Abstract
We developed a multiplex asymmetric PCR (MAPCR)-based DNA microarray assay for characterization of the clinically relevant antibiotic resistance genes leading to penicillin, methicillin, aminoglycoside, macrolide, lincosamide, and streptogramin B (MLSB) resistance in staphylococci. The DNA-based assay involves detection of specific conserved regions of the mecA, blaZ (methicillin and penicillin resistance), aac(6′)-Ie-aph(2‴) (aminoglycoside resistance), ermA and ermC genes (MLSB resistance), and the msrA gene (macrolide and streptogramin B resistance). The microarray uses a variable sequence region of the 16S rRNA gene to broadly differentiate between Staphylococcus aureus and other coagulase-negative staphylococci (CoNS). The performance of the microarray was validated with a total of 178 clinically important S. aureus and 237 CoNS isolates, with correlations of 100% for S. aureus to CoNS discrimination and more than 90% for antibiotic resistance between the genotypic analysis determined by the microarray and the phenotype determined by standard methods of species identification and susceptibility testing. The major discrepant results were 17 mecA-positive CoNS and 60 aac(6′)-Ie-aph(2‴)-positive CoNS isolates measured by microarray that were susceptible to the corresponding antibiotics based on disk diffusion assay. Overall, this microarray-based assay offers a simultaneous, fast (≤5 h), and accurate identification of antibiotic resistance genes from a single colony, as well as species classification. Our extensive validation of the microarray suggests that it may be a useful tool to complement phenotypic susceptibility testing in clinical laboratories and to survey the spread of antibiotic resistance determinants in epidemiological studies.
Three groups of important antibiotics commonly used in treatment of staphylococcal infections include beta-lactams, particularly lactamase-resistant oxacillin, aminoglycoside, and macrolide, lincosamide, and streptogramin B (MLSB). However, resistance to these antibiotics is increasingly prevalent among staphylococci (20, 31, 32). For instance, PBP 2a protein, encoded by the mecA gene, is responsible for oxacillin (methicillin) resistance in staphylococci (16, 34, 41). β-Lactamase encoded by the blaZ gene accounts for the resistance to penicillins. Schmitz et al. (31) have reported that the aac(6′)-Ie-aph(2‴) gene, encoding a bifunctional enzyme AAC(6′)/APH(2‴), is the most frequently encountered (70 to 90%) aminoglycoside resistance mechanism among staphylococcal isolates. Ribosome modification confers MLSB resistance (9, 39), principally by a single base change in the 23S rRNA by methylases encoded by erythromycin ribosomal methylase (erm) genes ermA or ermC. Resistance to macrolides and streptogramin B (MS resistance) can also occur in staphylococci with active efflux by a membrane-bound transporter protein (msrA gene) (30). These different antibiotic resistance genes are either chromosomally encoded (mecA) (34), or carried by transferable genetic elements such as transposons [blaZ, aac(6′)-Ie-aph(2‴), and ermA] (2, 23) and plasmids (ermC and msrA gene) (18, 30).
Accurate and rapid antibiotic susceptibility information is crucial for clinicians to make appropriate therapy decisions (3, 4). Microarray technology, which allows for the simultaneous analysis of a large amount of genetic information in a single assay (29), has recently been developed to analyze specific bacterial species or test for a few microbial antibiotic resistance determinants (5, 14, 27, 40). Here we have developed a new multiplex PCR-based DNA microarray for detection of six antibiotic resistance genes in staphylococci, including mecA, blaZ, aac(6′)-Ie-aph(2‴), ermA, ermC, and msrA, using one sequence-specific probe for each gene. The microarray also had probes specific to a variable region of 16S rRNA gene, simultaneously differentiating between S. aureus and other coagulase-negative staphylococci (CoNS) isolates. Validation of the microarray with 415 nonduplicate staphylococcal isolates has demonstrated that this platform will be a suitable complement for phenotypic susceptibility testing and will provide a rapid guide for appropriate antimicrobial therapy as well as infection control.
MATERIALS AND METHODS
Bacterial strains.
Table 1 lists the descriptions of the reference strains used in the present study. A total of 178 clinically important S. aureus and 237 CoNS isolates previously characterized (42) from blood (n = 145), pus (n = 110), respiratory tract (n = 92), urine (n = 44), and other tissue sources (n = 24) were used to validate the microarray. The isolates were collected from inpatients of the hospital departments and intensive care units of the Beijing Hospital, the Beijing Tiantan Hospital, the Beijing Tongren Hospital, and the Peking Union Medical College Hospital from January to May 2003. Duplicate samples from the same patient were excluded. The isolates were cultured at 35°C on blood agar (Jinzhang Co., Ltd., Tianjin, China) before testing. All isolates were confirmed as S. aureus or CoNS by colony morphology, Gram stain, catalase test, and coagulase test and by the Vitek 2 system (bioMérieux, France). For CoNS, the collection comprised 144 S. epidermidis, 41 S. haemolyticus, 18 S. auricularis, 13 S. simulans, 7 S. hominis, 4 S. capitis, 4 S. sciuri, and 6 other CoNS strains.
TABLE 1.
Reference strains used in this study
| Strain | Species | Origin | Relevant resistance gene(s)a | Inhibition zone diam (mm) withb:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| PEN | OX | FOX | GM | CM | ER | ||||
| TR146 | S. aureus | Pus | None | 41 | 19 | 27 | 25 | 25 | 25 |
| J143 | S. epidermidis | Sputum | mecA | 25 | 15 | 24 | 29 | 30 | 34 |
| R3k728 | S. epidermidis | Urine | msrA | 36 | 21 | 30 | 22 | 26 | 9 |
| TR1780 | S. epidermidis | Sputum | ermC | 29 | 24 | 29 | 29 | MLSBi | 6 |
| B150 | S. aureus | Pus | blaZ | 18 | 22 | 29 | 25 | 27 | 28 |
| TR1708 | S. aureus | Pus | aac6 | 32 | 18 | 26 | 14 | 21 | 24 |
| 7605 | S. simulans | Sputum | ermA, ermC | 40 | 26 | 27 | 23 | MLSBi | 6 |
| 6314 | S. aureus | Sputum | blaZ, aac6, msrA | 16 | 22 | 22 | 14 | 29 | 9 |
| TR558 | S. aureus | Sputum | blaZ, mecA, aac6, ermA, ermC | 6 | 6 | 11 | 10 | 6 | 6 |
| 7622 | S. auricularis | Sputum | blaZ, mecA, aac6, ermC, msrA | 6 | 6 | 6 | 6 | 6 | 6 |
aac6: aac(6′)-Ie-aph(2‴).
OX, oxacillin; FOX, cefoxitin; PEN, penicillin; GM, gentamicin; CM, clindamycin; ER, erythromycin.
Antimicrobial susceptibility tests.
Antimicrobial susceptibility was tested by the agar disk diffusion method on Muller-Hinton agar (Tiantan Biotechnology Co., Ltd., Beijing, China) according to the National Committee for Clinical Laboratory Standards guidelines (25). Disks (Tiantan Biotechnology) were preloaded with the following antibiotics at the specific absolute concentrations indicated in parentheses: penicillin G (10 U), oxacillin (1 μg), cefoxitin (30 μg), gentamicin (10 μg), erythromycin (15 μg), and clindamycin (2 μg). Plates with disks were incubated at 35°C for 24 h to measure the inhibition zone diameters. S. aureus ATCC 25923 was included for quality control. The diameters of the zones of inhibition (in millimeters) used for interpretation were as indicated for penicillin (resistant [R] ≤ 28, susceptible [S] ≥ 29) and oxacillin (R ≤ 10, I = 11 to 12, S ≥ 13 for S. aureus; R ≤ 17, S ≥ 18 for CoNS), gentamicin (R ≤ 12, I = 13 to 14, S ≥ 15), erythromycin (R ≤ 13, I = 14 to 22, S ≥ 23), clindamycin (R ≤ 14, I = 15 to 20, S ≥ 21), and cefoxitin (R ≤ 19, S ≥ 20 for S. aureus and S. lugdunensis; R ≤ 24, S ≥ 25 for CoNS) (26).
D-zone test.
The D-zone test was performed as described by Fiebelkorn et al. (11) for the detection of MLSBi (inducible macrolide, lincosamide, and streptogramin B resistance) strains. Quality control was performed with S. aureus ATCC 25923.
β-Lactamase assays.
The microtiter nitrocefin (Calbiochem, San Diego, CA) method was used (1). Quality controls included Staphylococcus aureus ATCC 29213 as a positive control and Staphylococcus aureus ATCC 25923 as a negative control.
Oligonucleotide primers and probes.
The complete list of oligonucleotide primers and probes is shown in Table 2. One primer of each primer pair was designed to be tagged with an unrelated universal sequence at its 5′ end (named the UT primer) for efficient multiplex asymmetric PCR amplification, and another was sequence specific. The fluorescent dye TAMRA, labeled at the 5′ end of the UT primer, was simultaneously incorporated into the PCR products for subsequent hybridization detection. Oligonucleotide probe sequences were designed by multiple-sequence alignment analysis of the sequences available in GenBank by using the DNAMAN (version 4.0) program. The probes were chosen according to the consensus sequences for the resistance genes and to several species-specific sequence regions of the 16S rRNA gene for differentiation of S. aureus from CoNS. The lengths of these probes were about 20 to 30 nucleotides, with melting temperatures (Tm) between 60 and 65°C. The 5′ end of each probe was modified by adding a spacer with 12 consecutive thymines and an amino-linker group (BioAsia Co., Ltd., Shanghai, China) for covalent immobilization on the aldehyde-coated glass surface.
TABLE 2.
Primers and probes used in this study
| Primer or probe | Sequence (5′-3′)a | Target (length in bp) |
|---|---|---|
| Primer | ||
| blaZ-f | CAACGTCTAAAAGAACTAGGAGA | blaZ (259) |
| blaZ-ur | TAMRA-Uni-TAGTCTTTTGGAACACCGTCT | |
| mecA-f | GATGGCTATCGTGTCACAATC | mecA (352) |
| mecA-ur | TAMRA-Uni-TGAGTTGAACCTGGTGAAGT | |
| aac6-f | AGCCTTGGGAAGATGAAGTT | aac(6′)-Ie-aph(2‴) (513) |
| aac6-ur | TAMRA-Uni-GCCACACTATCATAACCACTAC | |
| ermA-f | CCTGTCGGAATTGGTTTTTAG | ermA (452) |
| ermA-ur | TAMRA-Uni-CGGTAAACCCCTCTGAGAATA | |
| ermC-f | AGTAATGCCAATGAGCGTTTT | ermC (303) |
| ermC-ur | TAMRA-Uni-GGTGTAATTTCGTAACTGCCA | |
| msrA-f | TACTTGAAGCTATTTACCACCA | msrA (256) |
| msrA-ur | TAMRA-Uni-TAATTTCGTTCTTTCCCCACC | |
| 23S-uf | TAMRA-Uni-AACGGTCCTAAGGTAGCGAA | 23S rRNA gene (231) |
| 23S-r | GGCTCCTACCTATCCTGTACA | |
| 16S-uf | TAMRA-Uni-AGAGTTTGATCCTGGCTCAG (10)* | 16S rRNA gene (∼1,500) |
| 16S-r | AAGGAGGTGATCCAGCC (19)* | |
| Uni | GGTTTCGGATGTTACAGCGT | |
| Probes | ||
| 16S-U | NH2-T12-GCTGCCTCCCGTAGGAGT (7)* | Bacteria 16S universal |
| 16S-G- | NH2-T12-AGGGCCATGATGACTTGACG (13)* | Gram negative 16S specific |
| 16S-G+ | NH2-T12-AAGGGGCATGATGATTTGACGTC | Gram positive 16S specific |
| 16S-Str | NH2-T12-GTTAGCCGTCCCTTTCTGG (12)* | Streptococcus 16S genus specific |
| 16S-Ent | NH2-T12-GTTTCCAAGTGTTATCCC | Enterococcus 16S genus specific |
| 16S-Sta | NH2-T12-TCCTCCATATCTCTGCGCAT | Staphylococcus 16S genus specific |
| 16S-SA | NH2-T12-AGAAGCAAGCTTCTCGTCCG | S. aureus 16S species specific |
| 16S-CoNS | NH2-T12-GGAGCAAGCTCCTTRTCTGTTC | CoNS 16S specific |
| blaZ | NH2-T12-CTGCTTTCGGTAAGACTTTAAATAAACTT | blaZ |
| mecA | NH2-T12-TATCCACCCTCAAACAGGTGAATT | mecA |
| aac6 | NH2-T12-ATTGGAGTAAAGGAATTGGTACAAGAT | aac(6′)-Ie-aph(2‴) |
| ermA | NH2-T12-ATAGTAAACCCAAAGCTCGTTGC | ermA |
| ermC | NH2-T12-TTGGAAATTATCGTGATCAACAAGTT | ermC |
| msrA | NH2-T12-GCAAATGGCATACTATCGTCAACT | msrA |
| H | NH2-TCACTTGCTTCCGTTGAGG-HEX | Position control |
| IC | NH2-T12-AYGGGGTCTTTCCGTCCTGT | Internal control (23S rRNA gene) |
| NC | NH2-T12-CAAGCAGCCACGCCAGTAC | Negative control |
| EC | NH2-T12-CCTCAACGGAAGCAAGTGAT | External control |
| ECT | TAMRA-ATCACTTGCTTCCGTTGAGG | Target of external control |
*, The source reference is indicated in parentheses. All other primers and probes were designed in the present study. Y = T or C; R = A or G. TAMRA, 6-carboxy-tetramethyl-rhodamine; HEX, hexachloro-6-carboxy-fluorescine; T12, 12 consecutive thymines.
Eight-plex asymmetric PCR amplification.
Analysis of the antibiotic resistance genes and species identification was performed by eight-plex asymmetric PCR amplification. In brief, bacteria were lysed by vortexing a single fresh colony suspended in 100 μl of 1× TE (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]) with 50 mg of glass beads (Sigma). The resulting lysate was boiled for 5 min and then centrifuged to collect the supernatant as a crude template. Each PCR contained 250 μM concentrations of each deoxynucleoside triphosphate, 1 U of Taq DNA polymerase (Tianwei Times Technology Co., Ltd., Beijing, China), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, and 1 μl of lysate supernatant as a template source. The final primer mixture was optimized, where the concentration of each primer was 0.05 μM, except the concentrations of the blaZ-ur and aac6-ur primers were both 0.2 μM and the concentrations of the 16S-uf and 16S-r primers were 0.5 and 0.25 μM, respectively. Finally, a 1 μM final concentration of a universal primer (its sequence identical to the unrelated universal sequence at the 5′ end of the UT primer) was added to the PCR mixture to further balance the amplification efficiency for each gene. The reaction was performed by using a two-round amplification on the thermal cycler PTC-200 (MJ Research, Inc.). After an initial denaturation step for 3 min at 94°C, 20 cycles of the first-round amplification were performed as follows: denaturation at 94°C for 30 s, annealing at 55°C for 40 s, and extension at 72°C for 90 s. Then, the second-round amplification of 20 cycles was performed as follows: denaturation at 94°C for 30 s, followed by annealing and extension at 72°C for 120 s. The PCR products were visualized after electrophoresis through a 1.2% agarose gel and ethidium bromide staining.
Fabrication of DNA microarray.
Microarrays were produced by using a SmartArray-48 microarrayer (CapitalBio Co., Ltd., Beijing, China). The oligonucleotide probes were spotted onto the surface of the aldehyde-activated slides (CapitalBio) at a concentration of 10 μM in DNA spotting solution (CapitalBio) and covalently immobilized on the slides by the mediation of an amino group at their 5′ ends. In each array, four control probes were printed including a fluorescent dye HEX-labeled oligonucleotide as a spotting and position control, an oligonucleotide complementary to a synthetic template included in the hybridization mixture as a hybridization positive control to monitor the hybridization process, an oligonucleotide with the consensus sequence of 23S rRNA gene as a process control, and an oligonucleotide designed to not hybridize to any sequences present in the hybridization mixture as the negative control for background signal corrections.
DNA hybridization and detection.
The fluorescently labeled PCR products (8 μl) were resuspended in 10 μl of hybridization buffer (5× Denhardt solution, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.2% sodium dodecyl sulfate, and 10% dextran sulfate) containing 0.01 μM TAMRA-labeled oligonucleotide as the target of the hybridization positive control probe. The resulting hybridization mixture was heat denatured, cooled on ice immediately, and then applied to the microarray. Hybridization was performed for 1.5 h at 56°C. After hybridization, the slides were washed once with 2× SSC plus 0.1% sodium dodecyl sulfate at room temperature for 5 min and then washed twice with distilled water at room temperature for 1 min. Slides were dried by brief centrifugation and subsequently scanned with a LuxScan-10K scanner (CapitalBio). The setting used for the scanner was laser power 90 and PMT 70. The fluorescence intensities of the spots were quantified by the SpotData Pro 2.1 (CapitalBio). The mean of the fluorescence signals for the quadruplicate spots was calculated after subtraction of the fluorescence intensity of the negative control. A positive spot was defined as having a signal intensity of more than 1,000.
RESULTS
Eight-plex asymmetric PCR amplification.
Before undertaking the multiplex reaction, we confirmed that the single PCR amplifications yielded the expected amplicons. An optimized multiplex asymmetric PCR (MAPCR) was then used to simultaneously amplify the six antibiotic resistance genes [mecA, blaZ, aac(6′)-Ie-aph(2‴), ermA, ermC, and msrA], the 16S rRNA genes, and the 23S rRNA genes. This MAPCR reliably amplifies multiple targets and efficiently generates single-stranded products in a linear manner after the exponential phase by use of the UT primers and an elevated annealing temperature (72°C). Agarose gel electrophoresis results showed that a fragment about 1,500 bp corresponding to the double-stranded products of the 16S rRNA gene and a second fragment of <1,000 bp corresponding to the molecular size of its single-stranded products were seen in all of the isolates (data not shown), indicating that the MAPCR produced single-strand products efficiently. The amplification products of the antibiotic resistance genes and the 23S rRNA gene were between about 230 to 500 bp. Although the products of these genes would not be clearly observed and differentiated due to their short lengths, they hybridized on the microarray with high sensitivity and specificity.
Specificity and sensitivity of DNA microarray.
Resistance genes assayed by the microarray test were selected based on clinical considerations. Our DNA microarray contained six probes specific to the consensus region of some six prevalent antibiotic resistance genes associated with resistance of staphylococci to clinically relevant antibiotics. In addition, eight sequence-specific probes based on known 16S rRNA gene sequences were included for differentiation between S. aureus and CoNS of other staphylococcal species. Included in this set were universal genus probes for streptococcus and enterococcus targets which were used as the negative controls for identification of staphylococcal species. The printed panel is shown in Fig. 1A.
FIG. 1.
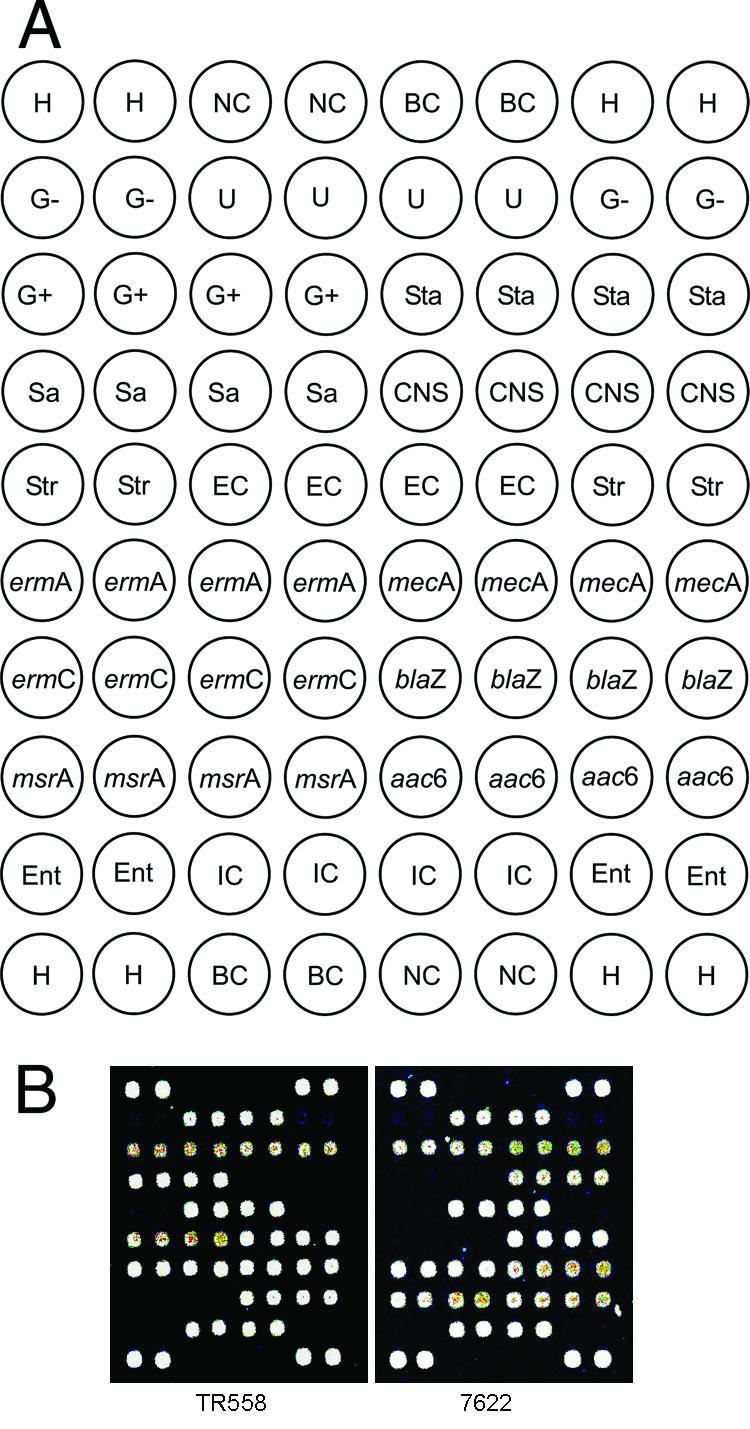
DNA microarray hybridization patterns of staphylococcal isolates. (A) Layout of the oligonucleotide probes on the array surface. H, position control; NC, negative control; BC, blank control (50% dimethyl sulfoxide); U, bacterial 16S rRNA gene universal; G−, gram-negative 16S rRNA gene specific; G+, gram-positive 16S rRNA gene specific; Str, Streptococcus 16S rRNA gene genus specific; Ent, Enterococcus 16S rRNA gene genus specific; Sta, Staphylococcus 16S rRNA gene genus specific; Sa, S. aureus 16S rRNA gene species specific; CNS, CoNS 16S rRNA gene specific; IC, internal control (23S rRNA gene); EC, external control. ermA, mecA, ermC, blaZ, msrA, and aac6 represented six antibiotic resistance genes prevalent in staphylococci. (B) Examples of DNA microarray hybridization patterns for S. aureus TR558 [with mecA, blaZ, ermA, ermC, and aac(6′)-Ie-aph(2‴) genes] and CoNS 7622 [with blaZ, mecA, aac(6′)-Ie-aph(2‴), ermC, and msrA genes].
To evaluate the specificity of the antibiotic resistance gene- and 16S rRNA gene-specific probes, we performed microarray hybridization with the MAPCR amplicons from five reference strains (Table 1), each of which harbored only a single antibiotic resistance gene and belonged either to S. aureus or to CoNS. Because no strain containing only the ermA gene was available, strain 7605 with the two genes ermA and ermC was used to assess the specificity of the ermA probe. Each of the probes hybridized specifically to its corresponding target resistance gene, and no obvious cross-hybridization with other targets was observed (data not shown). The 16S rRNA gene-specific probes also clearly differentiated S. aureus from CoNS. S. aureus TR146 did not contain any of the six tested resistance genes; thus, no signal could be detected on the probes of the tested resistance genes. Furthermore, the blank control (double-distilled H2O) gave no hybridization signals at the antibiotic resistance gene probes and 16S or 23S rRNA gene-specific probes, except for the expected signal at the external hybridization positive control. The hybridization signal intensities varied slightly due to the different thermal stabilities of the different probe sequences when bound to their targets and the variation in the input of PCR amplicons. Nonetheless, detection of antibiotic resistance profiles was not affected. To assess the detection limit of the multiplex PCR-based microarray assay, we performed a serial dilution test with S. aureus 6314, S. aureus TR558, and CoNS 7622. Reliable detection of each of the six specific resistance gene targets could be made with a minimum of 103 S. aureus or CoNS cells. This result indicates that the microarray assay could reliably and directly detect the antibiotic resistance genes from a single colony which contains about 107 to 108 cells (28).
In addition, tests using the reference strains showed that the signal intensities of the specific probes were only decreased marginally (20 to 30%) if the hybridization mixture was not denatured before hybridization (data not shown). The time of hybridization could also be shortened to 30 min with a similar small reduction of the signal intensities (a decrease of 10 to 30%) compared to the 1.5-h hybridization (data not shown).
Microarray testing of clinical isolates.
We validated the microarray assay using some 415 confirmed clinical staphylococcal isolates recovered from different patients by comparison with the phenotypic results by classical disk diffusion assay. The hybridization results showed that the microarray differentiated S. aureus from other CoNS isolates.
The accuracy of this differentiation between S. aureus and CoNS isolates was independently confirmed in all cases by analysis with the Vitek 2 system, which provides species identification (36). An overview of the resistance genotypes of all 415 clinical isolates determined with the DNA microarray is shown in Table 3, and two examples of the microarray hybridization patterns are shown in Fig. 1B. A total of 29 different resistance genotypes were detected in the clinical isolates by the DNA microarray. Interestingly, the resistance genotypes harbored in S. aureus and CoNS isolates were to some extent different. Many clinical isolates contained multiple antibiotic resistance genes. For example, 68 S. aureus isolates harbored blaZ, mecA, aac(6′)-Ie-aph(2‴), ermA, and ermC, and 35 isolates harbored blaZ, mecA, aac(6′)-Ie-aph(2‴), and ermA, whereas for CoNS, 41 isolates harbored blaZ, mecA, aac(6′)-Ie-aph(2‴), and ermC, and 34 isolates harbored blaZ, mecA, aac(6′)-Ie-aph(2‴), ermC, and msrA.
TABLE 3.
Resistance genotypes of the 415 different staphylococcal isolates tested
| Resistance genotypea | No. of strains
|
|
|---|---|---|
| CoNS | S. aureus | |
| Wild type | 12 | 8 |
| aac6 | 1 | |
| blaZ | 4 | 22 |
| ermC | 8 | 4 |
| mecA | 4 | |
| msrA | 9 | |
| blaZ aac6 | 1 | 6 |
| blaZ ermC | 16 | |
| blaZ mecA | 9 | |
| blaZ msrA | 1 | |
| ermA ermC | 1 | |
| mecA aac6 | 6 | |
| mecA ermC | 6 | |
| mecA msrA | 1 | |
| blaZ aac6 msrA | 3 | |
| blaZ mecA aac6 | 10 | 4 |
| blaZ mecA ermA | 1 | 3 |
| blaZ mecA ermC | 18 | |
| blaZ mecA msrA | 17 | |
| mecA aac6 ermA | 1 | |
| mecA aac6 ermC | 22 | |
| mecA aac6 msrA | 2 | |
| blaZ mecA aac6 ermA | 35 | |
| blaZ mecA aac6 ermC | 41 | 7 |
| blaZ mecA aac6 msrA | 22 | |
| blaZ mecA ermC msrA | 5 | |
| blaZ mecA aac6 ermA ermC | 1 | 68 |
| blaZ mecA aac6 ermA msrA | 1 | 1 |
| blaZ mecA aac6 ermC msrA | 34 | |
| Total | 237 | 178 |
aac6, aac(6′)-Ie-aph(2‴).
Relationship between the antibiotic resistance genes and the phenotypic resistance.
Table 4 shows the relationships between the microarray results and the phenotypic resistance determined by the disk diffusion methods and the microtiter nitrocefin method. The MAPCR-based microarray results correlated well with phenotypic antimicrobial susceptibility testing, and accurately differentiated the phenotypically resistant isolates from the phenotypically susceptible isolates, with a sensitivity of >90% for all of the tested antimicrobials. There were no significant differences between the genotypes and phenotypes for resistance to oxacillin, penicillin (detected by the nitrocefin method), and gentamicin in S. aureus and to penicillin (detected by the nitrocefin method), clindamycin, and erythromycin in CoNS (all P values were >0.05 as determined by the McNemar matched chi-square test; Table 4). Furthermore, the microarray results for mecA gene were found to be 100% consistent with the results of the “gold standard” mecA-PCR (26; data not shown), suggesting that the microarray assay would be acceptable for the detection of methicillin resistance in the patient care setting, such as a positive blood culture.
TABLE 4.
Relationship between resistance gene status determined by microarray and phenotypic resistance
| Parametera | Relationshipb
|
|||||
|---|---|---|---|---|---|---|
| mecA to OX | blaZ to nitrocefin | blaZ/mecA to PEN | aac(6′)-Ie-aph(2‴) to GM | ermA/ermC to CM | ermA/ermC/msr to ER | |
| S. aureus (n = 178) | ||||||
| No. of gene-positive isolates | 118 | 165 | 165 | 125 | 134 | 137 |
| % Sensitivity (%) | 98.3 (116/118) | 99.4 (164/165) | 95.4 (165/173) | 98.3 (119/121) | 91.1 (133/146c) | 92.5 (136/147) |
| % Specificity (%) | 96.7 (58/60) | 92.3 (12/13) | 100.0 (5/5) | 89.5 (51/57) | 96.9 (31/32) | 96.8 (30/31) |
| P | 1.000 | 1.000 | 0.008 | 0.289 | 0.002 | 0.006 |
| CoNS (n = 237) | ||||||
| No. of gene-positive isolates | 201 | 165 | 207 | 139 | 139 | 191 |
| % Sensitivity | 97.9 (184/188) | 98.1 (158/161) | 91.9 (203/221) | 100 (79/79) | 98.6 (139/141d) | 97.9 (191/195) |
| % Specificity | 65.3 (32/49) | 90.8 (69/76) | 75.0 (12/16) | 62.0 (98/158) | 100.0 (96/96) | 100.0 (42/42) |
| P | 0.007 | 0.344 | 0.004 | <0.001 | 0.500 | 0.125 |
| Total (n = 415) | ||||||
| % Sensitivity | 98.0 (300/306) | 98.8 (322/326) | 93.4 (368/394) | 99.0 (198/200) | 94.8 (272/287) | 95.6 (327/342) |
| % Specificity | 82.6 (90/109) | 91.0 (81/89) | 81.0 (17/21) | 69.3 (149/215) | 99.2 (127/128) | 98.6 (72/73) |
| P | 0.015 | 0.388 | <0.001 | <0.001 | 0.001 | 0.001 |
Sensitivity was calculated as the number of resistance gene-positive strains with phenotypic resistance/the number of strains with phenotypic resistance (indicated in parentheses). Specificity was calculated as the number of resistance gene-negative strains with phenotypic susceptibility/the number of strains with phenotypic susceptibility (indicated in parentheses). The P value was calculated using the McNemar matched chi-square test (SPSS 13.0; SPSS, Inc., Chicago, IL).
OX, oxacillin (phenotypic oxacillin results were detected by using a cefoxitin disk); PEN, penicillin; GM, gentamicin; CM, clindamycin; ER, erythromycin. The status of the related gene(s) was determined by microarray hybridization.
Includes 22 S. aureus isolates with MLSBi resistance.
Includes 45 CoNS isolates with MLSBi resistance.
The microarray assay could detect >95% of the phenotypically resistant S. aureus and CoNS isolates for methicillin, penicillin (detected by the nitrocefin method), and gentamicin; however, for penicillin resistance in CoNS the sensitivity was only 91.9% using the penicillin disks. For clindamycin and erythromycin, the microarray assay detected 98.6 and 97.9%, respectively, of the phenotypically resistant CoNS isolates, while its accuracy was only 91.1 and 92.5%, respectively, in S. aureus isolates. Overall, the specific probes on the microarray can detect ≥90% of S. aureus and CoNS isolates that are phenotypically resistant to the antibiotics most frequently used for staphylococci infections.
The specificity of the microarray results with the phenotypic susceptibility was good for S. aureus isolates, with >90% specificity for all of the tested antimicrobials except for gentamicin. This result further indicates that the absence of the corresponding antibiotic resistance gene(s) is highly correlated with the phenotypic susceptibility in S. aureus isolates. However, for CoNS, the microarray results were only highly related to the phenotypic susceptibility results for MLSB, macrolide, and penicillin (when detected by the nitrocefin method) with a specificity of >90%. For penicillin (detected by penicillin disk) and oxacillin (detected by cefoxitin disk), the specificity relationship was much lower at 75 and 65.3%, respectively. Furthermore, the specificity of gentamicin susceptibility was low both in S. aureus (89.5%) and in CoNS (62.0%) isolates. Many aac(6′)-Ie-aph(2‴)-positive isolates were phenotypically susceptible to gentamicin, especially among CoNS isolates (n = 60). For all strains with a discrepancy between phenotype and genotype, the MICs of the corresponding antibiotics were determined by the agar dilution method on Muller-Hinton agar with an inoculum of 104 CFU per spot according to the NCCLS (24), using the breakpoints of the MICs as follows (in mg/liter): oxacillin (R ≥ 4, S ≤ 2 for S. aureus; R ≥ 0.5, S ≤ 0.25 for CoNS), gentamicin (R ≥ 8, S ≤ 4), clindamycin (R ≥ 4, S ≤ 0.5), and erythromycin (R ≥ 8, S ≤ 0.5). The MIC results showed that most of the phenotypically susceptible CoNS strains containing related resistance genes [mecA or aac(6′)-Ie-aph(2‴)] were borderline resistant or susceptible [15 of 17 mecA-positive CoNS isolates and 43 of 60 aac(6′)-Ie-aph(2‴)-positive CoNS isolates]. The high percentage of oxacillin borderline-resistant CoNS isolates may be the cause of the lower specificity for CoNS and oxacillin resistance as evaluated using the cefoxitin disk compared to that reported by Swenson et al. (38). In addition, five (2.8%) S. aureus (MLSBc phenotypic resistance) contained the ermB gene (PCR and DNA sequence analysis [data not shown]), conferring the resistant MICs to clindamycin and erythromycin.
DISCUSSION
We describe here a MAPCR-based microarray assay that can be used to survey clinically relevant antibiotic resistance genes frequently encountered in staphylococci. One of the major advantages of our method over other multiplex PCR-based or microarray-based systems described previously (27, 37) is that it reliably detects the most prevalent different groups of antibiotic resistance genes in staphylococci, and simultaneously differentiates S. aureus from other CoNS. These properties are necessary for its potential use for clinical diagnosis of the antibiotic resistance of staphylococcal infections. Probes for some infrequently encountered antibiotic resistance genes, such as ermB, were not included in the microarray assay, with the consequence that about 0.6 to 3% of the MLSB resistance cannot be detected (32, 40). In addition, the genes aphA3 coding for APH(3′)III enzyme and aadC coding for ANT(4′,4‴) enzyme, which combined contribute only 10 to 30% of the resistance to aminoglycosides and are not considered clinically relevant because they mediate resistance to aminoglycosides not usually prescribed to treat staphylococcal infections (33), were also not present in the microarray assay. The low number of abundant resistance genes detected by the current MAPCR-based microarray assay was intentional, since it allows for sufficient numbers of samples of each genotype to be validated statistically. In the future, we may need to analyze more samples to ensure the accuracy of the microarray platform over a wider diversity of different resistance genotypes. We intend also to develop a second generation of MAPCR-based microarray by increasing the range of resistance genes to include less frequently encountered resistance mechanisms such as those encoded by the ermB, aphA3, and aadC genes and others.
Hamels et al. (15) also described a microarray for simultaneous identification of Staphylococcus species and methicillin resistance. However, other prevalent antibiotic resistance genes could not be detected in that assay. The MLSBi phenotype is not easily detected by standard susceptibility test methods, while failure to identify MLSBi resistance may lead to clinical failure when clindamycin therapy is used (8, 35). It is increasingly important to distinguish the MLSBi strains from MS-resistant strains that contain the msrA gene. Thus, the second benefit of our microarray is that it clearly identifies strains that remain susceptible to clindamycin but have MS resistance and the MLSBi phenotype. In the clinical setting, the simultaneous identification of the bacteria and determination of its susceptibility to antibiotics generally require 48 h (3), whereas in our method the detection time can be shortened to 5 h from a cultured isolate. In addition, unlike the traditional multiplex PCR which requires extensive optimizations (17), the MAPCR reaction used in the present study was easily optimized and reproducibly achieved efficient multiplex amplification by simple adjustment of the individual primer concentrations, without additional optimization of either the reaction components or annealing temperatures. Initially, we used the equimolar concentrations of each primer pair in the multiplex asymmetric amplification. However, the hybridization signals of the 16S rRNA, blaZ, and aac(6′)-Ie-aph(2‴) genes were weak or undetected (data not shown). When we increased the concentration ratios of these three primer pairs to 2:1 for the 16S rRNA gene and to 4:1 for the blaZ and aac(6′)-Ie-aph(2‴) genes, strong hybridization signals were seen for all genes without further optimization. Since the microarray was evaluated using isolated bacterial colonies, which required culture prior to analysis, further work will focus on increasing the sensitivity of the methods so that this assay could be adapted to use in direct detection from positive blood cultures and from normally stable clinical samples such as sputum or urine. In addition, the probes for detection of additional clinically relevant resistance genes in staphylococci such as vanA and vanB for glycopeptide resistance and the mutations of DNA topoisomerases for fluoroquinolone resistance, and the resistance genes of other clinically significant isolates, such as Streptococcus and Enterococcus, can be designed and incorporated into a system for extensive detection of possible infection and antibiotic resistance profiles of gram-positive bacteria.
We have compared this microarray assay for the detection of antibiotic resistance genes with traditional phenotypic methods for the determination of antibiotic susceptibility. Overall, we found correlations were more than 90% for detection of the phenotypic resistance and 100% for species differentiation. However, some of the discrepancies between the microarray results and the disk diffusion results are significant (P < 0.05 as determined by the McNemar matched chi-square test; Table 4) and were due mainly to the phenotypically susceptible but resistance gene-positive isolates. For example, we encountered oxacillin-susceptible but mecA-positive CoNS (n = 17), presumably because of the known heterogeneous expression of mecA gene in Staphylococcus in vitro (6). For more frequently encountered gentamicin-susceptible but aac(6′)-Ie-aph(2‴)-positive S. aureus (n = 6) and CoNS (n = 60) isolates, the discrepancy in genotype and phenotype might be attributed to the so-called silent antibiotic resistance gene that might become activated to express the resistance (21), and their presence may also facilitate the spread to other bacteria. From a clinical perspective, a susceptible strain harboring but not expressing an antibiotic resistance gene should be regarded as potentially resistant to that antibiotic (22). Thus, detection of the discrepancies between the presence of the resistance gene and the phenotypic susceptibility (false-positive) is important for physicians to guide prescription of appropriate functional antibiotic therapy so as to control the spread of the resistance due to antibiotic selection. Furthermore, there are also discrepancies between the absence of the antibiotic gene test on the microarray and the phenotypic resistance (false negative). For example, approximately 7 to 10% of MLSB resistance in S. aureus could not be detected by the microarray assay because of the presence of the ermB gene or other mechanisms, which results in a discrepancy of detection of clindamycin resistance in S. aureus between phenotype and genotype that is significant (P = 0.002 as determined by the McNemar matched chi-square test). We intend to develop the microarray to increase the genotypic testing of such related resistance genes so that it can also be readily adopted for detection of clindamycin resistance in samples such as skin or soft tissue isolates of S. aureus.
In conclusion, the MAPCR-based microarray assay provides a rapid, simple, and reliable tool for parallel detection of the prevalent antibiotic resistance genes in staphylococci and a definite discrimination between S. aureus and CoNS in a 5-h procedure after pure culture isolation. This approach appears to be highly robust and highly informative, can be adapted to analyze the clinically important staphylococcal isolates for diagnosis-based studies, and could supplement or provide an early indication of likely antibiotic resistances. This early detection would allow clinicians initially to avoid potentially inappropriate treatment options and allow prompt intervention in infection control issues. In the future, we will work on increasing the sensitivity of the microarray for nonculture diagnoses of bacterial infections and expanding the repertoire of the antibiotic resistance genes for detection of more phenotypic resistance.
Acknowledgments
This study was funded by grant 2006AA020701 from the Department of Science and Technology of China.
We thank Wei-Ping Yang for critical reading of the manuscript and Ning Du, Tian-Tian Cai, Xia Zhou, Qing-Mei Ma, and Na Zhang for proficient technical support.
Footnotes
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Appelbaum, P. C., S. K. Spangler, and M. R. Jacobs. 1990. Evaluation of two methods for rapid testing for β-lactamase production in Bacteroides and Fusobacterium. Eur. J. Clin. Microbiol. Infect. Dis. 9:47-50. [DOI] [PubMed] [Google Scholar]
- 2.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron, M. G., and M. Ouellette. 1998. Preventing antibiotic resistance through rapid genotypic identification of bacteria and of their antibiotic resistance genes in the clinical microbiology laboratory. J. Clin. Microbiol. 36:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byl, B., P. Clevenbergh, F. Jacobs, M. J. Struelens, F. Zech, A. Kentos, and J. P. Thys. 1999. Impact of infectious diseases specialists and microbiological data on the appropriateness of antimicrobial therapy for bacteremia. Clin. Infect. Dis. 29:60-66. [DOI] [PubMed] [Google Scholar]
- 5.Call, D. R., M. K. Bakko, M. J. Krug, and M. C. Roberts. 2003. Identifying antimicrobial resistance genes with DNA microarrays. Antimicrob. Agents Chemother. 47:3290-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daims, H., A. Brühl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 8.Drinkovic, D., E. R. Fuller, K. P. Shore, D. J. Holland, and R. Ellis-Pegler. 2001. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48:315-316. [DOI] [PubMed] [Google Scholar]
- 9.Eady, E. A., J. I. Ross, J. L. Tipper, C. E. Walters, J. H. Cove, and W. C. Noble. 1993. Distribution of genes encoding erythromycin ribosomal methylases and an erythromycin efflux pump in epidemiologically distinct groups of staphylococci. Antimicrob. Agents Chemother. 31:211-217. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct nucleotide determination of entire genes. Characterization of a gene coding for 16S rRNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiebelkorn, K. R., S. A. Crawford, M. L. McElmeel, and J. H. Jorgensen. 2003. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J. Clin. Microbiol. 41:4740-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human faeces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giammarinaro, P., S. Leroy, J. P. Chacornac, J. Delmas, and R. Talon. 2005. Development of a new oligonucleotide array to identify staphylococcal strains at species level. J. Clin. Microbiol. 43:3673-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamels, S., J.-L. Gala, S. Dufour, P. Vannuffel, N. Zammatteo, and J. Remacl. 2001. Consensus PCR and microarray for diagnosis of the genus Staphylococcus, species, and methicillin resistance. BioTechniques 31:1364-1366. 1368:1370-1372. [DOI] [PubMed] [Google Scholar]
- 16.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henegariu, O., N. A. Heerema, S. R. Dlouhy, G. H. Vance, and P. H. Vogt. 1997. Multiplex PCR: critical parameters and step-by-step protocol. BioTechniques 23:504-511. [DOI] [PubMed] [Google Scholar]
- 18.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 150:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroes, I., P. Lepp, and D. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall, S. A., W. W. Wilke, M. A. Pfaller, and R. N. Jones. 1998. Staphylococcus aureus and coagulase-negative staphylococci from blood stream infections: frequency of occurrence, antimicrobial susceptibility, and molecular (mecA) characterization of oxacillin resistance in the SCOPE program. Diagn. Microbiol. Infect. Dis. 30:205-214. [DOI] [PubMed] [Google Scholar]
- 21.Martineau, F., F. J. Picard, L. Grenier, P. H. Roy, M. Ouellette, M. G. Bergeron, et al. 2000. Multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci isolated from patients infected after cardiac surgery. J. Antimicrob. Chemother. 46:527-533. [DOI] [PubMed] [Google Scholar]
- 22.Martineau, F., F. J. Picard, N. Lansac, C. Ménard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2000. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy, E., L. Huwyler, and M. Freire-Bastos. 1985. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 4:3357-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 6th ed. M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 25.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disk susceptibility tests; approved standard, 8th ed. M2-A8. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 26.National Committee for Clinical Laboratory Standards. 2005. Performance standards for antimicrobial susceptibility testing; fifth informational supplement. M100-S15. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 27.Perreten, V., L. Vorlet-Fawer, P. Slickers, R. Ehricht, P. Kuhnert, and J. Frey. 2005. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. J. Clin. Microbiol. 43:2291-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramakrishnan, R., W. Buckingham, M. Domanus, L. Gieser, K. Klein, G. Kunkel, A. Prokhorova, and P. V. Riccelli. 2004. Sensitive assay for identification of methicillin-resistant Staphylococcus aureus, based on direct detection of genomic DNA by use of gold nanoparticle probes. Clin. Chem. 50:1949-1952. [DOI] [PubMed] [Google Scholar]
- 29.Ramsay, G. 1998. DNA chips: state-of-the art. Nat. Biotechnol. 16:40-44. [DOI] [PubMed] [Google Scholar]
- 30.Ross, J. I., E. A. Eady, J. H. Cove, W. J. Cunliffe, S. Baumberg, and J. C. Wootton. 1990. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 4:1207-1214. [DOI] [PubMed] [Google Scholar]
- 31.Schmitz, F. J., A. C. Fluit, M. Gondolf, R. Beyrau, E. Lindenlauf, J. Verhoef, H. P. Heinz, and M. E. Jones. 1999. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 43:253-259. [PubMed] [Google Scholar]
- 32.Schmitz, F. J., J. Petridou, A. C. Fluit, U. Hadding, G. Peters, C. von Eiff, et al. 2000. Distribution of macrolide-resistance genes in Staphylococcus aureus blood-culture isolates from fifteen German university hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 19:385-387. [DOI] [PubMed] [Google Scholar]
- 33.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shore, A., A. S. Rossney, C. T. Keane, M. C. Enright, and D. C. Coleman. 2005. Seven Novel Variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 49:2070-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siberry, G. K., T. Tekle, K. Carroll, and J. Dick. 2003. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin. Infect. Dis. 37:1257-1260. [DOI] [PubMed] [Google Scholar]
- 36.Spanu, T., M. Sanguinetti, D. Ciccaglione, T. D'Inzeo, L. Romano, F. Leone, and G. Fadda. 2003. Use of the VITEK 2 system for rapid identification of clinical isolates of staphylococci from bloodstream infections. J. Clin. Microbiol. 41:4259-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strommenger, B., C. Kettlitz, G. Werner, and W. Witte. 2003. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 41:4089-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swenson, J. M., F. C. Tenover, and the Cefoxitin Disk Study Group. 2005. The results of disk diffusion testing with cefoxitin correlate with presence of mecA in Staphylococcus spp. J. Clin. Microbiol. 43:3818-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakker-Varia, S., W. D. Jenssen, L. Moon-McDermott, M. P. Weinstein, and D. T. Dubin. 1987. Molecular epidemiology of macrolide-lincosamide-streptogramin B resistance in Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob. Agents Chemother. 31:735-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volokhov, D., V. Chizhikov, K. Chumakov, and A. Rasooly. 2003. Microarray analysis of erythromycin resistance determinants. J. Appl. Microbiol. 95:787-798. [DOI] [PubMed] [Google Scholar]
- 41.Wielders, C. L. C., A. C. Fluit, S. Brisse, J. Verhoef, and F. J. Schmitz. 2002. mecA gene is widely disseminated in Staphylococcus aureus population. J. Clin. Microbiol. 40:3970-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, L., Z. Zhang, C. Wang, H. Yang, Q. Zhang, and J. Cheng. 2006. Evaluation of the CLSI cefoxitin 30-μg disk-diffusion method for detecting methicillin resistance in staphylococci. Clin. Microbiol. Infect. 12:1039-1042. [DOI] [PubMed] [Google Scholar]


