Abstract
Respiratory tract infections can be caused by a heterogeneous group of viruses and bacteria that produce similar clinical presentations. Specific diagnosis therefore relies on laboratory investigation. This study developed and evaluated five groups of multiplex nested PCR assays that could simultaneously detect 21 different respiratory pathogens: influenza A virus (H1N1, H3N2, and H5N1); influenza B virus; parainfluenza virus types 1, 2, 3, 4a, and 4b; respiratory syncytial virus A and B; human rhinoviruses; human enteroviruses; human coronaviruses OC43 and 229E; severe acute respiratory syndrome coronavirus; human metapneumoviruses; Mycoplasma pneumoniae; Chlamydophila pneumoniae; Legionella pneumophila; and adenoviruses (A to F). These multiplex nested PCRs adopted fast PCR technology. The high speed of fast PCR (within 35 min) greatly improved the efficiency of these assays. The results show that these multiplex nested PCR assays are specific and more sensitive (100- to 1,000-fold) than conventional methods. Among the 303 clinical specimens tested, the multiplex nested PCR achieved an overall positive rate of 48.5% (95% confidence interval [CI], 42.9 to 54.1%), which was significantly higher than that of virus isolation (20.1% [95% CI, 15.6 to 24.6%]) and that of direct detection by immunofluorescence assay (13.5% [95% CI, 9.7 to 17.4%]). The improved sensitivity was partly due to the higher sensitivity of multiplex nested PCR than that of conventional methods in detecting cultivatable viruses. Moreover, the ability of the multiplex nested PCR to detect noncultivatable viruses, particularly rhinoviruses, coronavirus OC43, and metapneumoviruses, contributed a major gain (15.6%) in the overall positive rate. In conclusion, rapid multiplex nested PCR assays can improve the diagnostic yield for respiratory infections to allow prompt interventive actions to be taken.
Respiratory tract infection is a major cause of hospitalization. The 2003 outbreaks of severe acute respiratory syndrome (SARS) and the more recent human avian H5N1 influenza virus cases underscore the importance of a rapid and accurate laboratory diagnosis to investigate infections associated with severe individual or public health consequences (1).
Respiratory tract infections in humans can be a result of infection caused by a heterogeneous group of viruses and bacteria that produce similar clinical presentations (10, 18, 30). Specific diagnosis therefore relies almost entirely on laboratory investigation. Rapid antigen detection assays are now widely used in routine diagnostic laboratories, but these assays have been shown to be inferior in sensitivity and specificity to assays based on PCR (5, 12), which can also be designed to screen for a wider range of pathogens. Numerous studies have developed and evaluated multiplex nested PCR, reverse transcription (RT)-PCR, or real-time PCR for the detection of respiratory viruses (4, 11, 21, 24, 27). However, these studies are still limited by their turnaround time and/or the range of viruses being detected.
This study developed and evaluated five groups of multiplex nested PCR assays that included an RT step where necessary. These assays could simultaneously detect 21 different respiratory pathogens including influenza virus group A (FluA) (subtypes H1, H3, and H5), influenza virus group B (FluB), parainfluenza virus type 1 (PIV-1), PIV-2, PIV-3, PIV-4a, PIV-4b, human respiratory syncytial viruses (hRSV), all serotypes of human adenoviruses (ADVs) (A to F), human metapneumoviruses (hMPV), human coronaviruses (HCoVs) (HCoV-229E, HCoV-OC43, and SARS coronavirus [SARS-CoV]), all serotypes of human enteroviruses (hEVs), and all serotypes of human rhinoviruses (hRVs) as well as the fastidious respiratory bacteria Chlamydophila pneumoniae, Mycoplasma pneumoniae, and Legionella pneumophila.
MATERIALS AND METHODS
Primer design and preparation.
The primers used in this study were either modified from previously published primer sequences (2-4, 7-9, 11, 17, 19, 23, 24, 26, 28, 29, 31-34) or designed from consensus genome regions obtained from GenBank (http://www.ncbi.nlm.nih.gov/). Typically, sequences of 10 to 20 representative strains of each pathogen were downloaded. The sequences were aligned using Clustal X (http://bips.u-strasbg.fr/en/Documentation/ClustalX/) (25). The program GeneTool Lite 1.0 (BioTools Inc., Edmonton, Alberta, Canada) was used to predict the compatibility of primer pairs and to estimate the optimal annealing temperatures. Primer pairs were selected to ensure that the sizes of the amplicons of different pathogens could be easily differentiated by agarose gel electrophoresis. The primers used were 20 to 30 bp in length and had G+C contents less than or equal to 70%, thus having an annealing temperature of 50 to 66°C.
Multiplex PCR primer grouping.
Five groups of multiplex nested PCR assays, targeting 21 respiratory viruses and bacteria, were developed. Each multiplex nested PCR detected four to five viruses and/or bacteria: group 1 was comprised of FluA and FluB group-specific and subtype H1-, H3-, and H5-specific primers. Group 2 was comprised of PIV-1, PIV-2, PIV-3, PIV-4a, and PIV-4b. Group 3 was comprised of hRSV A and B, hRV, and hEV. Group 4 was comprised of HCoV (HCoV-OC43, HCoV-229E, and SARS-CoV) and hMPV. Group 5 was comprised of Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and ADV. The sequences and amplicon sizes of the outer and inner sets of primers are given in Tables 1 and 2.
TABLE 1.
Primers used in the first round of multiplex nested PCR
| Organism | Primer | Sequence (5′→3′)a | Primer concn (μM) | Product length (bp) | Target geneb | Source or reference |
|---|---|---|---|---|---|---|
| Group 1 | ||||||
| Influenza A virus | FluA-OF3 | TYGAGGCTCTCATGGARTGGCTAAAG | 0.052 | 412 | Matrix | This study |
| FluA-OR3 | GCTGGCCARMACCATTCTGTTYTCAT | 0.052 | ||||
| Influenza A virus H1 | H1-OF1 | CCCAGGRTATTTCKCCGAYTATGAGG | 0.104 | 760 | Hemagglutinin | This study |
| H1-OR1 | TACCATTCCAGTCCACCCCCCTTCA | 0.104 | ||||
| Influenza A virus H3 | H3-OF1 | ATGGGACCTTTTTRTYGAACGCAGCA | 0.052 | 519 | Hemagglutinin | This study |
| H3-OR1 | CCCCKAGGAGCAATTAGATTCCCTGT | 0.052 | ||||
| Influenza A virus H5 | H5N1-VIET-1B | ATCAAACAGATTAGTCCTTGCG | 0.208 | 265 | Hemagglutinin | This study |
| H5N1-VIET-2B | GGCCTCAAACTGAGTGTTCATT | 0.208 | ||||
| Influenza B virus | FluB-OF3 | AGGAAGRGCAATGGCAGAYAGAGG | 0.834 | 883 | Nucleoprotein | This study |
| FluB-OR2 | TGCTGTGTCCCTCCCAAAGAAGAAA | 0.834 | ||||
| Group 2 | ||||||
| PIV-1 | PIV-1-OF1 | TCTGGATCCACCACAATTTCAG | 0.933 | 848 | Hemagglutinin-neuraminidase | This study |
| PIV-1-OR1 | WACCAGTTGCAGTCTKGGTTTC | 0.933 | ||||
| PIV-2 | PIV2-F1 | CTTGCAGCATTTTCTGGGGAACTCC | 0.0373 | 716 | Hemagglutinin-neuraminidase | This study |
| PIV2-OR1 | GCATCATCATCCTGGGAGCCTCTGT | 0.0373 | ||||
| PIV-3 | PIV-3-OF1 | GATTTTTGGAGATGCACGTCTG | 0.187 | 1,118 | Hemagglutinin-neuraminidase | This study |
| PIV-3-OR1 | GAGAGTGTTYTGTTTCGGATGG | 0.187 | ||||
| PIV-4 | PIV-4AB-IF1 | AYGGATGCATTCGAATTCCATCATTC | 0.093 | 432 | Hemagglutinin-neuraminidase | This study |
| PIV-4AB-OR1 | TCCRTRAGRCCYCCATACAARGG | 0.093 | ||||
| Group 3 | ||||||
| hRSV A | RSV-A-OF2 | CAGCTCCGTTATCACATCTCTAGGAGCC | 0.278 | 576 | Fusion protein | This study |
| RSV-A-OR2 | TGGGTTGTCTATGAGCAGATAKKAAACCA | 0.278 | ||||
| hRSV B | RSV-B-OF2 | CGGGCCAGAAGAGAAGCACCACAGTA | 0.069 | 673 | Fusion protein | This study |
| RSV-B-OR2 | TGATCCTTCTTTGATGTTGGTGGTGC | 0.069 | ||||
| hRV | OL26-MOD-RV | CACTTCTGTTTCCCCGGAGCGAG | 0.139 | 388 | 5′ UTR | 14 |
| RV-OR2 | GAAACACGGACACCCAAAGTAGTCGGT | 0.139 | ||||
| hEV | EV-OF3 | CTGCGYTGGCGGCCYMCC | 0.139 | 481 | 5′ UTR | 14 |
| EV-OR2 | CCGGATGGCCAATCCAATAACTATATGGT | 0.139 | ||||
| Group 4 | ||||||
| HCoV-OC43 | HCoVOC43-OF3 | CGGTTACACTGTTCAGCCAATYGCA | 0.313 | 793 | Spike protein | This study |
| HCoVOC43-OR3 | CCAACCCAAAAATGCTTGTGGTYG | 0.313 | ||||
| SARS-CoV | COR1 | CACCGTTTCTACAGGTTAGCTAACGA | 0.313 | 310 | Polymerase | 32 |
| COR2 | AAATGTTTACGCAGGTAAGCGTAAAA | 0.313 | ||||
| HCoV-229E | COR229E-IF2 | TCACCCATTTGAAGAATTGGAATTTTGG | 0.313 | 566 | Matrix | This study |
| COR229E-IR2 | TCGTACGTAGAAAACCCAGCCTGTGC | 0.313 | ||||
| hMPV | Meta-M-OF2 | CAATATGGTTCCCTTTGTTTCAGGCCA | 0.313 | 462 | Matrix | This study |
| Meta-M-OR2 | TGGTCTGCTTCACTGCTTATWGCAGCTT | 0.313 | ||||
| Group5 | ||||||
| M. pneumoniae | Mpneumoniae-OF2 | GACCATTCCACCCAGCCCCAGC | 0.089 | 343 | Cytadhesin P1 gene | 17 |
| Mpneumoniae-OR2 | GTTCAGCGAGTGGGGTGCGTACAATA | 0.089 | ||||
| C. pneumoniae | Chlamy-pneum-OF4 | TGCGCTACTTGGTGCGACGCTA | 0.179 | 571 | Outer membrane protein A | 17 |
| Chlamy-penum-OR4 | CGCCTTTATAGCCCTTGGGTTTRTTT | 0.179 | ||||
| L. pneumophila | Legionella-OF1 | CGCTCAATTGGCTTTAACCGAACAG | 0.054 | 425 | Macrophage infectivity potentiator (“mip”) | This study |
| Legionella-OR1 | CGCTRCGTGGRCCATATGCARGAC | 0.054 | ||||
| ADV | ADVAtoF-OF3 | TACATGCACATCKCSGGVCAGGA | 0.179 | 983 | Hexogene | This study |
| ADVAtoF-OR3 | CCRGCCARHACHCCCATRTTDCCHGT | 0.179 |
Degenerate primer abbreviations are as follows: M, A/C; R, A/G; W, A/T; S, C/G; Y, C/T; K, G/T; V, A/C/G; H, A/C/T; D, A/G/T; N, A/C/G/T.
UTR, untranslated region.
TABLE 2.
Primers used in the second round of multiplex nested PCR
| Organism | Primer | Sequence (5′→3′)a | Primer concn (μM) | Product length (bp) | Target geneb | Source or reference |
|---|---|---|---|---|---|---|
| Group 1 | ||||||
| Influenza A virus | FluARe-F1 | AAGACCAATCCTGTCACCTCTGA | 0.103 | 104 | Matrix | 30 |
| FluARe-R1 | CAAAGCGTCTACGCTGCAGTCC | 0.103 | ||||
| Influenza A virus H1 | H1-IF2 | TCGCCGACTATGAGGAACTGAGGGA | 0.021 | 431 | Hemagglutinin | This study |
| H1-IR2 | TTGTATCCCCGGGTTCCAGCAGAGT | 0.021 | ||||
| Influenza A virus H3 | H3-IF2 | CCCTTATGATGTGCCGGATTATGCC | 0.205 | 259 | Hemagglutinin | This study |
| H3-IR2 | GGTGGTGAACCCCCCAAATGTACAA | 0.205 | ||||
| Influenza A virus H5 | H5N1-VIET-1A | TGCGACTGGRCTCAGAAATA | 0.512 | 172 | Hemagglutinin | This study |
| H5N1-VIET-4B | GGATTCTTTGTCTGCAGCGT | 0.512 | ||||
| Influenza B virus | FLUB-IF3 | AAAACAARTGCTCTGCRCCYCAAC | 0.41 | 516 | Nucleoprotein | This study |
| FLUB-IR3 | CRTCTCCACCTACTTCRTTYCCCCC | 0.41 | ||||
| Group 2 | ||||||
| PIV-1 | PIV-1-IF1 | AATTGGTGATGCAATATATGCKTATTC | 0.61 | 600 | Hemagglutinin-neuraminidase | This study |
| PIV-1-IR1 | TCGACAACAATYTTTGGCCTATC | 0.61 | ||||
| PIV-2 | PIV2-F2 | AGGACAGCAGAGGACCTCGGCATG | 0.0305 | 343 | Hemagglutinin-neuraminidase | This study |
| PIV2-R2 | ACCTGATGTTCTTTGCGGTATGGGG | 0.0305 | ||||
| PIV-3 | PIV-3-IF1 | CAACTGTGTTCRACTCCCAAAG | 0.457 | 717 | Hemagglutinin-neuraminidase | This study |
| PIV-3-IR1 | TGGGTTYACTCTCGATTTTTGY | 0.457 | ||||
| PIV-4 | PIV-4AB-IF2 | GACGGATGYYTRCKGWATTGTGT | 0.153 | 231 | Hemagglutinin-neuraminidase | This study |
| PIV-4AB-IR2 | CCRTRAGRCCYCCATACAARGGAA | 0.153 | ||||
| Group 3 | ||||||
| hRSV A | RSVA-IF2 | TGACCCATTAGTGTTCCCCTCTGATGAAT | 0.278 | 228 | Fusion protein | This study |
| RSVA-IR2 | CTTCTGGCCTTRCAGTATARGAGCAGT | 0.278 | ||||
| hRSV B | RSV-B-IF1 | GTCGCATCTCCAACATTGRAAC | 0.069 | 336 | Fusion protein | This study |
| RSV-B-IR1 | TGGTGCATAGAGGTGATGTGTG | 0.069 | ||||
| hRV | RV-OF2 | CACTTCTGTTTCCCCGGAGCGAGG | 0.139 | 283 | 5′ UTR | 14 |
| JWA-1B-MOD-RV | CCGCATTCAGGGGCCGGAG | 0.139 | ||||
| hEV | EV-IF3 | CCTCCGGCCCCTGAATGCG | 0.139 | 106 | 5′ UTR | 3, 14 |
| EV-IR3 | CCAAAGTAGTCGGTTCCGCYRCRGA | 0.139 | ||||
| Group 4 | ||||||
| HCoV-OC43 | HCoVOC43-IF2 | CKGTGCCCTCTCCATTAAATTGGG | 0.25 | 635 | Spike protein | This study |
| HCoVOC43-IR2 | GACCCGAACAGTGCTCACCTATGCC | 0.25 | ||||
| SARS-CoV | COR3 | AGTGAGATGGTCATGTGTGG | 0.5 | 210 | Polymerase | 32 |
| COR4 | CACTCATAGAGCCTGTGTTG | 0.5 | ||||
| HCoV-229E | COR229E-IF3 | TTGGGATTCTAATTGGGCCTTTGTTGC | 0.25 | 361 | Matrix | This study |
| COR229E-IR3 | GCTCGGCACGGCAACTGTCATGTAT | 0.25 | ||||
| hMPV | Meta-M-IF2 | CCCTTTGTTTCAGGCCAAYACACCACC | 0.25 | 431 | Matrix protein | 2 |
| Meta-M-IR2 | GCAGCTTCAACAGTRGCTGATTCACTCTC | 0.25 | ||||
| Group 5 | ||||||
| M. pneumoniae | Mpneumoniae-OF1 | AGGGGGTTCTTCAGGCTCAGGTCAA | 0.094 | 160 | Cytadhesin P1 gene | 17 |
| Mpnuemoniae-OR1 | CCCCACCACATCATTCCCCGTATTA | 0.094 | ||||
| C. pneumoniae | Chlamy-pneum-IF6 | RCCTACWGGATCCGCTRCTGCRAA | 0.313 | 317 | Outer membrane protein A | 17 |
| Chlamy-pneum-IR6 | GCRCCTACGCTCCAAGMRAAAGWRG | 0.313 | ||||
| L. pneumophila | Legionella-IF1 | TGAAAACAAAAACAAGCCAGGCGTTG | 0.063 | 232 | Macrophage infectivity potentiator (“mip”) | This study |
| Legionella-IR1 | TGGCATCAATTGYAAAGCYTCTGTCC | 0.063 | ||||
| ADV | ADVAtoF-IF3 | TGGCYWSCACNTWCTTTGACATYMG | 0.782 | 463 | Hexogene | This study |
| ADVAtoF-IR3 | GCRWAWGAHCCRTARCAKGGYTDCAT | 0.782 |
Degenerate primer abbreviations are as follows: M, A/C; R, A/G; W, A/T; S, C/G; Y, C/T; K, G/T; V, A/C/G; H, A/C/T; D, A/G/T; N, A/C/G/T.
UTR, untranslated region.
Nucleic acid extraction and cDNA synthesis.
Total RNA and DNA were extracted together by using the QIAamp MinElute Virus Spin kit (QIAGEN, Valencia, CA), and 60 μl of nucleic acids was eluted according to the manufacturer's recommendations.
The RNA template (8 μl) was mixed with 1 μl of random primers (2.5 ng/μl) and 1 μl of deoxynucleoside triphosphates (0.5 mM each) in a final volume of 10 μl for incubation at 65°C for 5 min. The solution was equilibrated at 4°C and completed with 2 units of RNaseOUT, 4 μl of 5× first-strand buffer, 0.5 mM dithiothreitol, and 10 U Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) in a final volume of 20 μl. RT was performed for 50 min at 50°C and then stopped by heating for 15 min at 70°C. The resulting cDNA products were used immediately for PCR.
Fast PCR conditions.
In order to provide the shortest possible turnaround time, the recently available “fast” thermal cycler (fast PCR machine; Applied Biosystems, Foster City, CA) was used. When coupled with the DNA polymerase contained in the fast PCR master mix (GeneAmp; Applied Biosystems, Foster City, CA), a 35-cycle PCR assay could be completed within 35 min, compared to the ∼180 min that would normally be required for standard thermocyclers. All multiplex nested PCR assays were optimized to fulfill the manufacturer's recommendation that two-step cycling with annealing at 64°C was used. Both the first and second round of PCRs were conducted in a 20-μl reaction mixture. Two microliters of the cDNA preparation was used as the template for the first round of PCR for groups 1 to 4, whereas 8 μl of the extracted preparation was used for group 5, which was comprised of bacteria and a DNA virus. In the second round of PCR, a 0.2-μl aliquot of the first-round PCR product was used as a template. The final concentration of each primer present in the reaction mixture is shown in Tables 1 and 2. The cycling conditions for the first and second round of PCRs were an initial denaturation step at 95°C for 10 s and then 30 cycles of denaturation at 95°C for 1 s and annealing/extension at 64°C for 40 s, followed by a final extension step at 72°C for 10 s. The cycling conditions were the same for groups 1 to 4, whereas 35 cycles of denaturation at 95°C for 5 s was used instead for group 5.
The PCR products were identified by electrophoresis in 2% agarose gels and stained by ethidium bromide.
Preparation of controls.
Cultured stocks of the target pathogens were used as positive controls for the corresponding sets of the multiplex nested PCR assays. For noncultivatable pathogens, clinical specimens known to contain the target agents were used. For enteroviruses, the primers were designed to detect all serotypes of enteroviruses. In this study, the most commonly encountered serotypes including coxsackievirus serotypes A9, B1, B2, B3, and B5; echovirus serotypes 3, 7, 11, and 30; enterovirus serotype 71; and poliovirus type 1 (vaccine strain) were selected for the evaluation process.
Prevention of PCR contamination.
Precautions were taken to prevent cross-contamination. The preparation of reagents, processing of samples, and nested PCR assays were carried out in separate rooms away from the area where amplified products were analyzed. Filtered pipette tips were used throughout the experiments.
Specificity of the assay.
The ability of the multiplex nested PCR assays to detect the presence of more than one pathogen in the same specimen was assessed by use of simulated specimens spiked with two or more pathogens.
After the initial primer selection and optimization using known positive samples, the specificity of each multiplex nested PCR assay was further evaluated by running the assays on 50 clinical specimens known to contain respiratory pathogens other than the intended targets. This was to reconfirm that the primer sets did not produce false-positive or nonspecific results.
Sensitivity of the assay.
For viruses with an RNA genome, the assay sensitivity was determined by using synthetic RNA standards. Synthetic RNA target standards were generated using T7 polymerase (Ambion, Austin, TX) with primers incorporating a T7 promoter sequence. The copy number of synthetic RNA molecules was determined by UV spectrometry and serially diluted in 2.5 μg/ml yeast tRNA (Ambion). Eight microliters of diluted RNA was reversely transcribed with random hexamers using Superscript III reverse transcriptase (Invitrogen) according to the above-mentioned protocol, and the output cDNA was used as a template for the multiplex nested PCR assays. For DNA pathogens, specific cDNA targets were also quantified by UV spectrometry. The DNA targets with known copy numbers were then serially diluted to serve as templates for the multiplex nested PCR assays.
Furthermore, for cultivable viruses, the method of limiting dilution was used to compare the analytical sensitivity of the multiplex nested PCR assays with that of virus isolation, and in the case of bacterial pathogens, the detection limit of the multiplex nested PCR assays was expressed as CFU/milliliter.
Evaluation of clinical specimens.
A total of 303 nasopharyngeal aspirate (NPA) specimens were collected for this study. These NPA samples were taken from patients who were admitted to the Prince of Wales Hospital for suspected respiratory tract infections. The specimens were kept at 4 to 10°C during transportation and temporary storage and were processed on the same day of collection. Two hundred thirty-five specimens were obtained from pediatric patients aged 1 month to 5 years (mean, 2 years of age). The other 68 specimens were obtained from elderly patients aged 65 to 107 years (mean, 65 years of age). Upon receipt, the specimens were separated into two halves, with one half submitted to routine virus isolation and antigen detection by immunofluorescence assay (IFA) and the other half used for the multiplex nested PCR.
Direct immunofluorescent test (IFA).
Specimens were tested for influenza A virus, influenza B virus, PIV (types 1 to 3), respiratory syncytial virus, and ADV by use of a direct immunofluorescence test to screen for the presence of respiratory viruses. Briefly, the respiratory specimens were washed with phosphate-buffered saline. One drop of cell suspension was coated onto a 12-well slide and allowed to dry. The slide was fixed in 100% acetone and incubated with virus-specific mouse fluorescein isothiocyanate-conjugated monoclonal antibodies (Chemicon, Temecula, CA) for 30 min at 37°C. Slides were washed with phosphate-buffered saline and read using a fluorescence microscope. The presence of bright green fluorescence within intact cells was considered to be a positive result. The results were confirmed by two experienced technicians.
Virus isolation.
A 200-μl aliquot of specimen was inoculated onto HEp-2, MDCK, and LLC-MK2 cell monolayers. After 1 h of adsorption at 37°C, maintenance medium was added, and the tubes were incubated at 37°C for HEp-2 cells and at 33°C for MDCK and LLC-MK2 cells. The HEp-2 tubes were incubated for 14 days and examined daily for a viral cytopathic effect. The hemadsorption assay was performed on day 10 for MDCK and LLC-MK2 cells. The positive growth of viruses was confirmed by IFA using virus-specific antibodies. All specimens were tested for FluA, FluB, PIV (types 1 to 3), hRSV, and ADV using IFA as previously described (13). Cells were collected and stained by use of standard methods (16). The results were confirmed by two experienced technicians.
RESULTS
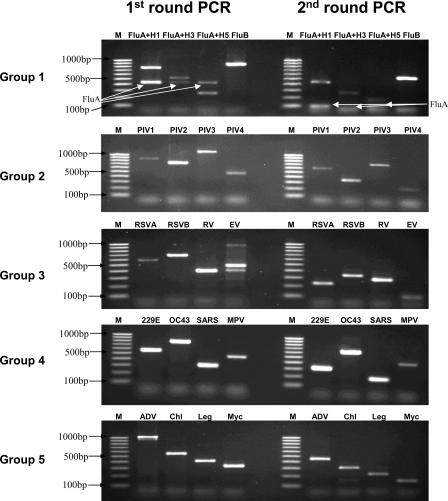
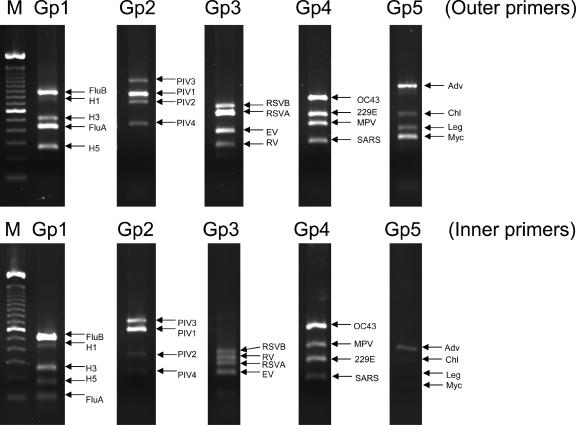
All five multiplex nested PCR assays produced amplification products with the expected sizes, which were clearly distinguishable by agarose gel electrophoresis (Fig. 1). The ability of the assays to detect multiple infections is shown in Fig. 2. It was found that multiple infections did not reduce the sensitivity of the assays. Testing of the 50 clinical specimens known to contain pathogens did not reveal any nonspecific cross-amplification.
FIG. 1.
Agarose gel (2%) electrophoresis showing the first and second rounds of multiplex nested PCR products. M, marker (100-bp ladder); RSV, hRSV; RV, hRV; EV, hEV; MPV, hMPV; Chl, C. pneumoniae; Leg, L. pneumophila; Myc, M. pneumoniae.
FIG. 2.
Agarose gel (2%) electrophoresis showing the first and second rounds of the group 1 to group 5 (Gp1 to Gp5) multiplex nested PCR products using a mixture of pathogens as a template. M, marker (100-bp ladder); RSV, hRSV; RV, hRV; EV, hEV; MPV, hMPV; Chl, C. pneumoniae; Leg, L. pneumophila; Myc, M. pneumoniae.
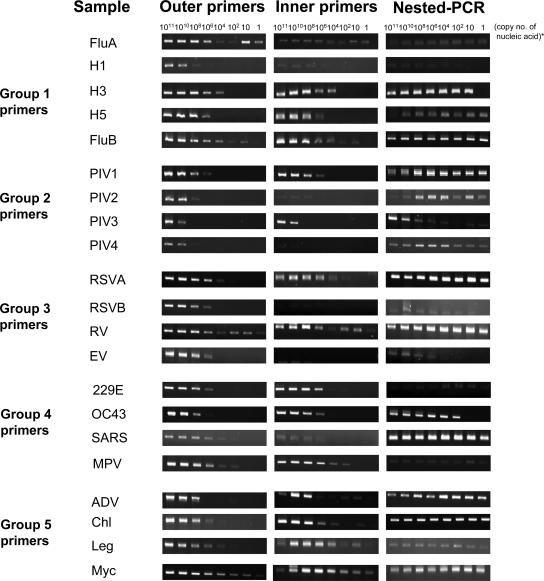
The analyses of analytic sensitivity showed that the multiplex nested PCR assays were highly sensitive, with a low detection limit of less than 10 copies of the target nucleic acids, with the exception of enteroviruses that could still reach a detection limit of 104 copies of nucleic acids (Fig. 3 and Table 3).
FIG. 3.
Agarose gel (2%) electrophoresis showing the performance of the five outer and inner primer groups as well as nested PCR of the multiplex assays. A single template was added to each corresponding group of primers to test the sensitivity of the assay in detecting that pathogen. For the nested PCR limit detection test, the PCR products from the first-round PCR are used as a template for the second-round PCR (copy number of nucleic acids − copy number of RNA or DNA template in the original samples). RSV, hRSV; RV, hRV; EV, hEV; MPV, hMPV; Chl, C. pneumoniae; Leg, L. pneumophila; Myc, M. pneumoniae.
TABLE 3.
Performance of individual and nested primer pairs in the corresponding multiplex nested PCR assay group
| Organism | Detection limit (copy no. of nucleic acids)
|
||
|---|---|---|---|
| Outer primera | Inner primera | Nested PCRb | |
| Group 1 | |||
| Influenza A virus | 1 | 1 | 1 |
| Influenza A virus H1 | 106 | 106 | 102 |
| Influenza A virus H3 | 103 | 104 | 10 |
| Influenza A virus H5 | 106 | 106 | 1 |
| Influenza B virus | 1 | 10 | 1 |
| Group 2 | |||
| PIV-1 | 106 | 106 | 1 |
| PIV-2 | 108 | 106 | 1 |
| PIV-3 | 1010 | 106 | 1 |
| PIV-4a,b | 108 | 1010 | 1 |
| Group 3 | |||
| hRSV A | 102 | 102 | 1 |
| hRSV B | 104 | 106 | 10 |
| hRV | 1 | 1 | 1 |
| hEV | 106 | 108 | 104 |
| Group 4 | |||
| HCoV-OC43 | 106 | 104 | 10 |
| SARS-CoV | 102 | 106 | 1 |
| HCoV-229E | 106 | 102 | 1 |
| hMPV | 102 | 102 | 1 |
| Group 5 | |||
| M. pneumoniae | 1 | 1 | 1 |
| C. pneumoniae | 104 | 1 | 1 |
| L. pneumophila | 102 | 1 | 1 |
| ADV (A to F) | 108 | 1010 | 1 |
The detection limits of outer and inner primers were tested in the form of multiplex primer mix.
The PCR products from the first-round PCR are used as a template for the second-round PCR in the nested PCR limit detection test.
The sensitivity of multiplex nested PCR assays to detect cultivable viruses was found to be 100- to 1,000-fold more sensitive than virus isolation by cell culture. The detection limit of group 5 multiplex nested PCR for Legionella pneumophila and Mycoplasma pneumoniae was 1,000 to 10,000 CFU/milliliter.
A total of 303 NPA specimens were tested using virus isolation, IFA, and multiplex nested PCR. Altogether, 61 specimens were positive by virus isolation, and 41 specimens were positive by IFA. All these isolation- or IFA-positive specimens were also found to be positive by multiplex nested PCR, with the same corresponding viruses detected (Table 4). The overall positive rate as determined by multiplex nested PCR was 48.5% (95% confidence interval [CI], 42.9 to 54.1%), which was significantly higher than those of virus isolation (20.1% [95% CI, 15.6 to 24.6%]) and IFA (13.5% [95% CI, 9.7 to 17.4%]). The positive rates for each pathogen with respect to detection method are shown in Table 4.
TABLE 4.
Performance of multiplex nested PCR assays compared to conventional methods
| Organism | No. (%) of positive specimens (n = 303)
|
||
|---|---|---|---|
| Multiplex nested PCR | Virus isolation | IFA | |
| Any infection | 147 (48.5) | 61 (20.1) | 41 (13.5) |
| Single infection | 140 (46.2) | 61 (20.1) | 41 (13.5) |
| Influenza A virus | 19 (6.3) | 15 (5.0) | 11 (3.6) |
| Influenza A virus H1 | 17 (5.6) | —a | —a |
| Influenza A virus H3 | 2 (0.7) | —a | —a |
| Influenza A virus H5 | 0 | —a | —a |
| Influenza B virus | 10 (3.3) | 9 (3.0) | 7 (2.3) |
| PIV-1 | 19 (6.3) | 14 (4.6) | 10 (3.3) |
| PIV-2 | 6 (2.0) | 1 (0.3) | 1 (0.3) |
| PIV-3 | 3 (1.0) | 2 (0.7) | 1 (0.3) |
| PIV-4 | 2 (0.7) | —a | —a |
| Respiratory syncytial virus | 8 (2.7) | 5 (1.7) | 6 (1.9) |
| Respiratory syncytial virus group A | 5 (1.7) | —a | —a |
| Respiratory syncytial virus group B | 3 (1.0) | —a | —a |
| Rhinovirus | 16 (5.3) | —a | —a |
| Enteroviruses | 3 (1.0) | 0 | 0 |
| HCoV-OC43 | 16 (5.3) | —a | —a |
| HCoV-229E | 3 (1.0) | —a | —a |
| SARS-CoV | 0 | 0 | 0 |
| hMPV | 15 (5.0) | —a | —a |
| M. pneumoniae | 5 (1.7) | —a | —a |
| L. pneumophila | 0 | —a | —a |
| C. pneumoniae | 0 | —a | —a |
| ADVs | 15 (5.0) | 15 (5.0) | 5 (1.7) |
| Coinfection | 7 (2.3) | 0 | 0 |
| Influenza A virus and M. pneumoniae | 1 (0.3) | —a | —a |
| Influenza A virus H1 and C. pneumoniae | 1 (0.3) | —a | —a |
| Influenza A virus H3 and HCoV-229E | 1 (0.3) | —a | —a |
| Influenza A virus H3 and PIV-2 | 1 (0.3) | —b | —b |
| hMPV and M. pneumoniae | 2 (0.7) | —a | —a |
| hMPV and PIV-4 | 1 (0.3) | —a | —a |
Organisms not isolated/differentiated by virus isolation.
PIV-2 was isolated.
A subgroup analysis was performed on viruses that were detected by virus isolation. All these cultivatable viruses showed a higher positive rate by multiplex nested PCR than by virus isolation, except for ADV. However, the differences were not statistically significant (the positive rate [95% CI] for FluA by PCR versus isolation was 6.3% [3.5 to 9.0%] versus 5.0% [2.5 to 7.4%], respectively; the positive rate for FluB by PCR versus isolation was 3.3% [1.3 to 5.3%] versus 3.0% [1.1 to 4.9%], respectively; the positive rate for PIV-1 by PCR versus isolation was 6.3% [3.5 to 9.0%] versus 4.6% [2.3 to 7.0%], respectively; the positive rate for PIV-2 by PCR versus isolation was 2.0% [0.4 to 3.5%] versus 0.3% [0 to 1.0%], respectively; the positive rate for PIV-3 by PCR versus isolation was 1.0% [0 to 2.1%] versus 0.7% [0 to 1.6%], respectively; the positive rate for hRSV by PCR versus isolation was 2.7% [0.8 to 4.4%] versus 1.7% [0.2 to 3.1%], respectively; the positive rate for hEV by PCR versus isolation was 1.0% [0 to 2.1%] versus 0% [0%], respectively; and the positive rate for ADV by PCR versus isolation was 5.0% [2.5 to 7.4%] versus 5.0% [2.5 to 7.4%], respectively). Compared to virus isolation, the overall gain in the positive rate for this group of cultivatable viruses as achieved by multiplex nested PCR was an increase from 20.1% to 29.7%.
When the group of viruses that can be diagnosed by direct detection using IFA was compared, the positive rate obtained by multiplex nested PCR was higher than that of IFA for all the viruses. However, again, the differences were not statistically significant. The positive rate (95% CI) for FluA by PCR versus IFA was 6.3% (3.5 to 9.0%) versus 3.6% (1.5 to 5.7%), respectively; that for FluB by PCR versus IFA was 3.3% (1.3 to 5.3%) versus 2.3% (0.6 to 4.0%), respectively; that for PIV-1 by PCR versus IFA was 6.3% (3.5 to 9.0%) versus 3.3% (1.3 to 5.3%), respectively; that for PIV-2 by PCR versus IFA was 2.0% (0.4 to 3.5%) versus 0.3% (0 to 1.0%), respectively; that for PIV-3 by PCR versus IFA was 1.0% (0 to 2.1%) versus 0.3% (0 to 1.0%), respectively; that for hRSV by PCR versus IFA was 2.7% (0.8 to 4.4%) versus 2.0% (0.4 to 3.5%), respectively; that for hEV by PCR versus IFA was 1.0% (0 to 2.1%) versus 0%, respectively; and that for ADV by PCR versus IFA was 5.0% (2.5 to 7.4%) versus 1.7% (0.2 to 3.1%), respectively. The overall gain in the positive rate achieved by multiplex nested PCR for this group of viruses was an increase from 13.5% to 27.7%.
Of the 21 pathogens included in the study, 7 were not detectable by isolation or IFA. Within this group, three were commonly found in our study samples, including rhinovirus, with a positive rate of 5.3%, HCoV-OC43 (5.3%), and hMPV (5.0%). Overall, these three viruses contributed 34.0% of the PCR-positive cases.
Multiple respiratory viruses were observed in 7 of the 303 (2.3%) specimens (Table 4). None of these coinfections were detected by virus isolation or IFA, as the majority of them contained a noncultivatable organism. These cases contributed 4.8% of PCR-positive cases.
Another advantage of this multiplex nested PCR was that it could be used to subtype pathogens in the same testing cycle. For the 19 FluA cases detected in multiplex nested PCR, 17 were H1 infections, and 2 were H3 infections. For the eight cases of hRSV identified, five were hRSV A infections and three were hRSV B infections. These subtyping results could be obtained directly by agarose gel electrophoresis without further testing.
DISCUSSION
Respiratory tract infection accounts for a majority of the admissions in acute care hospitals. While it has long been recognized that viruses contribute to a significant proportion of these cases, the urgency for laboratory diagnosis remains paradoxically low in most settings. One of the main reasons is the long turnaround time of conventional virus detection methods and their inability to detect fastidious viruses. The lack of specific treatment for most viral infections is another practical consideration when prioritizing laboratory resources. The outbreak of SARS and the threat of avian influenza virus reawakened the need for rapid diagnosis to enable the prompt and accurate diagnosis of index cases.
In this study, we sought to develop and evaluate multiplex nested PCR assays for the rapid and accurate diagnosis of respiratory tract infections. The rationale for primer groupings was as follows. Firstly, RNA pathogens and DNA pathogens were separated, i.e., groups 1 to 4 for RNA pathogens and group 5 for DNA pathogens. Secondly, pathogens of the same or similar family were grouped together, for example, FluA (H1 to H5) and FluB were grouped together into group 1. PIVs were grouped into group 2. In this way, each family member amplified within the same PCR could be easily differentiated. Thirdly, PCR product size was another factor affecting multiplex grouping. For example, the primers designed for hRSVA, hRSVB, hEV, and hRV were compatible to form a multiplex. Fourthly, only four pathogens were included in each group because, on one hand, the size of the PCR products being amplified would be very suitable for visual differentiation on agarose gel and, on the other hand, the amplification efficiency for each PCR would not be jeopardized too much by multiplexing. For example, if too many pathogens were included in a single multiplex reaction, in order to have sufficient visual differentiation of PCR products on an agarose gel, some of the PCR products would need to be very large, and that might lower the sensitivity of the pathogens being amplified.
Molecular techniques have increased the speed and sensitivity with which such pathogens can be detected and allow laboratories to identify organisms that do not grow or grow slowly in conventional viral culture. However, the gain in analytical sensitivity may not necessarily be reflected in clinical situations. For instance, in settings where clinical specimens are collected and maintained in good quality, the amount of virus present may well be enough for detection by the “less sensitive” conventional methods (culture and IFA). In fact, our data are in line with this. When cultivatable viruses were compared, despite the finding that a higher sensitivity for multiplex nested PCR was observed, the differences were not statistically significant. In particular, we observed the same positive rate for ADV using both PCR and conventional virus isolation methods. In a previous study, a discrepancy between direct detection and RT-PCR for ADV was also reported (19). Although the lack of a statistically significant improvement in a positive detection rate could be due to a low general prevalence of the individual organism, the overall results indicate that the main impact of this multiplex nested PCR was its broader spectrum of detection. Expanding the detection spectrum has also been the main focus of previous studies, and as many as nine different respiratory pathogens have been targeted (11, 15, 22, 27). In the current study, we included 21 respiratory pathogens and provided the widest spectrum ever reported. We found that the gain in the overall positive detection rate from clinical specimens was attributed mainly to the inclusion of hRV, HCoV-OC43, and hMPV detection. All these viruses are not detectable by conventional cell culture isolation or direct detection using IFA. The improvement in the diagnostic yield by adding hRV was also reported previously by Gruteke et al. (14). Given that these “trivial” respiratory viruses can cause severe illnesses (6, 20), they should be included in the development of multiplex assays. Another advantage of multiplex nested PCR as demonstrated in this study is the ability to detect coinfections, although the overall improvement in the positive rate was not substantial due to the relatively few instances of coinfection in our study cohort.
While multiplex nested PCR assays may be more economical due to the fact that multiple pathogens can be detected in a single assay without a proportional increase in reagent costs, they have their drawbacks. First, their detection sensitivities are often lower than those of equivalent monoplex PCR assays. In this study, only about 30% of the positive specimens showed a positive result from the first round of PCR. This finding indicates the need for a nested PCR, which may be associated with a higher risk of cross-contamination. Second, the presence of several pairs of primers in a PCR increases the probabilities of mispairing and nonspecific amplification, particularly the formation of primer-dimers.
In group 1 multiplex nested PCR, we incorporated specific primers for influenza A virus subtypes H1, H3, and H5. This is important in the context where rapid differentiation between H5 and non-H5 influenza virus is necessary. The group 1 multiplex nested PCR assay was also comprised of primer pairs targeting the consensus region of influenza A virus. This would allow the detection of non-H1/H3/H5 subtypes, which may occasionally cause human infections (e.g., H7 and H9 influenza viruses).
A nucleic acid extraction kit that can extract both viral DNA and RNA simultaneously was used in our study. This can minimize the amount of samples required for the detection of both DNA and RNA viruses. At the same time, this can minimize the time, labor, and materials involved in nucleic acid extraction.
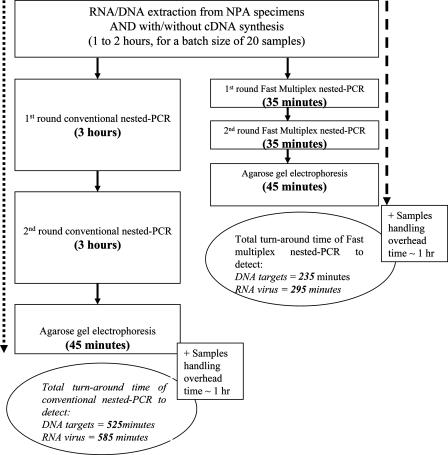
Also, a newly available fast thermal cycler was used in our study, which allowed rapid cycling, shortening the time required to complete the PCR. The thermal cycler's patented sample temperature control provided a quick and uniform thermal response. Therefore, the cycling times for the first- and second-round PCRs were considerably reduced. Furthermore, the GeneAmp fast PCR master mix allows a two-step PCR (same temperature holding for the annealing and extension steps) instead of the more conventional three-step PCR. With the use of this fast PCR system, the time required for a single round of PCR was reduced from 3 h to 35 min, i.e., saving a total of about 300 min in a nested PCR assay (Fig. 4). Therefore, the whole testing process can be completed within 1 day. This rapid turnaround not only is critical in urgent outbreak investigations but may potentially decrease the overall costs for the hospital, as has been shown in previous studies (1, 30). However, one disadvantage of using this fast PCR system is that primers used in standard multiplex PCR assays need to be redesigned with higher annealing temperatures.
FIG. 4.
Estimated turnaround times for the fast multiplex nested PCR protocol and conventional nested PCR protocol.
The multiplex nested PCR assays developed in this study improved the diagnostic yield in terms of the overall sensitivity as well as the spectrum of coverage for respiratory infections. Furthermore, the assay provided a rapid turnaround time, with results being available within the same day of specimen collection. The overall cost reduction may justify the routine use of these broader-spectrum, rapid molecular diagnostic assays.
Acknowledgments
This study was supported by the Research Fund for the Control of Infectious Diseases from the Health, Welfare, and Food Bureau of the Hong Kong Special Administrative Region Government (project no. 01030782).
Part of the work was performed at the Lo Kwee Cheong Research Laboratory.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Adcock, P. M., G. G. Stout, M. A. Hauck, and G. S. Marshall. 1997. Effect of rapid viral diagnosis on the management of children hospitalized with lower respiratory tract infection. Pediatr. Infect. Dis. J. 16:842-846. [DOI] [PubMed] [Google Scholar]
- 2.Bellau-Pujol, S., A. Vabret, L. Legrand, J. Dina, S. Gouarin, J. Petitjean-Lecherbonnier, B. Pozzetto, C. Ginevra, and F. Freymuth. 2005. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods 126:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billaud, G., S. Peny, V. Legay, B. Lina, and M. Valette. 2003. Detection of rhinovirus and enterovirus in upper respiratory tract samples using a multiplex nested PCR. J. Virol. Methods 108:223-228. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, G., S. Cote, P. Dery, G. De Serres, and M. G. Bergeron. 2004. Multiplex real-time PCR assay for detection of influenza and human respiratory syncytial viruses. J. Clin. Microbiol. 42:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casiano-Colon, A. E., B. B. Hulbert, T. K. Mayer, E. E. Walsh, and A. R. Falsey. 2003. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J. Clin. Virol. 28:169-174. [DOI] [PubMed] [Google Scholar]
- 6.Chidekel, A. S., C. L. Rosen, and A. R. Bazzy. 1997. Rhinovirus infection associated with serious lower respiratory illness in patients with bronchopulmonary dysplasia. Pediatr. Infect. Dis. J. 16:43-47. [DOI] [PubMed] [Google Scholar]
- 7.Coiras, M. T., P. Perez-Brena, M. L. Garcia, and I. Casas. 2003. Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested-PCR assay. J. Med. Virol. 69:132-144. [DOI] [PubMed] [Google Scholar]
- 8.Coiras, M. T., J. C. Aguilar, M. L. Garcia, I. Casas, and P. Perez-Brena. 2004. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J. Med. Virol. 72:484-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cote, S., Y. Abed, and G. Boivin. 2003. Comparative evaluation of real-time PCR assays for detection of the human metapneumovirus. J. Clin. Microbiol. 41:3631-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debbia, E. A., G. C. Schito, A. Zoratti, L. Gualco, E. Tonoli, and A. Marchese. 2001. Epidemiology of major respiratory pathogens. J. Chemother. 13:205-210. [DOI] [PubMed] [Google Scholar]
- 11.Fan, J., K. J. Henrickson, and L. L. Savatski. 1998. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex). Clin. Infect. Dis. 26:1397-1402. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Sabe, N., J. Carratala, B. Roson, J. Dorca, R. Verdaguer, F. Manresa, and F. Gudiol. 2003. Community-acquired pneumonia in very elderly patients: causative organisms, clinical characteristics, and outcomes. Medicine (Baltimore) 82:159-169. [DOI] [PubMed] [Google Scholar]
- 13.Freymuth, F., M. Quibriac, J. Petitjean, F. Daon, and M. L. Amiel. 1987. Viruses responsible for respiratory infections in pediatrics. Evaluation of 3,480 nasal aspirates performed in children over a 6-year period. Ann. Pediatr. (Paris) 34:493-501. (In French.) [PubMed] [Google Scholar]
- 14.Grondahl, B., W. Puppe, A. Hoppe, I. Kuhne, J. A. Weigl, and H. J. Schmitt. 1999. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 37:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruteke, P., A. S. Glas, M. Dierdorp, W. B. Vreede, J. W. Pilon, and S. M. Bruisten. 2004. Practical implementation of a multiplex PCR for acute respiratory tract infections in children. J. Clin. Microbiol. 42:5596-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehl, S. C., K. J. Henrickson, W. Hua, and J. Fan. 2001. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J. Clin. Microbiol. 39:1696-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kendal, A. P., M. S. Pereira, and J. J. Skehel. 1982. Concepts and procedures for laboratory-based influenza surveillance. WHO Collaborating Centers for Reference and Research on Influenza, Geneva, Switzerland.
- 18.McDonough, E. A., C. P. Barrozo, K. L. Russell, and D. Metzgar. 2005. A multiplex PCR for detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and Bordetella pertussis in clinical specimens. Mol. Cell. Probes 19:314-322. [DOI] [PubMed] [Google Scholar]
- 19.Monto, A. S. 2002. Epidemiology of viral respiratory infections. Am. J. Med. 112(Suppl. 6A):4S-12S. [DOI] [PubMed] [Google Scholar]
- 20.Osiowy, C. 1998. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J. Clin. Microbiol. 36:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papadopoulos, N. G., P. J. Bates, P. G. Bardin, A. Papi, S. H. Leir, D. J. Fraenkel, J. Meyer, P. M. Lackie, G. Sanderson, S. T. Holgate, and S. L. Johnston. 2000. Rhinoviruses infect the lower airways. J. Infect. Dis. 181:1875-1884. [DOI] [PubMed] [Google Scholar]
- 22.Santti, J., T. Hyypia, and P. Halonen. 1997. Comparison of PCR primer pairs in the detection of human rhinoviruses in nasopharyngeal aspirates. J. Virol. Methods 66:139-147. [DOI] [PubMed] [Google Scholar]
- 23.Stockton, J., J. S. Ellis, M. Saville, J. P. Clewley, and M. C. Zambon. 1998. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J. Clin. Microbiol. 36:2990-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stralin, K., A. Backman, H. Holmberg, H. Fredlund, and P. Olcen. 2005. Design of a multiplex PCR for Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae and Chlamydophila pneumoniae to be used on sputum samples. APMIS 113:99-111. [DOI] [PubMed] [Google Scholar]
- 25.Templeton, K. E., S. A. Scheltinga, M. F. Beersma, A. C. Kroes, and E. C. Claas. 2004. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 42:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vabret, A., F. Mouthon, T. Mourez, S. Gouarin, J. Petitjean, and F. Freymuth. 2001. Direct diagnosis of human respiratory coronaviruses 229E and OC43 by the polymerase chain reaction. J. Virol. Methods 97:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valassina, M., A. M. Cuppone, M. G. Cusi, and P. E. Valensin. 1997. Rapid detection of different RNA respiratory virus species by multiplex RT-PCR: application to clinical specimens. Clin. Diagn. Virol. 8:227-232. [DOI] [PubMed] [Google Scholar]
- 29.Vijgen, L., E. Keyaerts, E. Moes, P. Maes, G. Duson, and M. Van Ranst. 2005. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J. Clin. Microbiol. 43:5452-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward, C. L., M. H. Dempsey, C. J. Ring, R. E. Kempson, L. Zhang, D. Gor, B. W. Snowden, and M. Tisdale. 2004. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J. Clin. Virol. 29:179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo, P. C., S. S. Chiu, W. H. Seto, and M. Peiris. 1997. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J. Clin. Microbiol. 35:1579-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization. 5 July 2007, accession date. PCR primers for SARS developed by WHO Network Laboratories. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/sars/primers/en/print/html.
- 33.Xu, W., M. C. McDonough, and D. D. Erdman. 2000. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol. 38:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, W., and D. D. Erdman. 2001. Type-specific identification of human adenovirus 3, 7, and 21 by a multiplex PCR assay. J. Med. Virol. 64:537-542. [DOI] [PubMed] [Google Scholar]