Abstract
We report a case of pyomyositis due to Mycobacterium haemophilum in a renal transplant recipient. M. haemophilum was identified by PCR-mediated sequence analysis of the heat shock protein gene in the DNA of the specimen. The patient was successfully treated with repeated surgical debridement and prolonged antimycobacterial therapy.
CASE REPORT
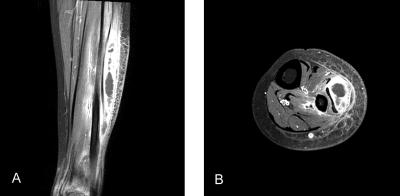
A 24-year-old female renal transplant recipient with a known history of chronic renal failure secondary to chronic rejection was admitted to our hospital for tender, erythematous, and palpably fluctuant swelling on the left calf. She had undergone kidney transplantation 8 years earlier, after which she had been on immunosuppressive treatment with cyclosporine A and mycophenolate mofetil. Contrast-enhanced, T1-weighted magnetic resonance imaging (MRI) showed a localized area of hypointensity with an ill-defined enhancing peripheral rim on the peroneus muscle and heterogeneous enhancement on the gastrocnemius muscle (Fig. 1). The patient underwent surgical debridement of the site, which revealed a collection of purulent fluid deep in the peroneus and gastrocnemius muscles that appeared to be a full-blown abscess. Histopathologic examination of tissue samples revealed granulated tissue with granulomatous inflammation, and Ziehl-Neelsen staining revealed numerous acid-fast bacilli (AFB). Antituberculosis therapy with isoniazid, rifampin, pyrazinamide, and ofloxacin was initiated for presumed tuberculous pyomyositis.
FIG. 1.
(A) Coronal T1-weighted MRI with contrast enhancement, showing a rim of increased signal intensity around a central hypointense region on the peroneus muscle and heterogeneously increased signal intensity on the gastrocnemius muscle. (B) Axial T1-weighted MRI with contrast enhancement, showing findings consistent with the coronal view.
The patient's condition remained stable for about 2 months after initiation of antituberculosis therapy, until she developed painful swelling and purulent drainage on the left calf. Because of the patient's clinical deterioration, mycophenolate mofetil was discontinued and the dosage of cyclosporine was reduced. The initial culture of the previous operative specimen in Ogawa egg medium, which required 8 weeks of incubation, was negative for mycobacteria. Considering the negative AFB culture and clinical aggravation despite 2 months of antituberculosis therapy, AFB-positive organisms other than Mycobacterium tuberculosis were suggested. Because the morphology of the AFB-positive organisms was totally different from that of Nocardia, nontuberculous mycobacterium (NTM) infection was strongly suspected.
To identify the species of mycobacteria, a 422-bp fragment of the heat shock protein gene (hsp65) was PCR amplified in DNA isolated from the AFB smear-positive pus, using the primers 5′-ATCGCCAAGGAGATCGAGCT-3′ (forward) and 5′-AAGGTGCCGCGGATCTTGTT-3′ (reverse), and the PCR product was directly sequenced (5). The resultant partial hsp65 DNA sequences were aligned and compared with those of mycobacterial reference strains. The similarity among sequences was determined using the multiple-alignment algorithm in the MEGALIGN software package (Windows version 3.12e; DNASTAR). The amplified sequence was 99% identical to that of Mycobacterium haemophilum hsp65. The patient's antimycobacterial therapy was changed to treatment with rifampin, clarithromycin, and ciprofloxacin.
Subsequently, the pus draining from her left calf was inoculated separately upon our request. An inoculation on chocolate agar was incubated at 30°C, and another inoculation in a BACTEC MGIT tube (BD Biosciences, Sparks, MD) was incubated on the BACTEC MGIT 960 system (BD Biosciences) at 37°C. Multiple rough, tiny, and nonpigmented colonies appeared after 10 days on chocolate agar. The positive signal was detected 12 days after MGIT culture, and the subculture of the broth medium yielded the same colonies on chocolate agar. DNA was extracted from these colonies, and PCR-restriction fragment length polymorphism analysis was carried out using Myco-ID (Molecules & Diagnostics, Wonju, Korea) (7). Digestion of the rpoB amplicon by MspI (Boehringer Mannheim Biochemicals, Mannheim, Germany) revealed restriction fragments compatible with the pattern of M. haemophilum.
Even after the change of antimycobacterial agents, however, three further debridements and explorations of the wound were required, after which the patient finally began to improve. M. haemophilum was isolated repeatedly from her operative biopsy specimen, but her final operative specimen, 6 weeks after the change of antimycobacterial therapy, was negative for M. haemophilum. Subsequently, her skin lesions gradually improved, and she was discharged on hospital day 130. During a follow-up period of 1 year, during which she continued treatment with antimycobacterial agents, no relapse was observed.
The recent increase in the incidence of NTM infections may reflect the increased number of immunocompromised hosts and/or the improved diagnostic capabilities of laboratories (9). In addition, the spectrum of disease caused by NTM species has expanded considerably and is likely to expand further over time (9). M. haemophilum has been identified primarily as a cause of cutaneous or subcutaneous lesions, although infections at extracutaneous sites, such as tenosynovitis, septic arthritis, osteomyelitis, pneumonitis, and septicemia, have also been described previously (11, 12).
In our search of the medical literature written in English, we identified three cases of pyomyositis caused by NTM (3, 6, 13), only one of which was caused by M. haemophilum. The other two cases were caused by Mycobacterium avium complex and were associated with AIDS (3, 6). In the case of pyomyositis due to M. haemophilum, the patient had been on long-term steroid treatment for polymyositis and presented with ulcerations over both thighs and the left arm, with M. haemophilum confirmed by a fatty acid profile generated by gas-liquid chromatography (13). This patient required repeated debridement and combination antimycobacterial therapy. Prior to clinical improvement, however, this patient died of fungemia due to Candida glabrata, and the clinical course and outcome could not be determined. Our patient, who also required repeated debridement and combination antimycobacterial therapy, showed complete clinical resolution without other serious complications.
M. haemophilum is a slowly growing NTM that requires distinctive culture conditions. For example, it grows optimally at cooler temperatures (30 to 32°C) and requires hemin for growth (hence its name) (11, 12). Therefore, the incidence of infection due to M. haemophilum is likely underestimated. In our case, the initial culture failed to isolate M. haemophilum because routine culture conditions were not suitable for this organism. When M. haemophilum infection is suspected, the microbiology laboratory must be informed so that the appropriate culture conditions can be prepared (11, 12). It would be more desirable to include routine culture for M. haemophilum with appropriate specimens. Also, it is reasonable to perform both solid- and liquid-based culture for mycobacterial cultures (10). In our case, if MGIT culture had been done from the beginning, early diagnosis could have been possible.
Although bacterial culture remains the standard method for the diagnosis of M. haemophilum infection, it is time-consuming and laborious, usually taking up to 3 to 4 weeks. Recent advances in various PCR-mediated methods have led to the rapid and sensitive differentiation of Mycobacterium species from clinical specimens or culture isolates. The target molecules include 16S rRNA, the 16S-23S internally transcribed spacer, and the rpoB and hsp65 genes. These molecular analyses are simple, rapid, and reliable methods for differentiating mycobacterial species (2, 4, 5, 7, 8). The lack of bacterial growth with routine culture methods led to the use of PCR-automated hsp65 DNA sequencing performed directly on pus of the patient to diagnose M. haemophilum infection. Identification was confirmed through PCR-restriction fragment length polymorphism analysis, based on the rpoB gene, of cultured isolates, using special culture conditions for M. haemophilum. The hsp65 and rpoB genes have been reported to better discriminate among mycobacteria than the 16S rRNA gene due to the limitations of the latter, including the presence of two different genes in a single organism and sequence identities within obviously different species (4, 5). The usefulness of applying this method as the primary means for identifying all mycobacteria in routine clinical laboratories remains to be determined (2, 8).
There is no consensus on the choice and duration of treatment with antimicrobial agents or on the role of surgical debridement for M. haemophilum infection. Based on limited data from individual cases, a combination of ciprofloxacin, clarithromycin, and one of the rifamycins appears effective (1, 11, 12). The duration of therapy should depend on the patient's underlying disease presentation, degree of immunosuppression, and response to therapy. Treatment should usually be continued for at least 1 year and perhaps for as long as 2 years (11, 12). Surgical drainage with debridement of necrotic tissue is required when extensive inflammation is present. Our patient required four surgical debridements.
Our case illustrates the importance of this species as the cause of opportunistic infection in transplant recipients. Clinicians should consider M. haemophilum in the differential diagnosis of pyomyositis in immunosuppressed patients. A prolonged course of multidrug therapy and intensive surgical intervention may be required to manage this disease.
Footnotes
Published ahead of print on 26 September 2007.
REFERENCES
- 1.Bernard, E. M., F. F. Edwards, T. E. Kiehn, S. T. Brown, and D. Armstrong. 1993. Activities of antimicrobial agents against clinical isolates of Mycobacterium haemophilum. Antimicrob. Agents Chemother. 37:2323-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheunoy, W., T. Prammananan, A. Chaiprasert, and S. Foongladda. 2005. Comparative evaluation of polymerase chain reaction and restriction enzyme analysis: two amplified targets, hsp65 and rpoB, for identification of cultured mycobacteria. Diagn. Microbiol. Infect. Dis. 51:165-171. [DOI] [PubMed] [Google Scholar]
- 3.Diego Miralles, G., and Z. Bregman. 1994. Necrotizing pyomyositis caused by Mycobacterium avium complex in a patient with AIDS. Clin. Infect. Dis. 18:833-834. [DOI] [PubMed] [Google Scholar]
- 4.Kim, B. J., S. H. Lee, M. A. Lyu, S. J. Kim, G. H. Bai, G. T. Chae, E. C. Kim, C. Y. Cha, and Y. H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 37:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim, H., S. H. Kim, T. S. Shim, M. N. Kim, G. H. Bai, Y. G. Park, S. H. Lee, G. T. Chae, C. Y. Cha, Y. H. Kook, and B. J. Kim. 2005. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int. J. Syst. Evol. Microbiol. 55:1649-1656. [DOI] [PubMed] [Google Scholar]
- 6.Lawn, S. D., T. A. Bicanic, and D. C. Macallan. 2004. Pyomyositis and cutaneous abscesses due to Mycobacterium avium: an immune reconstitution manifestation in a patient with AIDS. Clin. Infect. Dis. 38:461-463. [DOI] [PubMed] [Google Scholar]
- 7.Lee, H., H. J. Park, S. N. Cho, G. H. Bai, and S. J. Kim. 2000. Species identification of mycobacteria by PCR-restriction fragment length polymorphism of the rpoB gene. J. Clin. Microbiol. 38:2966-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNabb, A., D. Eisler, K. Adie, M. Amos, M. Rodrigues, G. Stephens, W. A. Black, and J. Isaac-Renton. 2004. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J. Clin. Microbiol. 42:3000-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrini, B. 2006. Non-tuberculous mycobacterial infections. Scand. J. Infect. Dis. 38:246-255. [DOI] [PubMed] [Google Scholar]
- 10.Pfyffer, G. E. 2007. Mycobacterium: general characteristics, laboratory detection, and staining procedures, p. 543-572. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 11.Saubolle, M. A., T. E. Kiehn, M. H. White, M. F. Rudinsky, and D. Armstrong. 1996. Mycobacterium haemophilum: microbiology and expanding clinical and geographic spectra of disease in humans. Clin. Microbiol. Rev. 9:435-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah, M. K., A. Sebti, T. E. Kiehn, S. A. Massarella, and K. A. Sepkowitz. 2001. Mycobacterium haemophilum in immunocompromised patients. Clin. Infect. Dis. 33:330-337. [DOI] [PubMed] [Google Scholar]
- 13.Shih, J. Y., P. R. Hsueh, Y. L. Chang, S. F. Lin, L. J. Teng, and K. T. Luh. 1998. Pyomyositis due to Mycobacterium haemophilum in a patient with polymyositis and long-term steroid use. Clin. Infect. Dis. 26:505-507. [DOI] [PubMed] [Google Scholar]