Abstract
Numerous members of the genus Exophiala are potential agents of human and animal mycoses. The majority of these infections are cutaneous and superficial, but also fatal systemic infections are known. We re-identified 188 clinical isolates from the United States, which had a preliminary morphological identification of Exophiala species, by sequencing internal transcribed spacer (ITS) region of the rRNA. Molecular identifications of the strains were as follows, in order of frequency: 55 E. dermatitidis (29.3%), 37 E. xenobiotica (19.7%), 35 E. oligosperma (18.6%), 13 E. lecanii-corni (6.9%), 12 E. phaeomuriformis (6.4%), 7 E. jeanselmei (3.7%), 7 E. bergeri (3.7%), 6 E. mesophila (3.2%), 5 E. spinifera (2.7%), 3 Exophiala sp. 1 (1.6%), 3 E. attenuata (1.6%), 3 Phialophora europaea (1.3%), 1 E. heteromorpha (0.5%), and 1 Exophiala sp. 2 (0.5%) strains. Exophiala strains were repeatedly isolated from deep infections (39.9%) involving lung, pleural fluid, sputum, digestive organs (stomach, intestines, bile), heart, brain, spleen, bone marrow, blood, dialysis fluid, lymph node, joint, breast, middle ear, throat, and intraocular tissues. About 38.3% of the Exophiala spp. strains were agents of cutaneous infections including skin, mucous membranes, nail, and corneal epithelium lesions. The other strains caused superficial infections (0.5%, including hair) or subcutaneous infection (12.0%, including paranasal sinusitis, mycetoma, and subcutaneous cyst). The systemic infections were preponderantly caused by E. dermatitidis, E. oligosperma, E. phaeomuriformis, E. xenobiotica, and E. lecanii-corni. Strains of E. bergeri, E. spinifera, E. jeanselmei, E. mesophila, and E. attenuata mainly induced cutaneous and subcutaneous infections. Since relatively few unknown ITS motifs were encountered, we suppose that the list of opportunistic Exophiala species in temperate climates is nearing completion, but a number of species still have to be described.
Black yeasts of the genus Exophiala are notoriously difficult to classify and identify. In the past, diagnostic schemes were morphological, and physiological parameters were soon added (10, 11). Several species indeed have marked phenetic characteristics, such as the large conidiophores of E. spinifera, or the thermotolerance and absence of nitrite assimilation in E. dermatitidis. The majority of species, however, are morphologically variable, due to their passage through complicated life cycles where diagnostic features are variably expressed (14) and, conversely, very similar microscopic structures can be expressed in phylogenetically remote species. In recent years diagnostic approaches have been supplemented by molecular tools, particularly sequence data of the rRNA internal transcribed spacer (ITS) regions (13, 15, 37).
A significant proportion of the known species are regularly encountered as causative agents of human mycoses (see, for example, references 4, 23, 28, 30, 32, 34, and 37). In harboring a wide array of clinically relevant species, the black yeasts and relatives are unique in the fungal kingdom. Because of the lack of tools for species distinction, Exophiala species have long been viewed as coincidental opportunists, having their prime occurrence as saprobes on plant material. However, when circumscribed according to modern criteria, some species have turned out to be consistent in their ecology and preferred sites of infection (15). This places the possibility of species-specific virulence and antifungal susceptibility in another light.
We analyzed retrospectively a large number of clinical strains preserved at the Fungus Testing Laboratory in the Department of Pathology at the University of Texas Health Science Center at San Antonio and determined their antifungal susceptibility profiles. Given the difficulty of morphological identification, final identifications were reached after sequencing. An overview of identification results showing discrepancies between morphological and molecular identifications can be sent upon request.
MATERIALS AND METHODS
Fungal strains.
A total of 188 clinical strains, previously submitted to the Fungus Testing Laboratory for identification, antifungal susceptibility testing, or both and added to the University of Texas Health Science Center at San Antonio collection were analyzed (Table 1). All isolates were stored at −80°C prior to study and had preliminary morphological identifications as Exophiala spp. Sequences were compared to the Centraalbureau voor Schimmelcultures database, which contains thousands of comparable sequences of environmental and clinical Exophiala species and related black yeast-like fungi (orders Chaetothyriales and Dothideales).
TABLE 1.
Source and identification of strains examined
| Identification | No. of strains
|
|||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deep
|
Subcutaneous
|
Cutaneous
|
||||||||||||||||||||||||||||||
| Lung or pleural fluid | Sputum | Blood | Heart | Stomach, intestine or bile | Stool | Dialysis fluid | Spleen | Bone marrow | Brain biopsy | Lymph node | Breast | Abdominal wound | Throat | Middle ear | Intraocular lesion | Joint | Total | Paranasal sinus | Mycetoma | Subcutaneous cyst | Total | Nail | Skin lesion | Corneal epithelium | Mucous membrane | Total | Superficial infection | Unknown (human) | Animal | Total | % | |
| E. dermatitidis | 16 | 8 | 2 | 1 | 1 | 1 | 1 | 3 | 1 | 2 | 36 | 3 | 1 | 4 | 9 | 2 | 1 | 12 | 3 | 55 | 29.3 | |||||||||||
| E. xenobiotica | 1 | 2 | 2 | 5 | 1 | 2 | 3 | 21 | 1 | 22 | 6 | 1 | 37 | 19.7 | ||||||||||||||||||
| E. oligosperma | 10 | 1 | 1 | 1 | 2 | 1 | 16 | 3 | 2 | 5 | 1 | 10 | 11 | 2 | 1 | 35 | 18.6 | |||||||||||||||
| E. lecanii-corni | 3 | 1 | 4 | 2 | 1 | 3 | 1 | 3 | 4 | 2 | 13 | 6.9 | ||||||||||||||||||||
| E. phaeomuriformis | 4 | 1 | 1 | 1 | 1 | 1 | 9 | 1 | 1 | 1 | 1 | 1 | 12 | 6.4 | ||||||||||||||||||
| E. jeanselmei | 0 | 1 | 1 | 6 | 6 | 7 | 3.7 | |||||||||||||||||||||||||
| E. bergeri | 0 | 1 | 1 | 5 | 1 | 6 | 7 | 3.7 | ||||||||||||||||||||||||
| E. mesophila | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 1 | 6 | 3.2 | ||||||||||||||||||||||
| E. spinifera | 0 | 1 | 1 | 2 | 1 | 1 | 2 | 5 | 2.7 | |||||||||||||||||||||||
| Exophiala sp. 1 | 1 | 1 | 0 | 1 | 1 | 2 | 3 | 1.6 | ||||||||||||||||||||||||
| E. attenuata | 0 | 0 | 3 | 3 | 3 | 1.6 | ||||||||||||||||||||||||||
| Exophiala sp. 2 | 0 | 0 | 0 | 1 | 1 | 0.5 | ||||||||||||||||||||||||||
| E. heteromorpha | 1 | 1 | 0 | 0 | 1 | 0.5 | ||||||||||||||||||||||||||
| P. europaea | 0 | 0 | 1 | 2 | 3 | 3 | 1.6 | |||||||||||||||||||||||||
| Total | 73 | 22 | 73 | 1 | 17 | 2 | 188 | 100 | ||||||||||||||||||||||||
DNA extraction.
About 1 cm2 of fungal material was transferred to a 2-ml Eppendorf tube containing a 2:1 (wt/wt) mixture of silica gel and Celite (silica gel H, Merck 7736/Kieselguhr Celite 545; Machery) and 300 μl of TES buffer [2 g Tris (hydroxymethyl)-aminomethane (Merck catalog no. 8382), 0.38 g Na-EDTA (Titriplex III; BioRad catalog no. 161-0729), and 2 g sodium dodecyl sulfate in 80 ml of ultrapure water (pH 8)]. The fungal material was ground with a micropestle for 1 to 2 min. The volume was adjusted by adding 200 μl of TES buffer. After vigorous shaking and the addition of 10 μl of a 10-mg/ml concentration of proteinase K to the tube, the mixture was incubated at 65°C for 10 min. The salt concentration was raised by adding 140 μl of 5 M NaCl solution. The mixture was combined with 1/10 volume (∼65 μl) of CTAB (cetyltrimethylammonium bromide) buffer 10%, followed by incubation for another 30 min at 65°C. One volume (∼700 μl) of chloroform-isoamyl alcohol (vol/vol = 24/l) was added and mixed carefully by hand. After incubation for 30 min at 0°C (on ice water) and centrifugation at 14,000 rpm at 4°C for 10 min, the top layer was transferred to a clean Eppendorf tube. The sample mixed with 225 μl of 5 M NH4-acetate was incubated for at least 30 min (on ice water) and spun again. The supernatant was transferred to a clean sterile Eppendorf tube and mixed with a 0.55 volume (∼510 μl) of ice-cold isopropanol. After being spun for 7 min at 14,000 rpm and 4°C (or room temperature), the supernatant was decanted. The pellet was washed with ice-cold ethanol 70% two times and dried by using a vacuum dryer. The powder was resuspended in 48.5 μl of Tris-EDTA buffer with 1.5 μl of 10 mg of RNase/ml, incubated at 37°C for 15 to 30 min, and stored at −20°C until used.
DNA amplification and sequencing.
PCR was performed in 50 μl of a reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2·6H2O, 0.01% gelatin, 200 mM concentrations of each deoxynucleotide triphosphate, 25 pmol of each primer, 10 to 100 ng of rRNA, and 0.5 U of Taq DNA polymerase (Sigma). ITS amplicons were generated for all strains by using the primers V9G (5′-TTA CGT CCC TGC CCT TTG TA-3′) and LS266 (5′-GCAT TCC CAA ACA ACT CGA CTC-3′) (20). Amplification was performed in a GenAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, CA) as follows: 95°C for 4 min, followed by 35 cycles consisting of 94°C for 45 s, 52°C for 30 s, and 72°C for 2 min, with a delay at 72°C for 7 min. Amplicons were cleaned with GFX columns (GE Healthcare UK, Ltd., Buckinghamshire, England). For each of the two primers separately, sequencing PCR using 1 μl of template DNA (1 to 10 ng), 3 μl of dilution buffer, 1 μl of BigDye v3.1, and 1 μl of 4 pmol primer filled with 4 μl of MilliQ water to a final volume of 10 μl was performed as follows: 95°C for 1 min, followed by 30 cycles consisting of 95°C for 10 s, 50°C for 5 s, and 60°C for 2 min. Reaction products were purified with Sephadex G-50 Fine (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) and analyzed by using an ABI Prism 3730xl DNA analyzer (Applied Biosystems).
Molecular identification.
The sequences were adjusted by using the Lasergene software program SeqMan Π (DNASTAR, Inc.) and aligned iteratively using Ward's averaging in the Bionumerics package v. 4.0 (Applied Maths, Kortrijk, Belgium). Nearest neighbors were found by local BLAST searches. The distance trees were based on a realigned file using the DCSE program (17) and calculated by the neighbor-joining method of the Treecon package (38) with Kimura-2 correction; only unambiguously aligned positions were taken into account. A total of 100 bootstrap replicates were used for analysis. Bootstrap values of >90 from 100 resampled datasets are shown. If the similarity of sequences of the ITS region was more than 99% between a studied strain and its nearest neighbor and they were distributed in same branch of the phylogenetic tree, the strain is regarded as belonging to the same species as its nearest neighbor.
Antifungal susceptibility.
Antifungal susceptibility testing of four currently available antifungal agents (amphotericin B, itraconazole, voriconazole, and posaconazole) was performed with all strains according to Clinical and Laboratory Standards Institute guidelines (M38-A) (8).
RESULTS
A total of 185 strains from the United States were determined to belong to the genus Exophiala as circumscribed by annellidic conidium production and phylogenetic affinity to the order Chaetothyriales. Budding cells are mostly present in any stage of the life cycle but are absent from some psychrophilic species. Three strains morphologically attributed to Exophiala appeared to be Phialophora europaea.
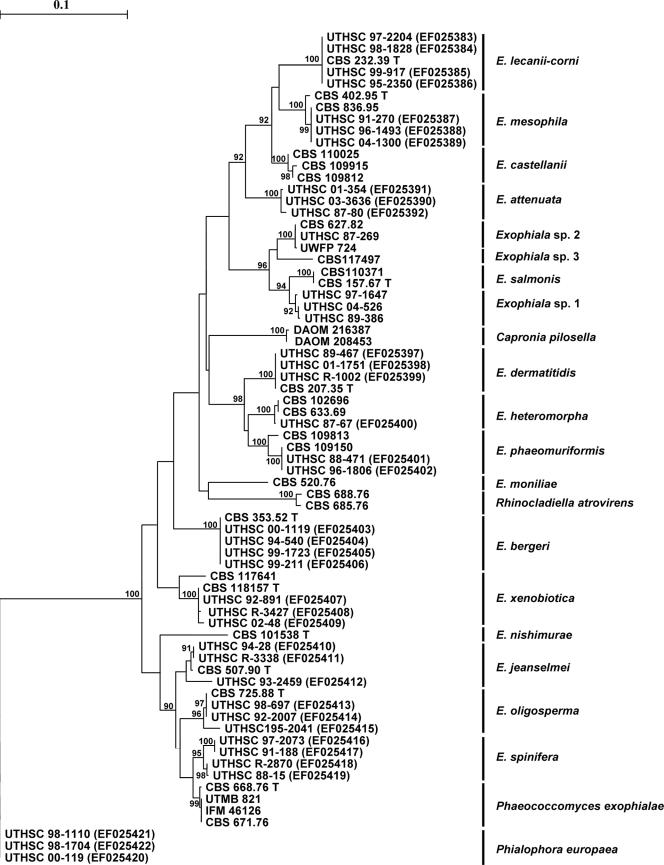
Figure 1 shows a distance tree of partial ITS rRNA of a selection of the strains identified, supplemented with some reference strains. In this tree each Exophiala species is clearly individualized in an independent branch supported by a high bootstrap value. Molecular identifications of all of the strains are shown in Table 1; a total of 14 species were identified, which included 2 undescribed novel Exophiala species. A comparison between morphological and genetic identifications can be found in Table 2. Only E. dermatitidis had a relatively high degree of congruent identifications with morphological and molecular approaches.
FIG. 1.
Consensus tree of ITS rRNA gene of 15 described clinical Exophiala and neighboring species, constructed by using the neighbor-joining algorithm in the Treecon package with Kimura-2 correction and 100 bootstrap replications (values of >90 are shown with the branches). P. europaea is selected as an outgroup. The numbers in parentheses are GenBank accession numbers for ITS sequences deposited in the GenBank database.
TABLE 2.
Comparison of morphological and genetic identifications of strains examined
| Genetic identification | Morphological identification (no. of isolates)
|
Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. castellanii | E. dermatitidis | E. jeanselmei |
E. jeanselmei
|
E. moniliae | E. spinifera | E. salmonis | Exophiala sp. | |||
| var. lecanii-corni | var. jeanselmei | |||||||||
| E. dermatitidis | 53 | 1 | 1 | 55 | ||||||
| E. xenobiotica | 3 | 12 | 16 | 6 | 37 | |||||
| E. oligosperma | 1 | 21 | 1 | 3 | 2 | 1 | 6 | 35 | ||
| E. lecanii-corni | 6 | 3 | 2 | 1 | 1 | 13 | ||||
| E. phaeomuriformis | 3 | 2 | 1 | 2 | 2 | 2 | 12 | |||
| E. jeanselmei | 4 | 1 | 2 | 7 | ||||||
| E. bergeri | 1 | 2 | 4 | 7 | ||||||
| E. mesophila | 2 | 1 | 3 | 6 | ||||||
| E. spinifera | 1 | 4 | 5 | |||||||
| Exophiala sp. 1 | 1 | 2 | 3 | |||||||
| E. attenuata | 1 | 2 | 3 | |||||||
| Exophiala sp. 2 | 1 | 1 | ||||||||
| E. heteromorpha | 1 | 1 | ||||||||
| P. europaea | 2 | 1 | 3 | |||||||
| Total | 5 | 60 | 53 | 27 | 3 | 10 | 7 | 1 | 22 | 188 |
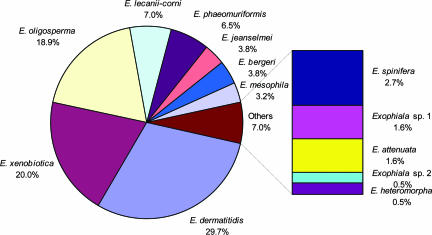
In order of frequency, the prevalent agents of Exophiala species were E. dermatitidis (29.7%), E. xenobiotica (20.0%), and E. oligosperma (18.9%), comprising more than two-thirds of isolates treated in the present study, followed by E. lecanii-corni (7.0%), E. phaeomuriformis (6.5%), E. jeanselmei (3.8%), E. bergeri (3.8%), E. mesophila (3.2%), and E. spinifera (2.7%) (Fig. 2). The total frequency of the second series of species was more than 25%. E. attenuata, E. heteromorpha, and two hitherto-undescribed species were seldom isolated.
FIG. 2.
Spectrum of clinical Exophiala species from the United States.
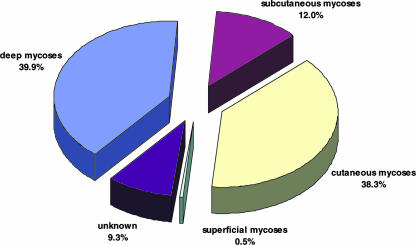
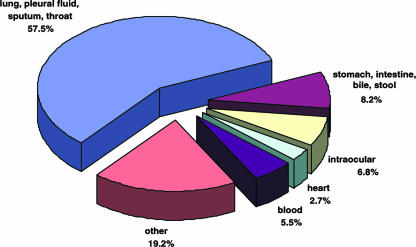
Exophiala strains were repeatedly isolated from human systemic, single-organ infections (39.9%), particularly those involving the lungs (Fig. 3 and 4). More than 50% of the systemic strains were isolated from the lungs, pleural fluid, or sputum (Fig. 4), whereas isolation from the digestive system and feces was uncommon. Cerebral infections were very rare. Strains from human cutaneous infections, including skin, mucous membranes, nail, and corneal epithelium, were equally common as agents from deep localizations (Fig. 3). Subcutaneous infections in humans were less common (12.0%, involving sinusitis, mycetoma, and subcutaneous cysts), whereas strains were exceptional as commensals (0.5%, involving hair). Two strains were isolated from animals. For a small number of strains no isolation data were available.
FIG. 3.
Localization of infections caused by Exophiala species in the United States.
FIG. 4.
Distribution of deep mycoses caused by Exophiala species in the United States.
The deep infections in human were preponderantly caused by E. dermatitidis (36 of 73 [49.3%]), E. oligosperma (16 of 73 [21.9%]), E. phaeomuriformis (9 of 73 [12.3%]), E. xenobiotica (5 of 73 [6.8%]), and E. lecanii-corni (4 of 73 [5.5%]) (Table 1). The three most common Exophiala agents of cutaneous and subcutaneous infection were E. xenobiotica (25 of 95 [27.2%]), E. dermatitidis (16 of 92 [17.4%]), and E. oligosperma (16 of 92 [17.4%]). E. jeanselmei, which in the literature has been regarded as a major agent of cutaneous and subcutaneous mycoses, was rarely observed (7 of 92 [7.6%]). Although E. dermatitidis, E. oligosperma, and E. phaeomuriformis caused different mycoses, they were more frequently isolated from deep infections than from cutaneous and subcutaneous lesions. Strains of the uncommon species E. jeanselmei, E. bergeri, E. spinifera, and E. attenuata were rarely systemic.
The results of antifungal susceptibility testing for nine species strains are shown in Table 3. Although there are no defined breakpoints for any species in the genus, MIC data correlated with safely achievable drug concentrations suggests clinical efficacy with each of the antifungal agents evaluated. Since the number of strains of less-common species is low, susceptibility data are not shown. Of note, however, E. attenuata (three strains) was found to be resistant to amphotericin B.
TABLE 3.
Results of in vitro antifungal susceptibility testinga
| Organism or type | MIC (mg/liter)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Amphotericin B
|
Itraconazole
|
Voriconazole
|
Posaconazole
|
|||||
| MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | |
| E. phaeomuriformis | 0.5 | 1 | ≤0.015 | 0.03 | 0.03 | 0.06 | ≤0.015 | ≤0.015 |
| E. xenobiotica | 0.25 | 0.5 | 0.03 | 0.125 | 0.125 | 0.5 | ≤0.015 | 0.03 |
| E. bergeri | 0.25 | 0.25 | 0.125 | 0.25 | 1.0 | 1.0 | 0.03 | 0.03 |
| E. lecanii-corni | 0.25 | 0.5 | 0.06 | 0.125 | 0.06 | 0.5 | ≤0.015 | 0.03 |
| E. spinifera | 0.5 | 0.5 | 0.03 | 0.06 | 0.125 | 0.5 | ≤0.015 | 0.03 |
| E. jeanselmei | 0.5 | 1 | 0.03 | 0.06 | 0.125 | 0.25 | ≤0.015 | 0.03 |
| E. mesophila | 0.25 | 0.5 | 0.125 | 0.25 | 0.5 | 0.25 | 0.06 | 0.06 |
| E. dermatitidis | 0.5 | 1.0 | 0.125 | 0.25 | 0.125 | 0.25 | 0.03 | 0.06 |
| E. oligosperma | 0.25 | 0.5 | 0.06 | 0.125 | 0.25 | 1.0 | 0.03 | 0.03 |
| Susceptibleb | ≤1.0 | ≤0.5 | ≤2.0 | ? | ||||
| Resistantc | >2.0 | ≥1.0 | >2.0 | ? | ||||
No defined breakpoints are available for the genus Exophiala, and the clinical correlates are primarily anecdotal. AMB, amphotericin B, ITRA.
Suggested MIC for clinical efficacy based upon in vitro data.
Suggested MIC for less than favorable clinical outcome based upon in vitro data.
DISCUSSION
Although some Exophiala species can be identified morphologically and with the help of physiological parameters, most taxa can only be recognized with sufficient certainty by using molecular methods. The sequence diversity of the ITS rRNA region has proven to be reliable for routine species distinction in the genus Exophiala (13, 37). For some of the main clinically relevant species (16), the conclusions in the present study have been confirmed by partial sequencing of the elongation factor 1-α and β-tubulin genes. Large distances were observed between nearly all species analyzed, even when these were highly similar morphologically and physiologically. Hence, the use of umbrella names such as “E. jeanselmei group,” which may be acceptable for daily routine, cannot be applied in the scientific literature. Publication of case reports should be accompanied by sequence data of at least the ITS rRNA. Since relatively few ITS motifs were encountered in the present study that were unknown to us, we suppose that the list of human-associated Exophiala species in temperate climates is nearing completion, although a few new species are still on the list to be described.
Species concepts in Exophiala have changed considerably after the large-scale application of molecular methods over the last 5 years. In particular, the common clinical species E. jeanselmei (9) appeared to comprise a number of cryptic species, such as E. heteromorpha (15), E. lecanii-corni (22), E. oligosperma (15), and E. xenobiotica (16), in addition to E. jeanselmei in a restricted sense (15). Most of these species have been reported from proven clinical cases, whereby slight, species-specific differences in preferred sites of infection were noted (15). Unfortunately, case reports continue to be published under obsolete concepts (33, 34) and thus create confusion. The present study utilizes species circumscriptions published after the latest overview of clinically relevant Exophiala species, which dates back to the year 2000 (9).
Thus far, 18 of 29 known Exophiala species have been proven or were suggested to cause infections in or colonization of humans and animals. The list includes the recently described taxa E. xenobiotica (16), E. oligosperma (1), and several mesophilic species (M. J. Harrak et al., unpublished data). No reliable data are available on the incidence of diseases, since the strains studied were those that were sent to reference laboratories for identification and thus may not be representative for their actual prevalence; the current data can only be taken as indicative of the frequencies of the main species. Unfortunately, only limited clinical data were available for the present set of strains (not shown). Proof of the clinical significance of a number of species thus has to be judged from future case reports.
Epidemics caused by these species have not been observed, but repeated reports of pseudoepidemics due to contaminated fluids have been published (2, 33, 34). In the large set of strains analyzed for the present study, 12 of the 17 known invasive species were encountered, with the exception of E. castellanii, E. moniliae, E. nishimurae, E. salmonis, and E. pisciphila. E. dermatitidis, and two segregants of E. jeanselmei, viz. E. xenobiotica and E. oligosperma, are the three major agents of mycoses caused by Exophiala species.
In the present overview, the frequency of deep mycoses in the studied set of isolates is almost two-fifths (39.9%) and thus significantly higher than that of the categories of subcutaneous and superficial mycoses and also slightly higher than that of cutaneous mycoses. Predisposing diseases or metabolic factors listed by clinicians at the submission of strains include solid organ or bone marrow transplant, hematologic or nonhematologic malignancy, diabetes mellitus, and, exceptionally, human immunodeficiency virus infection. Microbial contamination at injury also occurs repeatedly in immunocompetent individuals. The most frequent deep infections are those of the respiratory system caused by E. dermatitidis, E. oligosperma, E. phaeomuriformis, and E. lecanii-corni. Pulmonary infections are mostly not invasive, but probably subclinical colonization is involved, as observed in patients with cystic fibrosis (18, 24). In Europe, this is one of the preponderant clinical pictures by Exophiala species, particularly E. dermatitidis (18, 24). This fungus occurs with a frequency of 2 to 8% in the susceptible patient population (21); similar screening has thus far not been done in the United States.
Cerebral infections caused by Exophiala species are very rare in the United States. Until now only a single fatal case has been reported, caused by strain CDC B-6450. It concerned a contaminated steroid injection, the fungus being directly inoculated into the circulation (19). Strain R-1002 was listed as originating from human brain, but the clinical data and origin were not specified, neither whether it was a primary cerebral infection nor whether there were any predisposing conditions. In Asia, cerebral infection in healthy adolescents is a remarkable clinical syndrome. At least 11 fatal cases were reported (6, 23, 26). Primary cerebral infection caused by E. dermatitidis appears to occur nearly only in Asian patients, the possibility of race-dependent virulence has been suggested (24). Strains of E. spinifera were mainly involved in cutaneous and subcutaneous mycoses. Outside the United States several cases of disseminated infection caused by this species have been reported (5, 31, 36, 40) in individuals without known immune disorder. The reason for the absence of systemic disease in the United States is unknown.
In cutaneous sources, the recently described species E. xenobiotica appeared to be the most frequently detected black yeast. This species is a recent segregant of E. jeanselmei, differing at the molecular level and having a different predilected site of infection (16). Also, E. dermatitidis and E. oligosperma occur in cutaneous infections. The underdiagnosis of E. xenobiotica and E. oligosperma is certainly due to recent developments in the taxonomy of black yeasts, which led to the description of these taxa, after they had been deposited in reference collections, mostly either as “E. jeanselmei” or “Exophiala sp.”
In conclusion, we suggest that infections with black yeasts of the genus Exophiala are severely underdiagnosed in the United States. In the case of the occurrence of E. dermatitidis in cystic fibrosis and in stool, frequencies have been published in Europe (12, 21); data from the United States are unlikely to be different. Underdiagnosis of Exophiala infection in superficial and cutaneous disorders is a worldwide problem, and the clinical significance of individual species is therefore hard to establish. Systemic and disseminated cases may be severe, particularly because they can take a fatal course in young and otherwise-healthy individuals. The reason why the severe syndromes seem to be relatively rare in the United States is currently not understood.
Most strains of Exophiala species tested appeared to be susceptible in vitro to the four widely used antifungal agents evaluated in the present study, except that Exophiala attenuata was resistant to amphotericin B; no significant differences were noted with the different phylogenetic positions of the species concerned. Particularly low MICs were noted for posaconazole. Similar results have been shown previously (3, 25, 27, 33, 39). Infections by Exophiala species may require a combination of surgical and medical treatment. Although amphotericin B and itraconazole, with or without additional flucytosine, are currently regarded to be efficacious against cutaneous and subcutaneous lesions, the newer triazole agents, voriconazole and posaconazole, expand the therapeutic options for these mycoses. The clinical outcome in deep infection, however, is dismal (6, 7, 29, 23, 26, 33, 35). Improvement may be expected, since posaconazole has shown striking effects in treating a case of disseminated infection (31).
Acknowledgments
We thank A. H. G. Gerrits van den Ende and K. F. Luijsterburg for technical assistance and R. C. Summerbell for comments on the text. We also thank E. H. Thompson and J. Ruiz for morphological identification and antifungal susceptibility testing, respectively.
This study was supported by a grant from Pfizer, Inc.
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Al-Obaid, I., S. Ahmad, Z. U. Khan, B. Dinesh, and H. M. Hejab. 2006. Catheter-associated fungemia due to Exophiala oligosperma in a leukemic child and review of fungemia cases caused by Exophiala species. Eur. J. Clin. Microbiol. Infect. Dis. 25:729-732. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2002. Exophiala infection from contaminated injectable steroids prepared by a compounding pharmacy-United States, July-November 2002. JAMA 289:291-293. [PubMed] [Google Scholar]
- 3.Blaschke-Hellmessen, R. 1996. Fluconazole and itraconazole susceptibility testing with clinical yeast isolates and algae of the genus Prototheca by means of the Etest. Mycoses 39(Suppl. 2):39-43. [DOI] [PubMed] [Google Scholar]
- 4.Bossler, A. D., S. S. Richter, A. J. Chavez, S. A. Vogelgesang, D. A. Sutton, A. M. Grooters, M. G. Rinaldi, G. S. de Hoog, and M. A. Pfaller. 2003. Exophiala oligosperma causing olecranon bursitis. J. Clin. Microbiol. 41:4779-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos-Takaki, G. M., and M. L. Jardim. 1994. Report of chronic subcutaneous abscesses caused by Exophiala spinifera. Mycopathologia 127:73-76. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C. L., D. S. Kim, D. J. Park, H. J. Kim, C. H. Lee, and J. H. Shin. 2000. Acute cerebral phaeohyphomycosis due to Wangiella dermatitidis accompanied by cerebrospinal fluid eosinophilia. J. Clin. Microbiol. 38:1965-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clancy, C. J., J. R. Wingard, and M. Hong Nguyen. 2000. Subcutaneous phaeohyphomycosis in transplant recipients: review of the literature and demonstration of in vitro synergy between antifungal agents. Med. Mycol. 38:169-175. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2002. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard M38-A. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.De Hoog, G. S., J. Guarro, J. Gené, and M. J. Figueras. 2000. Atlas of clinical fungi, 2nd ed. Centraalbureau voor Schimmelcultures/Universitat Rovira i Virgili, Utrecht, The Netherlands.
- 10.De Hoog, G. S., A. H. G. Gerrits van den Ende, J. M. J. Uijthof, and W. A. Untereiner. 1995. Nutritional physiology of type isolates of currently accepted species of Exophiala and Phaeococcomyces. Antonie Leeuwenhoek 68:43-49. [DOI] [PubMed] [Google Scholar]
- 11.De Hoog, G. S., and G. Haase. 1993. Nutritional physiology and selective isolation of Exophiala dermatitidis. Antonie Leeuwenhoek 64:17-26. [DOI] [PubMed] [Google Scholar]
- 12.De Hoog, G. S., T. Matos, M. Sudhadham, K. F. Luijsterburg, and G. Haase. 2005. Intestinal prevalence of the neurotropic black yeast Exophiala (Wangiella) dermatitidis in healthy and impaired individuals. Mycoses 48:142-145. [DOI] [PubMed] [Google Scholar]
- 13.De Hoog, G. S., N. Poonwan, and A. H. G. Gerrits van den Ende. 1999. Taxonomy of Exophiala spinifera and its relationship to E. jeanselmei. Stud. Mycol. 43:133-142. [Google Scholar]
- 14.De Hoog, G. S., K. Takeo, S. Yoshida, E. Göttlich, K. Nishimura, and M. Miyaji. 1994. Pleoanamorphic life cycle of Exophiala (Wangiella) dermatitidis. Antonie Leeuwenhoek 65:143-153. [DOI] [PubMed] [Google Scholar]
- 15.De Hoog, G. S., V. Vicente, R. B. Caligiorne, S. Kantarcioglu, K. Tintelnot, A. H. G. Gerrits van den Ende, and G. Haase. 2003. Species diversity and polymorphism in the Exophiala spinifera clade containing opportunistic black yeast-like fungi. J. Clin. Microbiol. 41:4767-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Hoog, G. S., J. S. Zeng, M. J. Harrak, and D. A. Sutton. 2006. Exophiala xenobiotica sp. nov., an opportunistic black yeast inhabiting environments rich in hydrocarbons. Antonie Leeuwenhoek 90:257-268. [DOI] [PubMed] [Google Scholar]
- 17.De Rijk, P., and R. De Wachter. 1993. DCSE v. 2.54, an interactive tool for sequence alignment and secondary structure research. Comput. Appl. Biosci. 9:735-740. [DOI] [PubMed] [Google Scholar]
- 18.Diemert, D., D. Kunimoto, C. Sand, and R. Rennie. 2001. Sputum isolation of Wangiella dermatitidis in patients with cystic fibrosis. Scand. J. Infect. 33:777-779. [DOI] [PubMed] [Google Scholar]
- 19.Engemann, J., K. Kaye, G. Cox, J. Perfect, W. Schell, S. A. McGarry, K. Patterson, S. Edupuganti, P. Cook, W. A. Rutala, D. J. Weber, K. K. Hoffmann, J. Engel, S. Young, E. Durant, K. McKinnon, N. Cobb, L. Bell, J. Gibson, D. Jernigan, M. Arduino, S. Fridkin, L. Archibald, L. Sehulster, J. Morgan, R. Hajjeh, M. Brandt, D. Warnock, and W. A. Duffus. 2002. Exophiala infection from contaminated injectable steroids prepared by a compounding pharmacy-United States, July-November 2002. Morb. Mortal. Wkly. Rep. 51:1109-1112. [PubMed] [Google Scholar]
- 20.Gerrits van den Ende, A. H. G., and G. S. de Hoog. 1999. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Stud. Mycol. 43:151-162. [Google Scholar]
- 21.Haase, G., H. Skopnik, T. Groten, G. Kusenbach, and H. G. Posselt. 1991. Long-term fungal cultures from patients with cystic fibrosis. Mycoses 34:373-376. [DOI] [PubMed] [Google Scholar]
- 22.Haase, G., L. Sonntag, B. Melzer-Krick, and G. S. de Hoog. 1999. Phylogenetic interference by SSU-gene analysis of members of the Herpotrichiellaceae with special reference to human pathogenic species. Stud. Mycol. 43:80-97. [Google Scholar]
- 23.Hiruma, M., A. Kawada, H. Ohata, Y. Ohnishi, H. Takahashi, M. Yamazaki, A. Ishibashi, K. Hatsuse, M. Kakihara, and M. Yoshida. 1993. Systemic phaeohyphomycosis caused by Exophiala dermatitidis. Mycoses 36:1-7. [DOI] [PubMed] [Google Scholar]
- 24.Horré, R., K. P. Schaal, R. Siekmeier, B. Sterzik, G. S. de Hoog, and N. Schnitzler. 2004. Isolation of fungi, especially Exophiala dermatitidis, in patients suffering from cystic fibrosis: a prospective study. Respiration 71:360-366. [DOI] [PubMed] [Google Scholar]
- 25.Li, D., R. Li, D. Wang, and S. Ma. 1999. In vitro activities of five antifungal agents against pathogenic Exophiala species. Chin. Med. J. 112:484-488. [PubMed] [Google Scholar]
- 26.Matsumoto, T., T. Matsuda, M. R. McGinnis, and L. Ajello. 1993. Clinical and mycological spectra of Wangiella dermatitidis infections. Mycoses 36:145-155. [DOI] [PubMed] [Google Scholar]
- 27.Meletiadis, J., J. F. Meis, G. S. de Hoog, and P. E. Verweij. 2000. In vitro susceptibilities of 11 clinical isolates of Exophiala species to six antifungal drugs. Mycoses 43:309-312. [DOI] [PubMed] [Google Scholar]
- 28.Murray, I. G., G. E. Dunkerley, and K. E. A. Hughes. 1964. A case of Madura foot caused by Phialophora jeanselmei. Sabouraudia 3:175-177. [PubMed] [Google Scholar]
- 29.Myoken, Y., T. Sugata, Y. Fujita, T. Kyo, M. Fujihara, M. Katsu, and Y. Mikami. 2003. Successful treatment of invasive stomatitis due to Exophiala dermatitidis in a patient with acute myeloid leukemia. J. Oral Pathol. Med. 32:51-54. [DOI] [PubMed] [Google Scholar]
- 30.Naka, W., T. Harada, T. Nishikawa, and R. Fukushiro. 1986. A case of chromoblastomycosis: with special reference to the mycology of the isolated Exophiala jeanselmei. Mykosen 29:445-452. [DOI] [PubMed] [Google Scholar]
- 31.Negroni, R., S. H. Helou, N. Petri, A. M. Robles, A. Arechavala, and M. H. Bianchi. 2004. Case study: posaconazole treatment of disseminated phaeohyphomycosis due to Exophiala spinifera. Clin. Infect. Dis. 38:15-20. [DOI] [PubMed] [Google Scholar]
- 32.Neumeister, B., T. M. Zollner, D. Krieger, W. Sterry, and R. Marre. 1995. Mycetoma due to Exophiala jeanselmei and Mycobacterium chelonae in a 73-year-old man with idiopathic CD4+ T lymphocytopenia. Mycoses 38:271-276. [DOI] [PubMed] [Google Scholar]
- 33.Nucci, M., T. Akiti, G. Barreiros, F. Silveira, S. G. Revankar, D. A. Sutton, and T. F. Patterson. 2001. Nosocomial fungemia due to Exophiala jeanselmei var. jeanselmei and a Rhinocladiella species: newly described causes of bloodstream infection. J. Clin. Microbiol. 39:514-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nucci, M., T. Akiti, G. Barreiros, F. Silveira, S. G. Revankar, B. L. Wickes, D. A. Sutton, and T. F. Patterson. 2002. Nosocomial outbreak of Exophiala jeanselmei fungemia associated with contamination of hospital water. Clin. Infect. Dis. 34:1475-1480. [DOI] [PubMed] [Google Scholar]
- 35.Padhye, A. A., A. A. Hampton, M. T. Hampton, N. W. Hutton, E. Prevost-Smith, and M. S. Davis. 1996. Chromoblastomycosis caused by Exophiala spinifera. Clin. Infect. Dis. 22:331-335. [DOI] [PubMed] [Google Scholar]
- 36.Rajendran, C., B. K. Khaitan, R. Mittal, M. Ramam, M. Bhardwaj, and K. K. Datta. 2003. Phaeohyphomycosis caused by Exophiala spinifera in India. Med. Mycol. 41:437-441. [DOI] [PubMed] [Google Scholar]
- 37.Tintelnot, K., G. S. de Hoog, E. Thomas, W.-I. Steudel, K. Huebner, and H. P. R. Seeliger. 1991. Cerebral phaeohyphomycosis caused by an Exophiala species. Mycoses 34:239-244. [DOI] [PubMed] [Google Scholar]
- 38.Van de Peer, Y., and R. De Wachter. 1994. Treecon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 39.Vitale, R. G., and G. S. de Hoog. 2002. Molecular diversity, new species and antifungal susceptibilities in the Exophiala spinifera clade. Med. Mycol. 40:545-556. [DOI] [PubMed] [Google Scholar]
- 40.Wang, D., R. Li, X. Wang, and W. Dai. 1987. Studies on three strains of Exophiala spinifera. Acta Mycol. Sin. 6:229-232. [Google Scholar]