Abstract
A PCR-oligochromatography test for diagnosis of human and animal trypanosomiasis was evaluated through a multicenter ring trial with six laboratories testing a set of 21 blinded samples, resulting in qualitative data (positive or negative). Results showed an intralaboratory repeatability (accordance) of 88.7% (credible interval [CI], 84.4 to 92.5%) and an interlaboratory repeatability (concordance) of 88.1% (CI, 84.3 to 92.3%).
Recently, four antibody enzyme-linked immunosorbent assays (ELISAs) for animal trypanosomiasis (Trypanosoma congolense and T. vivax) were evaluated in a multicenter validation trial (5). Given that ELISAs generate quantitative data, modified Youden plots could be used to analyze the interlaboratory reproducibility of these assays. Unfortunately, this approach is unsuitable for the analysis of qualitative data (PCR, PCR-oligochromatography). For analyzing qualitative assays, different formulae were first described by Langton et al. (3) for a fixed number of laboratories and further modified for use in larger “population” trials by van der Voet and van Raamsdonk (6). Recently, these formulae were used to measure foot-and-mouth disease vaccine potency testing in cattle (2).
We developed a Trypanozoon-specific PCR-oligochromatography test of which the proof of principle is described by Deborggraeve et al. (1). In brief, Trypanosoma brucei 18S ribosomal DNA is amplified through PCR and amplicons are visualized on a dipstick through hybridization with a gold-conjugated probe (oligochromatography). Visualization is straightforward and takes only 5 min. Controls both for the PCR and for DNA migration are incorporated into the assay.
Before its adoption for diagnostic tests, we subjected it to a multicenter ring trial to evaluate its repeatability and reproducibility according to OIE (World Organization for Animal Health) validation criteria (4) and in accordance with the WHO/Special Programme for Research and Training in Tropical Diseases recommendation that “a multicenter validation of diagnostics based on molecular techniques (e.g., PCR) for epidemiological and clinical studies is strongly recommended” (7).
Six laboratories in Europe, Africa, and Asia (Belgium, Burkina-Faso, Germany, Kenya, Uganda, and Vietnam) participated in this multicenter trial, set up to evaluate the accordance (ACC; intralaboratory repeatability) and the concordance (CON; interlaboratory reproducibility) of the Trypanozoon PCR-oligochromatography test.
A sample set consisting of positive (T. brucei brucei DNA) and negative (human DNA) controls and 21 blinded test samples was sent to all participating laboratories together with the necessary test reagents, a test report sheet, and a standard operating procedure following the protocol described by Deborggraeve et al. (1). Thus, the only obvious sources of variability between the laboratories are the manipulator, the micropipettes, the water bath, and the PCR thermocycler. Each laboratory received sufficient material to perform PCR-oligochromatography in triplicate on each sample. The blinded samples included a twofold serial dilution series of T. brucei brucei DNA (seven samples with concentrations ranging from 1,280 fg/μl to 2.5 fg/μl) and DNA at 100 ng/μl from the five taxa within Trypanozoon (T. brucei brucei, T. brucei gambiense, T. brucei rhodesiense, T. evansi, and T. equiperdum) and from nine non-Trypanozoon DNA samples (Table 1).
TABLE 1.
Triplicate results of Trypanozoon PCR-oligochromatography on a set of 23 DNA samples
| Sample (amt [fg]) | Expected result | No. of positive results (performed in triplicate) at laboratory:
|
|||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| T. brucei brucei AnTat 2.2 | Positive | 3 | 3 | 3 | 3 | 3 | 3 |
| Human DNA | Negative | 0 | 0 | 0 | 1 | 0 | 0 |
| T. brucei brucei (2.5) | Positive | 3 | 3 | 0 | 3 | 3 | 3 |
| T. brucei brucei (5) | Positive | 3 | 3 | 0 | 3 | 2 | 3 |
| T. brucei brucei (10) | Positive | 3 | 3 | 0 | 3 | 0 | 3 |
| T. brucei brucei (20) | Positive | 3 | 3 | 3 | 3 | 2 | 3 |
| T. brucei brucei (80) | Positive | 3 | 3 | 3 | 3 | 3 | 3 |
| T. brucei brucei (320) | Positive | 3 | 3 | 3 | 3 | 3 | 3 |
| T. brucei brucei (1,280) | Positive | 3 | 3 | 3 | 3 | 3 | 3 |
| T. brucei gambiense LiTat 1.3 | Positive | 3 | 3 | 3 | 3 | 3 | 3 |
| T. brucei rhodesiense AnTat 25.1 | Positive | 3 | 3 | 3 | 3 | 3 | 3 |
| T. evansi RoTat 1.2 | Positive | 3 | 3 | 3 | 3 | 1 | 3 |
| T. equiperdum OVI | Positive | 3 | 3 | 3 | 3 | 3 | 3 |
| T. brucei gambiense AnTat 9.1 | Positive | 3 | 3 | 3 | 1 | 0 | 3 |
| T. congolense TRT 17 | Negative | 0 | 0 | 0 | 0 | 2 | 0 |
| T. vivax ILRAD 700 | Negative | 0 | 0 | 0 | 0 | 0 | 2 |
| Theileria parva | Negative | 0 | 0 | 0 | 0 | 1 | 0 |
| Leishmania sp. | Negative | 0 | 0 | 0 | 0 | 2 | 0 |
| Plasmodium sp. | Negative | 0 | 0 | 0 | 0 | 3 | 2 |
| Schistosoma sp. | Negative | 0 | 0 | 0 | 1 | 0 | 0 |
| Bovine DNA | Negative | 0 | 0 | 0 | 0 | 0 | 1 |
| Trypanosoma cruzi | Negative | 0 | 0 | 0 | 0 | 3 | 0 |
| Trypanosoma rangeli | Negative | 0 | 0 | 0 | 0 | 3 | 0 |
DNA was extracted using the QIAamp DNA minikit (QIAGEN, Germany) according to the manufacturer's instructions and was quantified by Nanodrop (Isogen, Belgium).
Two main parameters were analyzed in casu: the ACC or intralaboratory repeatability, which is defined as the average chance of finding the same result for two identical DNA samples analyzed in the same laboratory under standard operating conditions (independently of whether the result is correct or not), and the CON or interlaboratory reproducibility, which is defined as the average chance of finding the same result for two identical samples analyzed in different laboratories under standardized conditions. As the interest is in the performance of the test on the samples in an arbitrary laboratory (not only the laboratories participating in this ring trial) the results of this multicenter trial were analyzed in a random framework (6), for which the formulae are as follows: 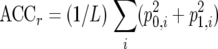 and
and 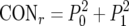 , in which p0,i2 and p1,i2 were defined as the squared proportion of PCR-oligochromatography-negative and -positive test results, respectively, for each analysis i and where P02 and P12 were defined by the equations
, in which p0,i2 and p1,i2 were defined as the squared proportion of PCR-oligochromatography-negative and -positive test results, respectively, for each analysis i and where P02 and P12 were defined by the equations 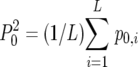 and
and 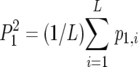 , with L the number of laboratories in the trial.
, with L the number of laboratories in the trial.
Uncertainty around p0,i and p1,i was quantified by Bayesian inference assuming that besides these data no other knowledge was available on the values of p0,i and p1,i. The uncertainty distributions of the ACC and CON were obtained by repeatedly (in this case 5,000 times) drawing randomly from these posterior distributions followed by, at each iteration the calculation of ACC and CON according to the formulae described above. The histograms of all these ACC and CON values, calculated at each iteration, represent the uncertainty of the estimates. By using this Bayesian “simulation from posterior” approach one does not have to rely on asymptotic-normality assumptions to derive credible intervals (CI) around the estimates.
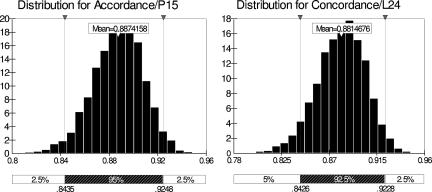
Statistical evaluation from the data set (Table 1), with 95% CI, gave the following results: an ACC of 88.7% (CI, 84.4 to 92.5%) and a CON of 88.1% (CI, 84.3 to 92.3%). These data and their distribution are presented in Fig. 1. Note that the results from laboratory 5 were excluded from this calculation due to multiple positive results in the negative-sample population, possibly due to cross-contamination or errors during test performance. Thus, the final analysis was performed on the data from the five remaining laboratories.
FIG. 1.
Distribution histograms for ACC (left) (intralaboratory “repeatability”) and CON (right) (interlaboratory “reproducibility”).
In this multicenter trial, the values for ACC and CON are high, indicating that this assay can be satisfactorily reproduced and applied in different laboratories.
As stated above the interest is in the performance of the test on the samples in an arbitrary laboratory (not only the laboratories participating in this ring trial); hence, ACC and CON are test-specific parameters. Uncertainty around these parameters can be reduced by increasing the sample size (the number of samples sent around). Sample sizes can be determined as a function of the minimal difference in ACC and CON that would likely be detected between different tests.
In four out of five laboratories, the PCR-oligochromatography assay could detect as low as 2.5 fg of DNA per PCR; laboratory 3 detected only 20 fg of DNA per reaction. This means that, in all laboratories evaluated, the assay can detect down to 1 parasite per reaction, since we assume that the genome of one trypanosome is 0.2 pg. The poor detection limit observed in laboratory 3 is probably due to the equipment used (thermocycler or calibration of pipettes) rather than the assay itself, since the repeatability of the results for the lower-quantity DNA samples is consistently negative in this laboratory.
The assay was developed to be Trypanozoon specific. Results from this trial show that this is indeed the case, except for some occasional false-positive results obtained in laboratories 4 and 6 (Table 1).
This is the first time that this statistical approach has been applied to evaluate the ACC and CON of a Trypanozoon-specific qualitative diagnostic assay. Results of this study show that (i) PCR-oligochromatography may serve as diagnostic test for human African trypanosomiasis and animal trypanosomiasis, alongside more-traditional diagnostic procedures, after further field evaluation and (ii) this statistical approach may be used in the future as a standard for multicenter trials to analyze other newly developed qualitative diagnostic tests.
Acknowledgments
We acknowledge V. K. Nguyen, I. Sidibe, P.-H. Clausen, J. Enyaru, and J. Kinuya for their collaboration in this ring trial.
This study received financial support from the International Atomic Energy Agency (Vienna, Austria; contract number 12851/RBF). Filip Claes is a Postdoctoral Fellow of the Research Foundation Flanders.
Footnotes
Published ahead of print on 19 September 2007.
REFERENCES
- 1.Deborggraeve, S., F. Claes, T. Laurent, P. Mertens, T. Leclipteux, J. C. Dujardin, P. Herdewijn, and P. Büscher. 2006. A molecular dipstick test for the diagnosis of sleeping sickness. J. Clin. Microbiol. 44:2884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goris, N., P. Merkelbach-Peters, V. I. Diev, D. Verloo, V. M. Zakharov, H. P. Kraft, and K. De Clercq. 2007. European Pharmacopoeia foot-and-mouth disease vaccine potency testing in cattle: between test variability and its consequences. Vaccine 25:3373-3379. [DOI] [PubMed] [Google Scholar]
- 3.Langton, S. D., R. Chevennement, N. Nagelkerke, and B. Lombard. 2002. Analysing collaborative trials for qualitative microbiological methods: accordance and concordance. Int. J. Food Microbiol. 79:175-181. [DOI] [PubMed] [Google Scholar]
- 4.OIE. 2002. Quality standard and guidelines for veterinary laboratories: infectious diseases. OIE Publications, Paris, France.
- 5.Rebeski, D. E., E. M. Winger, J. O. Ouma, S. Kong Pages, P. Buscher, Y. Sanogo, R. H. Dwinger, and J. R. Crowther. 2001. Charting methods to monitor the operational performance of ELISA method for the detection of antibodies against trypanosomes, Vet. Parasitol. 96:11-50. [DOI] [PubMed] [Google Scholar]
- 6.van der Voet, H., and L. W. van Raamsdonk. 2004. Estimation of accordance and concordance in interlaboratory trials of analytical methods with qualitative results. Int. J. Food Microbiol. 95:31-234. [DOI] [PubMed] [Google Scholar]
- 7.WHO/Special Programme for Research and Training in Tropical Diseases. 2001. Report of the Scientific Working Group Meeting on African Trypanosomiasis, 4 to 8 June 2001. WHO/Special Programme for Research and Training in Tropical Diseases, Geneva, Switzerland.