladesh that the emergence of an endemic new lineage of the large EAI superfamily designated as “Matlab type” can be defined. Indeed, our observation about the endemic nature of the “Matlab type” was also evident from an independent study conducted in the Sunamganj District of Bangladesh (19). TB patients of this district are mostly farmers having little contact with people from other parts of the world. The “Matlab type” strains were detected from 26% of TB patients of this district.
The W-Beijing family is the second most prevalent clade (prototype ST 1) and constitutes about 15.2% of the strains included in this study (n = 34: 7 from rural TB patients and 27 from urban TB patients) (Fig. 1). Its origin in Asia is not yet precisely dated, but it could be ancient, arguing that these strains belong to PGG 1, as assessed by katG-gyrA polymorphism (18). However, a recent study documented a single-nucleotide polymorphism located within the nitrate reductase (narGHJ) operon promoter that clusters Beijing together with PGG 2 and 3 organisms (10).
The CAS family represents 15.0% of the isolates (n = 34: 2 from rural and 32 from urban TB patients) (Fig. 1). Prototypic ST 26, predominant in this study, is popularly known as the Delhi type, which is prevalent in north India (3). Other well-represented variants like CAS 1 signatures, ST 794 (n = 7), ST 1120 and ST 288 (CAS2; n = 2), and the newly described ST 2149 (n = 3) were also detected in this study. This family is also prevalent in Pakistan, Sudan, and Libya (3, 9). The CAS 2 variant (ST 288) was also detected at a lower frequency in Bangladesh. No specific strains of the CAS clade localized to Bangladesh were detected.
PGG 2 and 3 (also designated as modern TB) represented one-quarter (25.0%) of the strains (Fig. 1). Twenty-one strains (4 from rural and 17 from urban TB patients) of this group could not be assigned to any clade designation, and, hence, were termed “T.” The Latin-American and Mediterranean (LAM) clade is the second most prevalent clade in modern TB isolates. Other spoligotype signatures within PGG 2 and 3 have been designated as “European low IS6110 copy” clade or “X clade” (21). This group is represented here as ST 137 (prototype of the X2) and could be related to T-PRT, also known as the “Portuguese type” (prototype, ST 244). It is predominant in Portugal and Brazil (5). The Portuguese entered the Indian subcontinent (part of which is current Bangladesh) at the end of the nineteenth century (13). Mycobacterial interspersed repetitive unit typing of ST 244 strains from Portugal and Bangladesh confirmed the likely existence of a common ancestor of today's clones (T. Zozio, N. Rastogi, and C. Sola, unpublished observations). Besides, a few other patterns, such as MANU 2, X3, S, and LAM3, were also found in this study (Fig 1).
Comparison of the 224 spoligotypes of MTC strains of this study with those in the SpolDB4 database is shown in Table 1, which contains a total of 720 spoligotyping data on isolates from Bangladesh from the present and previous studies (17; L. Rigouts and F. Portaels unpublished data). Most spoligotyping patterns described in this study suggest that the genotypes described here, indeed, represent true discovery of genetic diversity.
TABLE 1.
Detailed results on nomenclature and distribution of STs of MTC clinical isolates in this study and other studies in Bangladesha
| ST | Absolute count in SpolDB4 databaseb | % in SpolDB4 database | Relevant geographic origin(s) in SpolDB4 database | Absolute count in:
|
% in Bangladesh | ||
|---|---|---|---|---|---|---|---|
| This study (n = 224) | Other Bangladesh studies (n =510)
|
||||||
| SpolDB4 database alonec | After introduction of result file (increase)d | ||||||
| 1 | 3,839 | 9.37 | Far east Asia | 32 | 121 (L.R., 67; ZR, 54) | 121 | 26.59 |
| 292 | 74 | 0.18 | Bangladesh | 11 | 62 (L.R., 24; Z.R., 38) | 62 | 13.63 |
| 1406 | 2 | 0.00 | Bangladesh | 2 | 2 (L.R., 2) | 4 (+2) | 0.88 |
| 11 | 347 | 0.85 | India | 7 | 14 (L.R., 2; Z.R., 12) | 14 | 3.08 |
| 48 | 256 | 0.62 | India, Somalia, Bangladesh | 9 | 41 (L.R., 16; Z.R., 25) | 41 | 9.01 |
| 236 | 108 | 0.26 | India, southeast Asia | 5 | 6 (L.R., 6) | 11 (+5) | 2.42 |
| 1389 | 9 | 0.02 | Bangladesh | 2 | 7 (L.R., 7) | 9 (+2) | 1.98 |
| 882 | 18 | 0.04 | Bangladesh | 6 | 12 (L.R., 7; Z.R., 5) | 13 (+1) | 2.86 |
| 591 | 31 | 0.08 | India, Bangladesh, Malaysia | 3 | 1 (L.R., 1) | 4 (+3) | 0.88 |
| 138 | 76 | 0.19 | India, Bangladesh, Malaysia, Somalia | 8 | 26 (L.R., 5; Z.R., 21) | 26 | 5.71 |
| 1400 | 7 | 0.02 | Bangladesh | 3 | 7 (L.R., 2; Z.R., 5) | 7 | 1.54 |
| 26 | 543 | 1.32 | India, Pakistan | 13 | 54 (L.R., 8; Z.R., 46) | 54 | 11.87 |
| 794 | 3 | 0.01 | Pakistan | 9 | 0 | 9 (+9) | 1.98 |
| 288 | 61 | 0.15 | India, Bangladesh | 3 | 2 (L.R., 2) | 5 (+3) | 0.44 |
| 53 | 2,580 | 6.29 | Europe, Africa | 7 | 24 (L.R., 7; Z.R., 17) | 24 | 5.27 |
| 52 | 403 | 0.98 | Ubiquitous | 2 | 1 (L.R., 1) | 3 (+2) | 0.66 |
| 1560 | 2 | 0.00 | Europe, Bangladesh | 3 | 0 | 3 (+3) | 0.66 |
| 244 | 44 | 0.11 | Portugal, Brazil, Bangladesh | 5 | 9 (Z.R., 9) | 9 | 1.98 |
| 137 | 754 | 1.84 | North America, Great-Britain | 3 | 0 | 3 (+3) | 0.66 |
| 42 | 1,300 | 3.17 | South America, Mediterranean basin, Africa | 8 | 28 (L.R., 9; Z.R., 19) | 28 | 6.15 |
| 1176 | 11 | 0.03 | Indonesia, Georgia | 5 | 0 | 5 (+5) | 1.10 |
Comparison was done on 2004 November 27 2004 against 40,987 spoligotypes. Some spoligotypes appear to be present only in Bangladesh, whereas others are also found in different south Asian countries as well as in Europe.
Representing a total of 40,987 spoligotypes present on 24 November 2004.
L.R. and Z.R. represent data submitted by Leen Rigouts and Zeaur Rahim, respectively. n = 720 spoligotypes from Bangladesh recorded in the SpolDB4 database.
After introduction of the file of results from this study. (The total is 455.) File counts do not include part of the results of L. Rigouts and F. Portaels included in the SpolDB4 database but not analyzed here.
In conclusion, our study is the first to describe the population structure of MTC and also reports a new and specific subclade of the EAI family named “Matlab type,” which may be suggestive of new south or southeast Asian-linked emerging genotypes.
Acknowledgments
The International Centre for Diarrheal Disease Research, Bangladesh, and Gates-Government of Bangladesh provided necessary funds for this research.
We are grateful to David Moore for carefully reading the manuscript.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Banu, S., S. V. Gordon, S. Palmer, R. Islam, S. Ahmed, K. M. Alam, S. T. Cole, and R. Brosch. 2004. Genotypic analysis of Mycobacterium tuberculosis in Bangladesh and prevalence of the Beijing strain. J. Clin. Microbiol. 42:674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, J., Å. B. Andersen, K. Kremer, and H. Miörner. 1999. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J. Clin. Microbiol. 37:2602-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhanu, N. V., D. van Soolingen, J. D. A. van Embden, L. Dar, R. M. Pandey, and P. Seth. 2002. Predominance of a novel Mycobacterium tuberculosis genotype in the Delhi region of India. Tuberculosis 82:105-112. [DOI] [PubMed] [Google Scholar]
- 4.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brudey, K., J. Driscoll, L. Rigouts, W. Prodinger, A. Gori, S. A. M. Al-Hajoj, C. Allix, L. Aristimuno, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Gutierrez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, T. Lillebaek, H. M. Ly, C. Martin, C. Martin, I. Mokrousov, O. Narvsakya, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rüsch-Gerdes, A. Sajduda, S. Samper, P. Seth, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. Skuce, D. Van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. An appraisal of the geographic prevalence of major genotypic families of Mycobacterium tuberculosis complex through the updated SpolDB4 database. BMC Microbiol. 6:23.16519816 [Google Scholar]
- 6.Chowdhury, M. Z. U., and C. Palittapongarnpim. 1997. DNA fingerprinting of Mycobacterium tuberculosis isolates from Bangladesh by polymerase chain reaction. J. Infect. Dis. Antimicrob. Agent 14:1-3. [Google Scholar]
- 7.Douglas, J. T., L. Qian, J. C. Montoya, J. M. Musser, J. D. A. van Embden, D. van Soolingen, and K. Kremer. 2003. Characterization of the Manila family of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:2723-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferdinand, S., C. Sola, S. Chanteau, H. Ramarokoto, T. Rasolonavalona, V. Rasolofo-Razanamparany, and N. Rastogi. 2005. A study of spoligotyping-defined Mycobacterium tuberculosis clades in relation to the origin of people and the demographic history in Madagascar. Infect. Genet. Evol. 4:340-348. [DOI] [PubMed] [Google Scholar]
- 9.Gascoyne-Binzi, D. M., R. E. Barlow, A. Essex, R. Gelletlie, M. A. Khan, S. Hafiz, T. A. Collyns, R. Frizzell, and P. M. Hawkey. 2002. Predominant VNTR family of strains of Mycobacterium tuberculosis isolated from South Asian patients. Int. J. Tuberc. Lung Dis. 6:492-496. [DOI] [PubMed] [Google Scholar]
- 10.Goh, K. S., N. Rastogi, M. Berchel, R. C. Huard, and C. Sola. 2005. Molecular evolutionary history of tubercle bacilli assessed by study of the polymorphic nucleotide within the nitrate reductase (narGHJI) operon promoter. J. Clin. Microbiol. 43:4010-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimont, P. A. D. 2000. TAXOTRON, 4th update edition. Institut Pasteur, Paris, France.
- 12.Groenen, P. M., A. E. Bunschoten, D. van Soolingen, and J. D. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis: application from strain differentiation by a novel typing method. Mol. Microbiol. 10:1057-1065. [DOI] [PubMed] [Google Scholar]
- 13.Islam, S. 2003. The Portuguese, p. 142-146. In S. Islam (ed.), Banglapedia: national encyclopaedia of Bangladesh. Asiatic Society Press, Dhaka, Bangladesh.
- 14.Jaccard, P. 1908. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 44:223-270. [Google Scholar]
- 15.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. M. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. A. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shamputa, I. C., L. Rigouts, L. A. Eyongeta, N. A. El Aila, A. van Deun, A. H. Salim, E. Willery, C. Locht, P. Supply, and F. Portaels. 2004. Genotypic and phenotypic heterogeneity among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients. J. Clin. Microbiol. 42:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sreevatsan, S., X. Pan, K. Stockbauer, N. Connell, B. Kreiswirth, T. Whittam, and J. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storla, D. G., Z. Rahim, M. A. Islam, S. Plettner, V. Begum, T. Manssaker, B. Myrvang, G. Bjune, and U. R. Dahle. 2006. Heterogeneity of Mycobacterium tuberculosis isolates in Sunamganj District, Bangladesh. Scand. J. Infect. Dis. 38:593-596. [DOI] [PubMed] [Google Scholar]
- 20.van der Zanden, A. G. M., E. M. T. Koppele-Vije, N. V. Bhanu, D. van Soolingen, and L. M. Schouls. 2003. Use of DNA extracts from Ziehl-Neelsen-stained slides for molecular detection of rifampin resistance and spoligotyping of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren, R. M., T. C. Victor, E. M. Streicher, M. Richardson, G. D. van der Spuy, R. Johnson, V. N. Chihota, C. Locht, P. Supply, and P. D. van Helden. 2004. Clonal expansion of a globally disseminated lineage of Mycobacterium tuberculosis with low IS6110 copy numbers. J. Clin. Microbiol. 42:5774-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]