Abstract
We have evaluated the COBAS TaqMan hepatitis C virus (HCV) test (version 2.0) for use with the High Pure system (HCVHPS V2), a new, revised real-time reverse transcription-PCR assay developed to improve the genotype quantitation of version 1.0 (HCVHPS V1). Revisions were made in the wash buffer and in the reverse transcription temperature. The genotype inclusivity of HCVHPS V2 was evaluated at three different sites, using HCVHPS V2, HCVHPS V1, and the COBAS AMPLICOR HCV MONITOR test (version 2.0) (CAHCM). The fully automated COBAS Ampliprep/COBAS TaqMan HCV test was also used in one of the participating laboratories. The mean differences in HCV RNA values between HCVHPS V2 and CAHCM and between HCVHPS V2 and HCVHPS V1 ranged from −0.21 to 0.13 log and from 0.24 to 1.27 log, respectively, with >0.5-log differences for genotypes 2, 3, 4, and 5. With a NIBSC panel of HCV genotypes 1 through 6, the measured HCVHPS V2 values were within 0.25 log of the nominal values for all 6 genotypes. When serial dilutions of genotype-specific clinical HCV specimens were tested, the assay showed a limit of detection between 10 and 20 IU/ml and a linear range of 25 IU/ml to 3.91 × 108 IU/ml. Clinical and analytical specificities of 100% were demonstrated with 100 HCV-seronegative specimens as well as with 12 non-HCV members of Flaviviridae and 22 additional microorganisms. These data indicate that HCVHPS V2 is a robust and accurate test for the quantitation of all six HCV genotypes and useful in monitoring viral load in all clinical HCV specimens.
Viral load monitoring is an important part of the disease management strategies that have been developed for chronic hepatitis C. Changes in hepatitis C virus (HCV) RNA levels occurring during the early phase of antiviral therapy have been derived from complex models of viral kinetics and applied to the prediction of treatment outcomes (14, 24-25). The high negative predictive value of a <2-logarithm decrease at week 12 of pegylated interferon/ribavirin combination therapy compared to the baseline has led to the introduction in treatment guidelines of a stopping rule which helps shorten therapy duration for nonresponsive patients (2, 11). More-recent studies suggest that this approach may be applied at earlier time points during treatment and that the predictive value of viral load at week 4 may help in individualizing treatment duration (3, 7, 12-13). Research and guidelines for treatment in this field will be better supported by the development and use of more-accurate quantitative assays that allow for precise measurement and monitoring of viral kinetics (8). Real-time PCR assays have been recently introduced to offer higher sensitivity and broader dynamic ranges than end point PCR tests (6, 15). One of these assays, the semiautomated, real-time PCR system COBAS TaqMan HCV test (version 2.0) for use with the High Pure system (HCVHPS V1), has been limited to genotypes 1 and 6 because of an inconsistent performance with the other genotypes (20). In this respect, we have evaluated in a multisite study an improved test, HCVHPS V2, by thoroughly investigating its genotype inclusivity, sensitivity, specificity, and linear range in comparison to those of the licensed COBAS AMPLICOR HCV MONITOR test (version 2.0) (CAHCM) and HCVHPS V1 systems.
MATERIALS AND METHODS
Samples.
Clinical specimens from patients with chronic HCV infection were obtained from various vendors: Saarland University Hospital (SUH; Homburg, Germany), TriCore Reference Laboratories (TRL; Albuquerque, NM), and Roche Diagnostics K.K. (RDKK; Tokyo, Japan). HCV genotypes and subtypes of each sample were determined either by sequencing of the 5′ untranslated region or by HCV genotyping line probe/blot assays. Armored HCV RNA was designed to contain a region of the 5′ untranslated region of an HCV genotype 1 specimen and prepared using processes described by WalkerPeach et al. (23). Armored HCV RNA was diluted with pools of HCV-negative human EDTA plasma or serum prior to testing. Due to the large volume required for testing in multiple replicates in multiple tests at multiple sites, some of the clinical specimens were also diluted with pools of HCV-negative human EDTA plasma or serum prior to testing. For additional studies on genotype inclusivity, a panel of all six HCV genotypes (NIBSC code 02/202) was obtained from the National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, United Kingdom.
Test procedures.
The specimens were tested by the COBAS TaqMan HCVHPS V2, the COBAS TaqMan HCVHPS V1, the COBAS AmpliPrep/COBAS TaqMan HCV test (HCMCAP), and CAHCM, according to instructions described in their respective package inserts. The differences between HCVHPS V2 and HCVHPS V1 are briefly discussed below.
(i) Extractions by the High Pure system.
In HCVHPS V2, the wash buffer reagent is reconstituted with 30 ml of deionized water and 50 ml of 96 to 100% ethanol, resulting in a final ethanol concentration of 48 to 50%. In HCVHPS V1, the wash buffer reagent is reconstituted with 80 ml of 96 to 100% ethanol, resulting in a final ethanol concentration of 77 to 80%.
(ii) Amplification and detection.
The temperature for the reverse transcription step is 64°C for HCVHPS V2 and 59°C for HCVHPS V1.
HCV genotype inclusivity.
The accuracy of quantitation for all HCV genotypes obtained using HCVHPS V2 was demonstrated by testing multiple sets of clinical specimens representing HCV genotypes 1 through 6 by three different tests (HCVHPS V2, CAHCM, and HCVHPS V1) in triplicate at three different sites: SUH (Homburg, Germany), TRL (Albuquerque, NM), and Roche Molecular Systems (RMS; Pleasanton, CA). Approximately 10 clinical specimens of each of the six HCV genotypes were tested at each of the three sites, with some of the clinical specimens overlapping among the three sites. All values were log10 transformed, and each value for each specimen at each site for each test was compared to the corresponding values obtained from the other tests. The mean difference between the values for any two tests for all specimens within each genotype at each site was then calculated. The site-to-site difference for the subset of specimens shared by more than one site was also calculated for HCVHPS V2.
At one of the sites (RMS), an additional set of 114 clinical specimens of all HCV genotypes was tested by four different tests: HCVHPS V2, HCVHPS V1, CAHCM, and HCMCAP. Furthermore, a panel of all six HCV genotypes from NIBSC (code 02/202) was also evaluated using HCVHPS V2.
Additional studies were performed at the fourth site, RDKK (Tokyo, Japan), using 30 genotyped HCV serum specimens. The specimens were tested in triplicate by each of the three tests: HCVHPS V2, CAHCM, and HCVHPS V1.
LOD (sensitivity) for all HCV genotypes.
To establish that HCVHPS V2 exhibited similar limits of detection (LOD) for all HCV genotypes, the HCV secondary standard (lot 0002, genotype 1a; 20,900 IU/ml) and available characterized clinical HCV specimens representing genotypes (2a, 2b, 3, 4, 5, and 6) were utilized. Serial dilutions to final concentrations of 30, 20, 10, 5, and 2.5 IU/ml were made in HCV-negative human EDTA plasma and serum. The concentrations of the source materials for the dilutions of HCV genotypes 2a, 2b, 3, 4, 5, and 6 were determined by CAHCM. The HCV secondary standard was prepared using a clinical genotype 1a HCV specimen calibrated to the first WHO international standard for HCV RNA nucleic acid amplification technology assays (NIBSC code 96/790) (10, 18). For each of the two different lots of the HCVHPS V2 kit, at least three independent dilution series were tested for each specimen matrix and each genotype. A minimum number of 12 replicates per concentration were tested with each HCVHPS V2 kit lot, for a total of 24 replicates per concentration. For each genotype, the lowest tested concentration that yielded a minimum of 95% positive results as well as the concentration for 95% positive results as calculated by PROBIT analysis was determined.
Linear range.
Two linearity panels consisting of 15 members of Armored HCV RNA (genotype 1) in EDTA plasma or serum were tested using three unique lot combinations of COBAS TaqMan HCV test (version 2.0) and High Pure system viral nucleic acid kits. The 15 members of the panels covered a range from 7.83 HCV RNA IU/ml to 7.83 × 108 HCV RNA IU/ml based on the source material whose concentration was determined by comparison to the HCV secondary standard (lot GTR015), prepared using a genotype 1a clinical HCV specimen calibrated to the second WHO international standard for HCV RNA nucleic acid amplification technology assays (NIBSC code 96/798) (10, 19). Twelve replicates per level for each of the three lot combinations were tested, for a total of 36 replicates per level. Linear range was determined by following the CLSI EP6-A guideline, using a maximum allowable bias of 0.2 log10 (22).
Precision.
The total imprecision of HCVHPS V2 for EDTA plasma specimens was assessed by testing each of six different concentrations of a genotype 1 specimen in EDTA plasma 48 times in 24 runs, using two lots of the COBAS TaqMan HCV test (version 2.0) kits, three analysts, and two COBAS TaqMan 48 analyzer instruments. The total imprecision of the HCVHPS V2 test for serum specimens was assessed by testing each of seven different concentrations of a genotype 1 specimen in serum 72 times in 36 runs, using two lots of the COBAS TaqMan HCV test (version 2.0) kits, three analysts, and three COBAS TaqMan 48 analyzer instruments. Lot-to-lot, instrument-to-instrument, operator-to-operator, between-run, within-run, and total imprecision were evaluated using a nested analysis of variance of log10-transformed HCV RNA values. The percent coefficient of variation (%CV) was calculated using the formula  , where SD and ln 10 represent the standard deviation of the log10-transformed value and the natural log of 10, respectively (4).
, where SD and ln 10 represent the standard deviation of the log10-transformed value and the natural log of 10, respectively (4).
Specificity.
The clinical specificity of HCVHPS V2 was determined by testing 50 HCV-seronegative EDTA plasma and 50 HCV-seronegative serum specimens with two different COBAS TaqMan HCV test (version 2.0) kit lots. The analytical specificity was evaluated by adding microorganisms into HCV-negative human EDTA plasma and serum. Twelve non-HCV members of Flaviviridae (West Nile virus, St. Louis encephalitis virus, dengue virus types 1 through 4, yellow fever virus, Zika virus, Banzi virus, Ilheus virus, Murray Valley virus, and hepatitis G virus) and 22 additional microorganisms (Staphylococcus aureus, Staphylococcus epidermis, Epstein-Barr virus, Propionibacterium acnes, human adenovirus type 3, cytomegalovirus Davis and Towne, herpes simplex virus type 1 MacIntyre and type 2G, hepatitis A virus, hepatits B virus genotypes A and B, human papilloma virus types 11, 18, and 6B, varicella-zoster virus, Candida albicans, human T-cell leukemia virus types 1 and 2, influenza B virus, Mycobacterium avium, and human immunodeficiency virus) were tested for possible cross-reactivity using HCVHPS V2.
Interference.
Twenty-three different drug compounds (tenofovir disoproxil fumarate, enfuvirtide, nevirapine, efavirenz, lamivudine, zidovudine, zalcitabine, stavudine, ribavirin, abacavir sulfate, didanosine, pegylated alpha interferon 2a, alpha interferon 2b, alpha interferon 2a, indinavir sulfate, ritonavir, nelfinavir sesylate, saquinavir, amprenavir, lopinavir/ritonavir, paroxetine, fluoxetine, and sertraline) at concentrations three times their respective peak plasma concentrations in EDTA plasma and serum were tested by HCVHPS V2. An HCV specimen of approximately 2 × 104 IU/ml with or without the spiked drug was examined by HCVHPS V2 in triplicate. The differences in mean log HCV RNA values (IU/ml) between the drug-spiked and the control (no-drug) specimens were calculated. A mean difference within 0.3 log10 was considered to indicate no interference for the drug tested.
RESULTS
HCV genotype inclusivity.
The results from HCV genotype inclusivity studies performed with HCVHPS V2, HCVHPS V1, and CAHCM at three sites (TRL, SUH, and RMS) are shown in Table 1. All three sites generated results with similar mean differences in log HCV RNA (IU/ml) values for the comparison between HCVHPS V2 and HCVHPS V1 or HCVHPS V2 and CAHCM for all genotypes except genotype 2 at SUH for the comparison between HCVHPS V2 and HCVHPS V1. The mean differences in log HCV RNA (IU/ml) between HCVHPS V2 and HCVHPS V1 for all three sites combined were 0.24, 0.56, 1.27, 1.05, 0.73, and 0.25 log10 for genotypes 1 through 6, respectively. These results showed greatly enhanced genotype inclusivity for HCVHPS V2 in comparison to that for HCVHPS V1 for genotypes 2, 3, 4, and 5. For the comparison of HCVHPS V2 to CAHCM, the mean differences in log HCV RNA (IU/ml) between the two tests for all three sites combined were much smaller, at −0.21, −0.11, 0.13, −0.02, −0.13, and −0.16 log10 for genotypes 1 through 6, respectively. The results demonstrated that the genotype inclusivity of HCVHPS V2 is very similar to that of CAHCM for all six genotypes.
TABLE 1.
Genotype inclusivity: mean differences in log HCV RNA value for each genotype at three different sites (TRL, SUH, and RMS) among three tests (HCVHPS V2, HCVHPS V1, and CAHCM)
| Site | Methods compared | Difference in log HCV RNA value (IU/ml) for indicated genotype(s)a
|
||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | All | ||
| TRL | HCVHPS V2, HCVHPS V1 | 0.19 (8) | 0.36 (9) | 1.37 (13) | 0.99 (9) | 0.64 (11) | 0.30 (10) | 0.69 (60) |
| SUH | HCVHPS V2, HCVHPS V1 | 0.30 (10) | 0.88 (10) | 1.09 (10) | 1.15 (9) | 0.87 (8) | 0.25 (10) | 0.75 (57) |
| RMS | HCVHPS V2, HCVHPS V1 | 0.23 (7) | 0.43 (9) | 1.35 (13) | 0.99 (6) | 0.68 (11) | 0.20 (10) | 0.69 (56) |
| All 3 sites | HCVHPS V2, HCVHPS V1 | 0.24 (25) | 0.56 (28) | 1.27 (36) | 1.05 (24) | 0.73 (30) | 0.25 (30) | 0.71 (173) |
| TRL | HCVHPS V2, CAHCM | −0.25 (8) | −0.18 (9) | 0.07 (13) | −0.06 (9) | −0.16 (11) | −0.18 (10) | −0.11 (60) |
| SUH | HCVHPS V2, CAHCM | −0.23 (10) | −0.04 (10) | −0.01 (10) | 0.04 (9) | 0.02 (8) | −0.20 (10) | −0.07 (57) |
| RMS | HCVHPS V2, CAHCM | −0.316 (7) | −0.12 (9) | 0.32 (13) | −0.03 (6) | −0.24 (11) | −0.12 (10) | −0.04 (56) |
| All 3 sites | HCVHPS V2, CAHCM | −0.21 (25) | −0.11 (28) | 0.13 (36) | −0.02 (24) | −0.13 (30) | −0.16 (30) | −0.07 (173) |
Numbers in parentheses denote the numbers of clinical specimens tested.
When analyzing differences among the three sites for the subset of shared specimens tested by HCVHPS V2 at more than one site, we observed mean differences in log HCV RNA (IU/ml) between any two sites of 0.10 log10 or less for all specimens and 0.21 log10 or less for each of the six HCV genotypes (data not shown).
The variable differences between the HCVHPS V2 test and the HCVHPS V1 test among the three sites for specimens of HCV genotype 2, possibly due to site-specific subtype distribution, were examined at a fourth site, RDKK (data not shown). The mean differences in log HCV RNA values (IU/ml) between HCVHPS V2 and HCVHPS V1 were −0.03, 0.67, and −0.09 log10 for genotypes 1b, 2a, and 2b, respectively. For the comparison of HCVHPS V2 to CAHCM, the mean differences in log HCV RNA values (IU/ml) between the two tests were −0.12, −0.34, and 0.14 log10 for genotypes 1b, 2a, and 2b, respectively. The results at RDKK demonstrated greatly enhanced genotype inclusivity for HCVHPS V2 for subtype 2a in comparison to that for HCVHPS V1 and are consistent with the above-mentioned possibility for variable composition of subtypes for the genotype 2 specimens tested at the three sites.
In order to further demonstrate the equivalent quantitation of HCV genotypes by HCVHPS V2, two more studies were carried out at one of the sites (RMS). In the first study, 114 clinical specimens of various HCV genotypes were examined by four different tests: HCVHPS V2, HCVHPS V1, CAHCM, and HCMCAP. The results shown in Table 2 demonstrated that the mean log HCV RNA values (IU/ml) for genotypes 2a, 3, 4, and 5 obtained by HCVHPS V2 are 0.48 to 0.90 log10 higher than those obtained by HCVHPS V1. In addition, the mean log HCV RNA values (IU/ml) obtained by HCVHPS V2 were similar to those obtained by CAHCM and HCMCAP for all HCV genotypes. Also in this series, genotype 2a, although representing a dramatic improvement over HCVHPS V1, was somewhat less efficiently quantified than the other genotypes. The result for quantification of this genotype was within 0.5 log10 of the HCMCAP result. In the second study, the panel of HCV genotypes 1 through 6 from NIBSC (code 02/202) was tested once using HCVHPS V2. As shown in Table 3, all six of the HCV genotype specimens in the NIBSC panel yielded values that were within 0.25 log of the expected nominal value.
TABLE 2.
Genotype inclusivity: mean differences in log HCV RNA value for each genotype at RMS among four assays (HCVHPS V2, HCVHPS V1, CAHCM, and HCMCAP)
| Methods compared | Difference in log HCV RNA value (IU/ml) for indicated genotype(s)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2a | 2b | 3 | 4 | 5 | 6 | All | |
| HCVHPS V2, HCVHPS V1 | −0.10 (25) | 0.77 (9) | −0.06 (14) | 0.90 (22) | 0.78 (22) | 0.48 (9) | 0.02 (13) | 0.40 (114) |
| HCVHPS V2, CAHCM | −0.12 (25) | −0.21 (7) | −0.04 (14) | 0.23 (22) | −0.07 (16) | −0.03 (9) | −0.15 (13) | −0.03 (106) |
| HCVHPS V2, HCMCAP | −0.15 (23) | −0.41 (9) | −0.01 (14) | 0.21 (20) | 0.12 (19) | 0.02 (9) | 0.01 (11) | −0.01 (105) |
Numbers in parentheses denote the numbers of clinical specimens tested.
TABLE 3.
Genotype inclusivity: difference between observed and expected log HCV RNA values for HCV genotypes 1 through 6 from the NIBSC HCV genotype panel (NIBSC code 02/202)
| HCV genotype | Log HCV RNA value (IU/ml)
|
Observed value − expected value | |
|---|---|---|---|
| Expected | Observed | ||
| 1 | 3.00 | 2.76 | −0.24 |
| 2 | 3.00 | 3.15 | 0.15 |
| 3 | 3.00 | 3.01 | 0.01 |
| 4 | 3.00 | 2.78 | −0.22 |
| 5 | 3.00 | 3.00 | 0.00 |
| 6 | 3.00 | 3.15 | 0.15 |
LOD (sensitivity) for all HCV genotypes.
To verify that HCVHPS V2 had similar LOD values for all HCV genotypes, limiting dilution experiments were performed. For each genotype, the lowest concentration required to yield positive results in at least 95% of replicates as well as the concentration for 95% positive results obtained by PROBIT analysis are shown in Table 4. The LOD values for all HCV genotypes for both specimen matrices were between 10 and 20 HCV RNA IU/ml based on the observed lowest level required to yield at least a 95% positivity rate. PROBIT analysis of these data showed similar LOD values that ranged from 6.3 (genotype 3 in EDTA plasma) to 19.4 (genotype 2a in serum) HCV RNA IU/ml. No trend was observed for consistently lower or higher quantitation results for a given specimen matrix.
TABLE 4.
LOD: HCV genotype detection for HCVHPS V2
| Genotype | Actually observed lowest concn (IU/ml) yielding at least 95% positive results
|
Concn (IU/ml) for 95% positive results as calculated by PROBIT analysis
|
||
|---|---|---|---|---|
| EDTA plasma | Serum | EDTA plasma | Serum | |
| 1 | 20 | 20 | 16.7 | 11.3 |
| 2a | 20 | 20 | 11.8 | 19.4 |
| 2b | 20 | 10 | 9.6 | 11.0 |
| 3 | 10 | 10 | 6.3 | 11.1 |
| 4 | 20 | 20 | 14.9 | 13.5 |
| 5 | 10 | 20 | 14.3 | 13.4 |
| 6 | 10 | 10 | 7.0 | 10.7 |
Linear range.
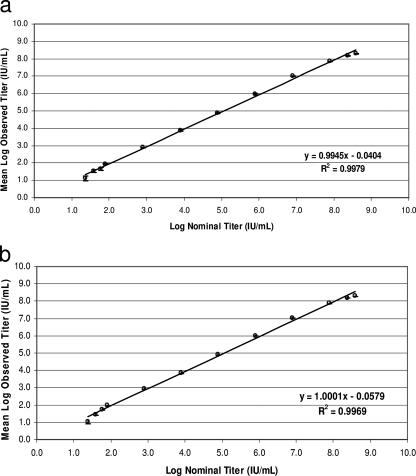
The analysis of linearity study results showed that the linear range for EDTA plasma matrix for HCVHPS V2 was from 23.5 HCV RNA IU/ml to 3.91 × 108 HCV RNA IU/ml (Fig. 1a). The largest difference between the linear fit and the third-order polynomial fit for all levels within the linear range was 0.16 log10. This value was within the preset 0.2 log10 allowable bias (data not shown). For serum matrix, the linear range was from 15.7 HCV RNA IU/ml to 3.91 × 108 HCV RNA IU/ml (Fig. 1b), with the largest difference (0.20 log10) between the linear fit and the third-order polynomial fit (data not shown).
FIG. 1.
Linear range of the COBAS TaqMan HCV test (version 2.0) in EDTA plasma (a) and serum (b). Error bars denote 2 standard errors of the mean values.
Precision.
Tables 5 and 6 show the %CV of each component of the variance analysis and the total %CV for HCV genotype 1 for EDTA plasma and serum, respectively. For EDTA plasma, the total %CV values for the six levels ranged from 32.2% to 35.8% and were fairly constant. For serum, the total %CV values for the seven levels ranged from 25.1% to 57.5%. The total %CV values for serum matrix were dependent on the concentrations of the levels tested. For the five levels with concentrations ranging from 13.1 to 1.17 × 106 HCV RNA IU/ml, the total %CV values were less than 33%. For the lowest two concentrations tested (65.6 HCV RNA IU/ml and 32.8 HCV RNA IU/ml), the total %CVs were equal to or greater than 49%.
TABLE 5.
Precision of the COBAS TaqMan HCV test (version 2.0) in EDTA plasma
| Component of variance | %CV value for indicated concn (IU/ml) of HCV genotype 1 in EDTA plasmaa
|
|||||
|---|---|---|---|---|---|---|
| 4.44 × 106 | 4.44 × 105 | 4.44 × 104 | 4.44 × 103 | 4.44 × 102 | 4.44 × 101 | |
| Lot to lot | 4.8 | 0.0 | 0.0 | 0.0 | 0.0 | 15.4 |
| Instrument to instrument | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 |
| Operator to operator | 18.4 | 17.7 | 18.7 | 16.8 | 14.8 | 12.7 |
| Between run | 18.6 | 20.7 | 17.1 | 12.7 | 23.0 | 9.8 |
| Within run | 18.8 | 17.6 | 20.0 | 24.4 | 17.2 | 28.0 |
| Total | 32.6 | 32.5 | 32.3 | 32.2 | 32.3 | 35.8 |
The %CV values for each component of the variance analysis and for the total variance are shown for each of the six levels tested. n was 48 for each level.
TABLE 6.
Precision of the COBAS TaqMan HCV test (version 2.0) in serum
| Component of variance | %CV value for indicated concn (IU/ml) of HCV genotype 1 in seruma
|
||||||
|---|---|---|---|---|---|---|---|
| 1.17 × 106 | 1.31 × 105 | 1.31 × 104 | 1.31 × 103 | 1.31 × 102 | 6.56 × 101 | 3.28 × 101 | |
| Lot to lot | 2.7 | 0.0 | 12.3 | 13.9 | 11.1 | 0.0 | 10.4 |
| Instrument to instrument | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Operator to operator | 12.0 | 12.8 | 0.0 | 0.0 | 6.6 | 0.0 | 9.7 |
| Between run | 16.5 | 17.5 | 16.2 | 16.5 | 16.9 | 32.9 | 31.2 |
| Within run | 15.2 | 13.9 | 18.9 | 12.9 | 24.3 | 36.5 | 46.1 |
| Total | 25.6 | 25.8 | 27.7 | 25.1 | 32.3 | 49.1 | 57.5 |
The %CV values for each component of the variance analysis and for the total variance are shown for each of the seven levels tested. n was 72 for each level.
In general, for both EDTA plasma and serum samples, relatively little or no variance was attributable to “lot-to-lot” or “instrument-to-instrument” components. For most of the samples tested, each of the rest of the components of the variance examined (operator to operator, between run, and within run) showed approximately equal contributions to the total variance. An exception was for the low-concentration samples, for which the contributions by the within-run and between-run components were much greater than those by the rest of the components.
Specificity.
All of the 200 test results obtained by HCVHPS V2 for the 50 seronegative EDTA plasma specimens and the 50 seronegative serum specimens using two different kit lots were negative, yielding 100% clinical specificity (data not shown). None of the 12 non-HCV members of the Flaviviridae or 22 additional microorganisms tested were positive for HCV RNA by HCVHPS V2 (data not shown). Based on these data, the analytical specificity is also 100%.
Interference.
All of the drug-spiked samples yielded results that were within 0.2 log10 of the results for their respective controls. Therefore, none of the 23 different drugs (that are commonly prescribed to HCV patients) tested by HCVHPS V2 were shown to interfere with the quantitation of HCV RNA (data not shown).
DISCUSSION
In recent years, quantitative HCV RNA tests have been developed to address the increasing demand for sensitive and accurate assays that are useful for monitoring and predicting treatment outcomes in chronic hepatitis C (1, 16). In the present study, we evaluated HCVHPS V2, a real-time PCR test that has been developed to overcome certain genotype-specific limitations shown by its previous version, HCVHPS V1. Because of significant underestimation of genotypes 2 through 5, HCVHPS V1 has been restricted to the quantitation of genotypes 1 and 6 only. Changes in the ethanol concentration of the wash buffer in sample preparation and the temperature of the reverse transcription step in reverse transcription-PCR were then introduced in the new version, HCVHPS V2. The performance of HCVHPS V2 was then evaluated by utilizing all HCV genotypes. Indeed, our results show that HCVHPS V2 provides high sensitivity and accurate quantitation for all six HCV genotypes compared to CAHCM. In all participating laboratories, with a variety of clinical specimens and standards used, the mean differences between the HCVHPS V2 and CAHCM results were within ± 0.3 log10. As expected, the mean log HCV RNA values (IU/ml) obtained by HCVHPS V2 were significantly greater for genotypes 2 through 5 than those obtained by HCVHPS V1. Furthermore, the mean differences in log HCV RNA values (IU/ml) for each of the six genotypes for the comparison between HCVHPS V2 and HCVHPS V1 or HCVHPS V2 and CAHCM were very similar for all three laboratories, with the sole exception of the result for genotype 2 at one of the three laboratories. Additional experiments performed on subtype-specific clinical specimens showed that the difference between the two HCVHPS versions is higher for genotype 2a than for 2b, suggesting a variable subtype composition for genotype 2 samples at the four study sites. Furthermore, HCVHPS V2 gave very similar results for all HCV genotypes compared to a fully automated HCMCAP (5, 20-21). These experimental results support the assumption that the decrease in the ethanol concentration of the wash buffer and the increased temperature of the reverse transcription step significantly improved quantitation across HCV genotypes 2 to 5.
The remainder of the experiments presented in this study address the performance of the improved version of the assay. The linear range of HCVHPS V2 is as would be expected based on the previous version of the assay. In both specimen matrices, EDTA plasma or serum, the assay was shown to have a broad linear range, from 25 IU/ml to 3.91 × 108 IU/ml. Extensive precision analysis, including lot-to-lot, operator-to-operator, within-run, and between-run variability, showed the assay to be robust and reproducible for both EDTA plasma and serum samples. In general, between-run and within-run variances and, to a lesser extent, the operator-to-operator variance contribute the most to the total variance seen with HCVHPS V2. The higher variability observed at the lower limit of the dynamic range in serum is to be expected because of analyte distribution in the sample matrix at the 10- to 50-IU/ml level (16). This variation at the lower dilutions was not observed in the EDTA plasma sample analysis.
Analysis of LOD was performed for all HCV genotypes in both EDTA plasma and serum specimen matrices. The results of this analysis show that the LOD values of HCVHPS V2 based on PROBIT analysis are between 6 and 17 HCV RNA IU/ml for EDTA plasma and 11 and 19 HCV RNA IU/ml for serum. The LOD for each genotype is similar, and there is no apparent bias for any genotype.
HCVHPS V2 appeared to be highly specific. No cross-reactivity was observed when closely related flaviviruses or other unrelated viruses, bacteria, or fungal agents that could be present in serum or plasma were tested in the assay. In addition, HCVHPS V2 was not affected by 23 different drugs that may be administered to patients with chronic hepatitis C, often presented with concurrent diseases or coinfections (9, 17).
Taken together, these data indicate that HCVHPS V2 has significantly improved the quantitation of genotypes 2 through 5 over the previous version. In addition, the changes in the extraction and thermal cycling parameters have not compromised the performance characteristics of the assay. The data demonstrate that HCVHPS V2 has a broad linear range, good precision through the linear range, low LOD values across all HCV genotypes, an excellent specificity for HCV, and a lack of interference by drugs administered to HCV-infected patients. We conclude that HCVHPS V2 can be used reliably to detect and quantitate viral load in HCV-infected patients.
Footnotes
Published ahead of print on 26 September 2007.
REFERENCES
- 1.Chevaliez, S., and J. M. Pawlotsky. 2006. Hepatitis C virus serologic and virologic test and clinical diagnosis of HCV related liver disease. Int. J. Med. Sci. 3:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis, G. L., J. B. Wong, J. G. McHutchinson, M. P. Manns, J. Harvey, and J. Albrecht. 2003. 2003. Early virologic response to treatment with peginterferon alpha 2b plus ribavirin in patients with chronic hepatitis C. Hepatology 38:646-652. [DOI] [PubMed] [Google Scholar]
- 3.Davis, G. 2006. Tailoring in anti-viral therapy in hepatitis C. Hepatology 43:909-911. [DOI] [PubMed] [Google Scholar]
- 4.Gleaves, C. A., J. Welle, M. Campbell, et al. 2002. Multicenter evaluation of the Bayer VERSANT HIV-1 RNA 3.0 assay: analytical and clinical performance. J. Clin. Virol. 25:205-216. [DOI] [PubMed] [Google Scholar]
- 5.Halfon, P., M. Bourliere, G. Penaranda, H. Khiri, and D. N. Ouzan. 2006. Real-time PCR for hepatitis C virus RNA quantification are adequate for clinical management of patients with chronic HCV infection. J. Clin. Microbiol. 44:2507-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′—-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen, D. M., T. R. Morgan, P. Marcellin, P. J. Pockrus, K. R. Reddy, and S. Hadzyiannis. 2006. Early identification of HCV genotype 1 patients responding to 24 weeks peginterferon alpha 2a/ribavirin therapy. Hepatology 43:954-960. [DOI] [PubMed] [Google Scholar]
- 8.Jungkind, D. 2001. Molecular testing for infectious disease. Science 294:1553-1555. [DOI] [PubMed] [Google Scholar]
- 9.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 35:41-52. [DOI] [PubMed] [Google Scholar]
- 10.Lee, S., A. Anthony, N. Lee, et al. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, S. S., E. J. Heathcote, K. R. Reddy, et al. 2002. Prognostic factor and early predictability of sustained viral response with peginterferon alfa-2a (40KD). J. Hepatol. 37:500-506. [DOI] [PubMed] [Google Scholar]
- 12.Liu, L. W., J. Tomlinson, T. Mazzulli, A. Murray, and J. Heathcote. 2003. Early predictors of non responders to treatment with interferon alpha 2b and ribavirin in patients with chronic hepatitis C. Can. J. Gastroenterol. 17:483-487. [DOI] [PubMed] [Google Scholar]
- 13.Mangia, A., R. Santoro, N. Minerva, G. L. Ricci, V. Carretta, M. Persico, et al. 2005. Peginterferon alpha 2a and ribavirin for 12 vs. 24 weeks in HCV genotype 2 or 3. N. Engl. J. Med. 352:2609-2617. [DOI] [PubMed] [Google Scholar]
- 14.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 15.Niesters, H. G. M. 2004. Molecular and diagnostic clinical virology in real time. Clin. Microbiol. Infect. 10:5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pawlotsky, J. M. 2002. Use and interpretation of virological tests for hepatitis C. Hepatology 36:S65-S73. [DOI] [PubMed] [Google Scholar]
- 17.Raimondo, G., M. Brunetto, P. Pontisso, A. Smedile, A. M. Maina, C. Saitta, et al. 2006. Longitudinal evaluation reveals a complex spectrum of virological profiles in hepatitis B/hepatitis C virus co-infected patients. Hepatology 43:100-107. [DOI] [PubMed] [Google Scholar]
- 18.Saldanha, J., N. Lelie, and A. Heath. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 19.Saldanha, J., A. Heath, C. Aberham, J. Albrech, G. Gentili, M. Gessner, and G. Pisani. 2005. World Heath Organization collaborative study to establish a replacement WHO international standard for hepatitis C virus RNA nucleic acid amplification technology assays. Vox Sang. 88:202-204. [DOI] [PubMed] [Google Scholar]
- 20.Sarrazin, C., B. C. Gärtner, D. Sizmann, R. Babiel, U. Mihm, W. P. Hofmann, M. von Wagner, and S. Zeuzem. 2006. Comparison of conventional PCR with real-time PCR and branched DNA based assays for hepatitis C virus RNA quantification and clinical significance for genotypes 1 to 5. J. Clin. Microbiol. 44:729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stelzl, E., A. Kormann-Klement, J. Haas, J., E. Draghofer, B. I. Santner, E. Marth, and H. H. Kessler. 2002. Evaluation of an automated sample preparation protocol for quantitative detection of hepatitis C virus RNA. J. Clin. Microbiol. 40:1447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tholen, D. W., M. Kroll, J. R. Astles, A. L. Caffo, T. M. Happe, J. Krouwer, and F. Lasky. 2003. Evaluation of the linearity of quantitative measurement procedures: a statistical approach; approved guideline, CLSI EP6-A. Clinical and Laboratory Standards Institute, Wayne, PA.
- 23.WalkerPeach, C. R., M. Winkler, D. B. Dubois, and B. L. Pasloske. 1999. Ribonuclease-resistant RNA controls (Armored RNA) for reverse transcription-PCR, branched DNA, and genotyping assays for hepatitis C virus. Clin. Chem. 45:2079-2085. [PubMed] [Google Scholar]
- 24.Zeuzem, S. 1999. Clinical implications of hepatitis C viral kinetics. J. Hepatol. 31(Suppl. 1):61-64. [DOI] [PubMed] [Google Scholar]
- 25.Zeuzem, S., E. Herrmann, J.-H. Lee, J. Fricke, A. Neumann, M. Modi, G. Colucci, and W. K. Roth. 2001. Viral kinetics in patients with chronic hepatitis C treated with standard or peginterferon alpha 2a. Gastroenterology 120:1438-1447. [DOI] [PubMed] [Google Scholar]