Abstract
There are numerous viral and bacterial causes of respiratory disease. To enable rapid and sensitive detection of even the most prevalent causes, there is a need for more-simplified testing systems that enable researchers and clinicians to perform multiplexed molecular diagnostics quickly and easily. To this end, a new multiplexed molecular test called the MultiCode-PLx respiratory virus panel (PLx-RVP) was developed and then implemented in a public health laboratory setting. A total of 687 respiratory samples were analyzed for the presence of 17 viruses that commonly cause respiratory disease. As a comparator, the samples were also tested using a standard testing algorithm that included the use of a real-time influenza virus A and B reverse transcription-PCR test and routine viral culture identification. The standard testing algorithm identified 503 (73%) samples as positive and 184 as negative. Analyzing the same 687 samples, the PLx-RVP assay detected one or more targets in 528 (77%) samples and found 159 samples negative for all targets. There were 25 discordant results between the two systems; 14 samples were positive for viruses not routinely tested for by the Wisconsin State Laboratory of Hygiene, and 13 of these were confirmed by real-time PCR. When the results of the standard testing algorithm were considered “true positives,” the PLx-RVP assay showed an overall sensitivity of 99% and an overall specificity of 87%. In total, the PLx-RVP assay detected an additional 40 viral infections, of which 11 were mixed infections.
The traditional viral agents that cause respiratory infections include influenza virus types A and B, respiratory syncytial viruses (RSVs), parainfluenza virus types 1 to 4 (PIV1 to PIV4), human rhinoviruses (HRVs), and a number of adenovirus (Ad) types (12, 33). Yet over the past decade, an unprecedented number of novel viral agents capable of causing respiratory disease have been identified (14, 17, 26, 45). This trend is sure to continue given today's molecular toolbox. Beyond the common etiological viral agents, we now know there exist a number of recently identified and additional unidentified agents that may cause respiratory disease (15). This fact, in conjunction with the limitations of current culture and staining methods, is now leading physicians and clinicians to rethink how they will diagnose patients with respiratory diseases in the future.
To help prevent respiratory disease, which between 1989 and 2002 ranked second in the number of days for which inpatients received hospital care in the United States, vaccination programs, along with surveillance and rapid identification strategies, are critical (25, 32, 35, 39, 42). When new, potentially lethal pathogens are identified, one of the first lines of action is to begin surveillance testing (11, 18). In recent years, outbreak surveillance programs have brought into play molecular testing to quickly and accurately diagnose regional patient populations (6, 13, 22, 31, 49). This is because molecular testing requires only the agent's genetic fingerprint, and assay development cycles are short (1 to 4 weeks). Recent data even suggest that when properly developed, molecular testing for viral agents can be more sensitive and more specific than viral culture and/or immunoassays (16, 19, 41). The molecular tests typically target the anticipated conserved regions of one or a few viral genomes (1, 47). Yet with so many agents capable of causing respiratory infections and with the characteristic genetic variability within viral types, multiple-target assay systems will be needed to provide comprehensive diagnoses.
In efforts to move closer to more-comprehensive disease scanning and laboratory-based surveillance, a number of multiplexed assays have been developed (2, 4, 8, 9, 20, 23, 24, 30, 38, 43, 44, 46, 48). Many use a combination of techniques, such as enzyme linkage formats for detection or two or more multiplexed reactions, to obtain full coverage. Others use arrays with the ability to scan for thousands of potential agents and subtypes. Yet even with the availability of these multiplexed assays and with molecular testing becoming more commonplace, the majority of clinical testing laboratories use culture, staining, and single-target PCR techniques. Minimal field implementation of the newer molecular multiplexed tests may be due to reasons and/or misperceptions that include training concerns brought on by assay complexity, amplicon carryover/contamination, regulatory approval, expense, and sensitivity/specificity limits. Before they will make the shift to molecular multiplex testing, clinicians and laboratory directors need to be convinced that these new technologies and assays can address their concerns.
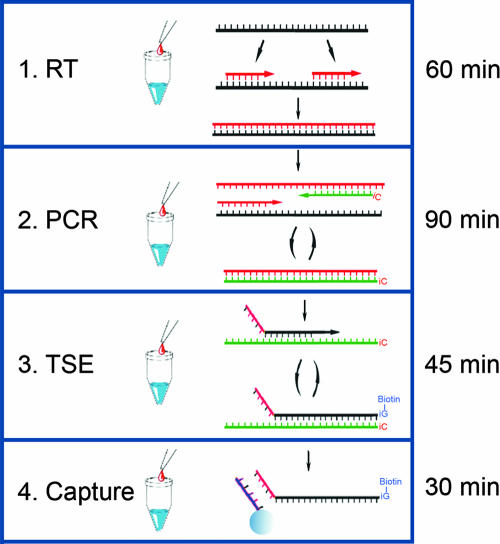
In an attempt to simplify the multiplexed molecular testing of respiratory viral agents, the PLx respiratory virus panel (RVP) assay was developed using the MultiCode-PLx system (21, 36, 37). The MultiCode-PLx system uses an expanded genetic alphabet to simplify the procedure and entails four major steps once the target nucleic acid is isolated (see Fig. 1). Once developed, the assay was tested over a 4-month period at the Wisconsin State Laboratory of Hygiene (WSLH) as part of its laboratory-based surveillance for influenza virus and other respiratory viruses. The PLx-RVP assay was compared to the WSLH standard testing algorithm, which included the use of a real-time influenza virus A and B reverse transcriptase PCR (RT-PCR) test protocol provided to state public health laboratories by the Centers for Disease Control and Prevention (CDC), as well as routine viral culture and identification.
FIG. 1.
Schematic of the PLx-RVP system. Four major steps are required to perform the PLx-RVP assay. PLx is enabled by one additional base pair constructed from the complementary bases isoguanosine and 5′-methyl-isocytosine (iC). These additional bases are used in steps 2 to 4. The average time required for each step is shown on the right. (Step 1) Reverse transcription (RT). Random 6-mer primers are used to produce cDNA from extracted RNA. (Step 2) Multiplexed PCR. PCR primers are designed to be target specific, and at least one primer of each pair contains a single iC on its 5′ end. (Step 3) TSE. The iC-containing strands of the PCR products act as labeling templates for the TSE step. During this step, labels attached to 2′-deoxy-iso-GTP are incorporated site-specifically onto EraCode-tagged target-specific extenders directed by the complementary iC contained in the template. (Step 4) Capture. EraCode tags are short sequences (typically 8 bases) assembled using a mix of natural and nonnatural bases. The tags are designed to hybridize only to their perfect complements encoded on spectrally addressed microspheres. The TSE reaction products are decoded by room temperature hybridization to complementary codes on spectrally addressed microspheres, followed by reading of the reporter signal.
The PLx-RVP assay was specifically formatted for the detection of 17 viruses implicated in causing upper respiratory tract disease and included internal positive controls. Infective agents that could be specifically identified included influenza viruses A and B, PIV1, -2, -3, -4a, and -4b, RSV A and B, HRV, coronaviruses (CoV) 229E, NL63, and OC43, Ad B, C, and E, and human metapneumovirus (HMPV). Respiratory specimens, including throat and/or nasopharyngeal swabs, were submitted to the WSLH by rapid influenza test sites (largely hospital clinical laboratories and primary care clinic laboratories) and sentinel clinician virus surveillance sites throughout Wisconsin. In total, 687 samples were analyzed by the PLx-RVP assay.
MATERIALS AND METHODS
Respiratory specimen acquisition.
During the 2005-to-2006 respiratory virus season (December 1 to April 1), throat and/or nasopharyngeal swabs were submitted to the WSLH in viral transport medium (VTM) (M4 or M5; volume, 3.0 ml; Remel, Inc., Lenexa, KS) by sentinel clinician sites or clinical laboratories throughout Wisconsin. Specimens were obtained from patients of all ages and genders who presented with signs and symptoms of respiratory infection. A number of these specimens were tested at the clinical sites for influenza virus and/or RSV using rapid antigen detection tests and were forwarded to the WSLH for confirmatory testing. The following specimen information was tracked: specimen identifier, date collected, specimen type (throat or nasopharyngeal swab), and testing results. Samples were received on wet ice and held at 4°C prior to processing within 24 h of receipt.
Sample preparation.
Specimens in VTM were vortexed for 1 min. The samples were divided into two aliquots: one used for virus culture and direct immunofluorescence assay (DFA) staining for RSV and one used for molecular testing. For molecular testing, prior to sample extraction, 1.0 ml of the specimen was centrifuged for 10 min at 15,000 × g at room temperature, and the cell pellet was resuspended in approximately 200 μl of supernatant. Nucleic acid extraction was performed with a MagNaPure LC (Roche Diagnostics) instrument by using the total-nucleic-acid kit (catalog no. 3038505; Roche). The sample and elution volumes were 200 and 50 μl, respectively. Sample extracts were held at −20°C prior to batch testing.
Influenza virus A and B real-time RT-PCR.
A 5.0-μl volume of extracted nucleic acid was tested by real-time PCR with the ABI 7500 Fast system (Applera) using CDC protocols for influenza A and B viruses. These protocols were validated at the WSLH in 2005 by comparison to virus culture.
Virus culture and DFA.
Specimens that tested negative for influenza viruses A and B by real-time PCR were inoculated into MDCK, A549, PRMK, and WI-38 cells using routine methods for culture of common cultivable viruses. Specimens were also tested by direct immunofluorescence for RSV using a routine method.
Reverse transcription.
Six microliters of extracted nucleic acid was added to 6 μl of reverse transcription solution, which included 15 μM random hexamers and 0.9 U of avian myeloblastosis virus reverse transcriptase. The reaction mixtures were incubated at 25°C for 5 min, 42°C for 10 min, 50°C for 20 min, and 85°C for 5 min and were then held at 4°C until amplification.
MultiCode-PLx assay.
Two versions of the PLx-RVP assay were used in the study: version 1, the RVP beta prototype; and version 2, the updated MultiCode-PLx Respiratory Virus Panel Core Reagents (EraGen catalog no. 9080). All reactions were carried out according to the MultiCode-PLx Respiratory Virus Panel Core Reagents protocol. The procedure is outlined as follows. Primers were used at the concentrations shown in Table S1 in the supplemental material. The genome regions targeted by PLx-RVP versions 1 and 2 have been discussed previously (27). Samples for testing were batched. The batch size was between 50 and 80 samples. All batched reactions were performed in 96-well PCR plates (MLL9601; Bio-Rad). The following analytes were tested for simultaneously: influenza viruses A and B, RSV A and B, Ad subgroups B (types 3, 7, 11, 14, 16, 21, 34, and 35), C (types 1, 2, 5, and 6), and E (type 4), PIV1, -2, -3, -4a, and -4b, HRV, CoV 229E, OC43, and NL63, and HMPV. All reactions included an internal positive DNA control (IPC) added to the amplification mix at a level of 1,200 copies per reaction. Primer sets specific to the IPC yield a positive IPC channel signal. The PLx-RVP assay procedure shown in Fig. 1 consists of the following steps, which all occur in the same well: amplification of viral cDNAs by PCR, target-specific extension (TSE) of EraCode-tagged primers, capture of TSE products to EraCode-labeled microspheres, and readout of fluorescent signals on each microsphere using the Bio-Rad BioPlex system (21, 34). Before reaction mixtures were heated, plates were sealed with Microseal A film (MSA5001; Bio-Rad) to prevent evaporation.
Amplification.
Reactions were initiated by combining 5 μl of the reverse transcription reaction mixture, prepared as described above, with 5 μl of PCR master mix, which includes 0.2 μl of Titanium Taq polymerase (catalog no. 639208; BD Bioscience) along with all PCR primer pairs. The PCR step was carried using the following conditions: 2 min at 95°C, followed by 30 cycles of 10 s at 95°C, 30 s at 55°C, and 30 s at 72°C. Then the mixture was held at 4°C.
TSE.
Following the PCR, 5 μl of a TSE master mix that includes the TSE primers along with 6 μM biotin-2′-deoxy-iso-GTP was added to the PCR product. The TSE reaction was carried out using the following conditions: 30 s at 95°C; 10 cycles of 5 s at 95°C and 2 min at 65°C; and 65°C for 5 min. Then the mixture was held at 4°C.
Capture and readout.
After the TSE reaction, 35 μl of microspheres/hybridization solution (catalog no. PN9550/9570; EraGen) was added to the TSE products. The resulting mixture was incubated at room temperature for 10 min in the dark to allow hybridization of TSE products to the tag-specific microspheres. Then 35 μl of sheath fluid (catalog no. 40-50000; Luminex) containing 2 μg of streptavidin R-phycoerythrin (catalog no. PJ31S; Prozyme) was added and allowed to incubate 15 min in the dark. Next, the fluorescent signal associated with each microsphere was measured in a Bio-Rad BioPlex instrument (96-well-plate flow cytometer). The signal is expressed as median fluorescence intensity (MFI). Samples with average signals of >6 standard deviations (SD) above the average negative-control signals (typically 400 to 500 MFI) were regarded as positive.
PLx data analysis.
Template setup within the Bio-Rad BioPlex 3.0/4.0 software was required, and the data export file was imported into MultiCode-PLx analysis software (catalog no. 9150; EraGen). Data files were parsed, and the resulting raw MFI values were organized by target and sample. Following data acquisition from all clinical samples tested, default cutoff windows for each target were empirically determined and set in a blinded fashion. Once determinations were made, reports were generated for offline analysis.
LOD.
Clones that contain the target genomic regions of the following viruses were used to assess assay sensitivity: Ad subgroups C (serotypes 1 and 5), B (serotypes 3, 7, 11, 21, and 34), and E (serotype 4); HRV (serotypes 1a, 2, 13, 14, 17, 59, 86, and 91); CoV NL63, 229E, and OC43; influenza virus B; HMPV; RSV A and B; PIV1, PIV2, PIV3, PIV4a, and PIV4b; and influenza virus A serotypes H3N2 and H5N1 (27). Purified plasmid DNA was quantified with a Quant-iT PicoGreen double-stranded DNA assay kit (Invitrogen) (40). All plasmids except those containing PIV4a and PIV4b sequences were linearized by restriction enzyme digestion of a 100-ng/μl solution of each target. The templates were then diluted to the concentrations used in the limit-of-detection (LOD) experiment. The assay sensitivity for each of the plasmid templates was demonstrated by analyzing duplicate aliquots containing 0.25, 2.5, 25, and 2,500 template copies by the PLx-RVP assay. Results were considered positive when the MFI output was at least 6 SD above the average background. The lowest dilution that gave a positive result for both replicates was considered the LOD.
Negative controls.
During the comparison testing, crossover contamination was analyzed using negative-control reactions spaced out throughout the plate. Each batched run conducted included negative controls in order to monitor possible cross-contamination. The negative controls were performed using an extraction elution buffer in 1 out of every 16 samples per batch.
Comparator RT-PCR testing.
Samples positive by the PLx-RVP assay and not detectable using the WSLH standard testing algorithm were further tested by the Washington University School of Medicine, St. Louis, MO, with real-time PCR assays. Real-time RT-PCR assays for CoV and HMPV were carried out in 50-μl reaction volumes in ABI optical-quality 96-well plates. Each reaction used 1× Qiagen Quantitect probe RT-PCR kit reaction mixture (catalog no. 204443; Qiagen, Valencia, CA). Thermal cycling was performed on an ABI 7300 RT-PCR instrument using “absolute quantitation” software. The conditions were as follows: for CoV, 20 min at 50°C, 15 min at 95°C, and 45 cycles of 15 s at 94°C and 60 s at 60°C; for HMPV, 30 min at 50°C, 15 min at 95°C, and 45 cycles of 15 s at 94°C and 60 s at 55°C. Threshold-crossing cycles were computed by the ABI 7300 RT-PCR system sequence detection software, version 1.2.1. RT-PCR for HMPV was performed using primers and minor groove binding (MGB) modifications of the probes described previously (29). Probes were labeled with 6-carboxyfluorescein at the 5′ end and had the MGB peptide at the 3′ end. For the CoV assay, primers and probes that amplify segments of the N genes of human CoV OC43, 229E, and NL63/New Haven were employed. CoV strains were obtained from the ATCC (OC43 and 229E) or from Lia van der Hoek (NL63). For each CoV, in vitro RNA transcripts from cloned segments that contained the assay target for each strain were created and quantified. The analytical sensitivity (the LOD is defined as the RNA level at which 95% of replicates were positive) of each assay was determined to be 10 copies/reaction (unpublished data). All assays were optimized for primer and probe concentrations and annealing temperature (unpublished data).
RESULTS
Analytical data.
The PLx-RVP assay developed for this study was tested for the LOD by using cloned target sequences for each virus tested. Reactions were performed using 10-fold differences in copy number (0.25 to 2,500 copies per reaction) of linearized and purified plasmid DNA containing target sequences. All plasmid targets consisted of linearized plasmid DNA except for the PIV4a and PIVb DNAs, which were nonlinearized. Target numbers were determined by the optical densities at 260 nm of purified plasmid extracts. The LOD was defined as the lowest target level at which the two replicates were called correctly and was determined to be 2.5 copies for every cut plasmid target tested and 100 copies for the nonlinearized plasmids. Due to concerns about sequence variability, multiple duplicate tests were performed using various serotypes for Ad and rhinovirus. Since all targets could be detected within each single multiplexed reaction and only one target was added to any given reaction mixture, analytical specificity for the other targets could also be determined. Overall, the analytical specificity for the assay was determined to be 100%. That is, no false-positive results occurred during any of the 62 reactions. Interference testing was not conducted.
During the 2005-to-2006 influenza season, the WSLH received more than 700 samples through the influenza sentinel surveillance network, 687 of which were included in the present study. For all the samples tested and retested, we used the signal generated from the IPC (added to all reaction mixtures) to determine the reproducibility of the assay. Over the course of 4 months, two operators performed nine batched runs. Each batched run included positive and negative controls. The signals generated in the IPC channel from 806 separate reactions averaged 7,746 MFI units, with an SD of 628 MFI units. Of the 806 reactions, 8 reactions (∼1%) did not produce an IPC signal above 2,500 MFI units. For these eight failed reactions, the average IPC signal was 1,125 (SD, 628) MFI units.
Positive-target run controls using a subset of the cloned target sequences described above were implemented in each batch reaction set. In the eight reactions of every batch set, all 17 targets except for PIV4b were tested (2 targets per reaction). Of the 70 reactions run, with a possible 140 positive signals, 1 reaction reported a failed IPC and 1 reaction failed to detect PIV1. All other reactions reported the correct positives and negatives. The coefficient of variation (CV) of positive tests (target detected) ranged from 7% for PIV1 to 34% for influenza virus B. Eleven of 17 targets yielded CVs below 20%, demonstrating excellent assay reproducibility (Table 1). Positive-call cutoffs were set by the following calculation: (average signal from negative samples) + (6 × SD).
TABLE 1.
Reproducibility of the PLx-RVP assay with positive controlsa
| Target | Call | MFI (SD) | CV (%) | No. of reactions analyzed |
|---|---|---|---|---|
| IPC | Absent | NA (NA) | NA | 1 |
| Present | 7,800 (1,260) | 16 | 69 | |
| HRV | Absent | 169 (106) | 63 | 61 |
| Present | 7,550 (702) | 9 | 9 | |
| Influenza virus A | Absent | 168 (122) | 73 | 61 |
| Present | 6,400 (714) | 11 | 9 | |
| Influenza virus B | Absent | 191 (110) | 58 | 61 |
| Present | 2,740 (927) | 34 | 9 | |
| PIV1 | Absent | 168 (104) | 62 | 63 |
| Present | 5,890 (391) | 7 | 7 | |
| PIV2 | Absent | 181 (122) | 67 | 61 |
| Present | 4,670 (1,100) | 24 | 9 | |
| PIV3 | Absent | 175 (96.3) | 55 | 61 |
| Present | 10,500 (843) | 8 | 9 | |
| PIV4a | Absent | 166 (121) | 73 | 61 |
| Present | 4,560 (473) | 10 | 9 | |
| PIV4b | Absent | 161 (118) | 73 | 70 |
| Present | NA (NA) | NA | 0 | |
| RSV A | Absent | 167 (126) | 75 | 61 |
| Present | 9,650 (954) | 10 | 9 | |
| RSV B | Absent | 185 (109) | 59 | 62 |
| Present | 2,830 (768) | 27 | 8 | |
| HMPV | Absent | 193 (106) | 55 | 61 |
| Present | 8,310 (1,160) | 14 | 9 | |
| Ad B | Absent | 156 (112) | 72 | 53 |
| Present | 4,630 (612) | 13 | 8 | |
| Ad C | Absent | 220 (153) | 70 | 62 |
| Present | 6,240 (819) | 13 | 8 | |
| Ad E | Absent | 206 (129) | 63 | 61 |
| Present | 8,620 (1,400) | 16 | 9 | |
| CoV OC43 | Absent | 189 (88.9) | 47 | 61 |
| Present | 13,300 (2,760) | 21 | 9 | |
| CoV NL63 | Absent | 184 (123) | 67 | 62 |
| Present | 3,230 (772) | 24 | 8 | |
| CoV 229E | Absent | 187 (104) | 56 | 61 |
| Present | 6,450 (1,850) | 29 | 9 |
Results were obtained over the course of 4 months, and a total of 70 reactions were performed; each reaction mixture was spiked with 2 of the 17 targets (PIV4b not included).
Clinical data.
Analysis of PLx-RVP, version 1, data from the initial surveillance network sample runs indicated that of the 687 samples, 442 were positive for one or more targets. The average positive signals were at least 17-fold above the background noise for any given target (Table 2). As expected, the targets with the highest percentages of sample positive calls were influenza viruses A and B. For these two targets, the average signal-to-noise ratios were 47-fold and 25-fold, respectively. The average target-specific signals for these targets were 5,680 and 3,450 MFI units, respectively (Table 2).
TABLE 2.
Results of the PLx-RVP assay, version 1, and signal-to-noise analysis for all extracted nucleic acids from clinical samples
| Target | MFI (SD)a for:
|
S/N ratiob
|
No. of positive test results | ||
|---|---|---|---|---|---|
| Negative samples | Positive samples | Mean | Minimum | ||
| HRV | 169 (106) | 5,990 (2,780) | 54.9 | 8.4 | 21 |
| Influenza virus A | 168 (117) | 5,680 (1,190) | 47.1 | 7.9 | 301 |
| Influenza virus B | 196 (132) | 3,450 (1,420) | 24.7 | 7.1 | 74 |
| PIV1 | 175 (119) | 4,920 (1,320) | 39.9 | 23.0 | 9 |
| PIV3 | 207 (123) | 11,000 (N/A) | 87.7 | 87.7 | 1 |
| RSV A | 182 (120) | 5,900 (2,590) | 47.7 | 6.8 | 13 |
| RSV B | 175 (112) | 2,230 (154) | 18.3 | 17.3 | 4 |
| HMPV | 181 (132) | 5,150 (3,100) | 37.6 | 8.7 | 5 |
| Ad B | 173 (107) | 1,990 (1,380) | 17.0 | 7.8 | 8 |
| Ad C | 180 (119) | 4,710 (3,270) | 38.1 | 7.6 | 9 |
| Ad E | 175 (124) | 2,340 (N/A) | 17.5 | 17.5 | 1 |
| CoV NL63 | 185 (119) | 2,350 (679) | 18.2 | 6.4 | 9 |
On a per-test basis.
S/N, signal to noise. Mean, ratio of the average signal generated from all samples positive for a specific target to the average noise generated from all samples negative for the same target; minimum, ratio of the lowest signal generated from a sample positive for a specific target to the highest noise generated from a sample negative for the same target.
Contamination potential and/or amplicon carryover was monitored by observing signal generation in blank reactions. VTM was added to the blank reaction mixtures prior to extraction. The blanks were distributed throughout the extraction plate (2 per 32-well plate) and throughout the PLx-RVP batched reaction plates (5 to 6 per 96-well plate). A total of 50 blank reactions were performed, of which 49 produced negative results and 1 produced a negative result but the IPC failed.
For direct comparison to the WSLH testing algorithm, a “true-positive” specimen was defined as a specimen that was positive by virus culture, RSV DFA, or influenza virus PCR. Of 687 specimens tested, 73% (n = 503) were positive by virus culture, RSV DFA, or influenza virus PCR, compared to 65% (n = 442) positive by the PLx-RVP assay, version 1. Of the 442 PLx-RVP assay-positive samples, 9 (2%) were identified as CoV NL63, 5 (1%) were identified as HMPV (agents not detected by WSLH methods), and 9 were identified as coinfections. There were 92 viruses detected by one or more of the reference methods but not detected by the PLx-RVP assay, version 1. Table 3 shows a direct comparison on a per-analyte basis of the results of the PLx-RVP assay, version 1, with the results obtained using the WSLH testing algorithm. The sensitivity of the PLx-RVP assay, version 1, ranged from 51% (for influenza virus B) to 100% (for adenoviruses and rhinoviruses). The PLx-RVP assay was able to detect 97% of influenza A virus samples positive by CDC RT-PCR; however, it also failed to detect an influenza A virus/H1 swine strain. The PLx-RVP assay failed to detect 49% of the influenza virus B CDC RT-PCR-positive samples. The CDC RT-PCR cycle thresholds ranged from 15 to 40, indicating a wide viral target input copy range.
TABLE 3.
Comparison on a per-analyte basis of the results of the PLx-RVP assay, version 1, with those obtained using the WSLH testing algorithm for all 687 samples
| Virus | No. of specimensa
|
Sensitivity (%) (95% CI)b | Specificity (%) (95% CI)b | |||
|---|---|---|---|---|---|---|
| W+ P+ | W− P+ | W+ P− | W− P− | |||
| Influenza A virus | 298 | 2 | 10 | 377 | 96.8 (94.1-98.2) | 99.2 (98.1-99.9) |
| Influenza B virus | 74 | 0 | 71 | 542 | 51 (43.0-59.0) | 100 (99.3-100) |
| RSV | 12 | 5 | 7 | 663 | 63.2 (41.0-80.9) | 99.3 (98.3-99.7) |
| PIV | 9 | 1 | 4 | 673 | 69.2 (42.4-87.3) | 99.9 (99.2-100) |
| Ad | 12 | 6 | 0 | 669 | 100 (75.7-100) | 99.1 (98.1-99.6) |
| Rhinovirus | 6 | 15 | 0 | 666 | 100 (61.0-100) | 97.8 (96.4-98.7) |
W+ and W−, positive and negative, respectively, by the WSLH testing algorithm; P+ and P−, positive and negative, respectively, by the PLx-RVP test.
Sensitivity and specificity calculations were based on “true positives” and “true negatives,” defined as specimens giving positive or negative results, respectively, by the WSLH testing algorithm. Sensitivity, specificity, and confidence intervals (CI) were calculated according to CLSI recommendations (7a).
Since the initial test data for influenza virus B sensitivity were not suitable for clinical practice, the PLx-RVP assay was updated to version 2. We sequenced the targeted regions of 63 of 71 discordant influenza virus B isolates and determined that the influenza virus B strains present in the missed samples were not in GenBank when the PLx-RVP primers were designed. Therefore, a new, broader-coverage influenza virus B design was implemented. In addition, due to pandemic concerns that resulted in a rapid increase in GenBank sequence deposits, we investigated and updated the other designs. To increase influenza virus A coverage, an additional set of primers (PCR and TSE) was added that enabled detection of swine and avian strains as well as of other sequences not previously deposited in GenBank. Also, to increase RSV A, RSV B, and PIV2 coverage, new primer sets (PCR and TSE) were substituted that enabled sequences not previously deposited in GenBank to be detected. The other target primer designs were not changed.
We retested a subset of 155 samples using the new PLx-RVP, version 2, assay. The PLx-RVP, version 1, assay had yielded the following results for these 155 samples: 5 concordant negative results, 22 concordant positive results, 6 partially concordant positive results for dually infected samples where the reference methods were capable of detecting both viruses but the PLx-RVP assay detected only one (HRV/Ad, influenza virus A/HRV, influenza virus B/influenza virus A, influenza virus A/RSV, influenza virus A/Ad, and influenza virus B/Ad), 2 concordant results for dually infected samples where the reference methods did not test for one of the viruses (influenza virus A/HMPV and influenza virus B/HMPV), 1 discordant result for a dually infected sample where the reference methods failed to detect one virus and did not test for the other virus (HRV/CoV NL63), 10 results positive for a single virus not tested for by the reference methods (CoV NL63 or HMPV), and 109 discordant results (90 samples negative by the PLx-RVP assay and positive by the reference method; 19 samples positive by the PLx-RVP assay and negative by the reference method). The PLx-RVP, version 2, assay agreed completely with the version 1 assay on the 27 concordant samples, the 9 dually infected samples, and the 10 samples positive for viruses not tested by the reference methods. Additionally, the version 2 assay detected two additional coinfections (influenza virus B/HRV and PIV1/CoV NL63). The PLx-RVP, version 2, assay correctly called 84 of 90 singly infected samples that were previously discordant (negative by the PLx-RVP, version 1, assay and positive by the reference method). These included one specimen that was called RSV positive by the reference method but Ad positive by the PLx-RVP, version 2, assay. Of the 19 samples positive by the PLx-RVP, version 1, assay and negative by the reference method, all were positive for the same viruses by version 2 of the PLx-RVP assay. Table 4 shows a direct comparison on a per-analyte basis between the PLx-RVP, version 2, assay results and those obtained using the WSLH testing algorithm for retested samples.
TABLE 4.
Direct comparison on a per-analyte basis of results of the PLx-RVP, version 2, assay with redesigned primers and results obtained using the WSLH testing algorithm for retested samples
| Virus | No. of specimensa
|
|||
|---|---|---|---|---|
| W+ P+ | W− P+ | W+ P− | W− P− | |
| Influenza A virus | 15 | 2 | 1 | 137 |
| Influenza B virus | 78 | 0 | 1 | 76 |
| RSV | 18 | 5 | 1 | 131 |
| PIV | 6 | 0 | 2 | 147 |
| Ad | 4 | 3 | 0 | 148 |
| Rhinovirus | 2 | 15 | 0 | 138 |
W+ and W−, positive and negative, respectively, by the WSLH testing algorithm; P+ and P−, positive and negative, respectively, by the PLx-RVP test.
When these new data were used to replace the initial data for the 155 retested samples, the sensitivity of the PLx-RVP assay for influenza virus B, RSV, and PIV improved considerably (Table 5). The redesigned influenza virus A primers were able to detect several positive specimens missed by version 1 of the PLx-RVP assay. As expected, the performance of the PLx-RVP assay for Ad and HRV did not change, since primers for these viruses were not redesigned.
TABLE 5.
Direct comparison of the PLx-RVP assay to the WSLH testing algorithm, incorporating data from 155 samples retested using version 2 of the PLx-RVP assay
| Virus | No. of specimensa
|
Sensitivity (%) (95% CI)b | Specificity (%) (95% CI)b | |||
|---|---|---|---|---|---|---|
| W+ P+ | W− P+ | W+ P− | W− P− | |||
| Influenza A virus | 307 | 2 | 1 | 377 | 99.7 (98.2-99.9) | 99.5 (98.1-99.9) |
| Influenza B virus | 144 | 0 | 1 | 542 | 99.3 (96.2-99.9) | 100 (99.3-100) |
| RSV | 18 | 4 | 1 | 664 | 97.4 (75.4-99.1) | 99.4 (98.5-99.8) |
| PIV | 11 | 0 | 2 | 674 | 84.6 (57.8-95.7) | 100 (99.4-100) |
| Ad | 12 | 6 | 0 | 669 | 100 (75.7-100) | 99.6 (98.1-99.6) |
| Rhinovirus | 6 | 15 | 0 | 666 | 100 (61.0-100) | 98 (96.4-98.7) |
W+ and W−, positive and negative, respectively, by the WSLH testing algorithm; P+ and P−, positive and negative, respectively, by the PLx-RVP test.
Sensitivity and specificity calculations were based on “true positives” and “true negatives,” defined as specimens giving positive and negative results, respectively, by the WSLH testing algorithm. Sensitivity, specificity, and confidence intervals (CI) were calculated according to CLSI recommendations (7a).
The samples that tested positive by the PLx-RVP assay for targets that current WSLH methods do not detect (9 CoV NL63- and 5 HMPV-positive samples) were tested by single-target real-time RT-PCR assays. Of all tests completed, real-time PCR data showed all but one (a sample negative for HMPV by real-time RT-PCR) to be concordant with the PLx-RVP assay results.
DISCUSSION
Multiple viruses contribute to the burden of respiratory viral disease, and diverse patient groups are often severely affected. These include infants, children, those with underlying cardiopulmonary disorders, the elderly, and immunocompromised hosts, particularly hematopoietic stem cell and lung transplant recipients. Early regional detection of pathogens that cause respiratory illness helps to prepare hospitals. Laboratory-based surveillance programs for influenza-like illnesses are critical to providing information concerning the presence of the virus. Surveillance and early diagnosis have been shown to result in timely administration of antiviral drugs to decrease the duration of outbreaks and lower total costs due to illness (7). They also prevent the inappropriate use of antibiotics (3).
Molecular technology provides a number of benefits for surveillance networks, including increased specificity and sensitivity, and quicker results (5, 50). When multiplexed molecular techniques are considered, these benefits expand to include broad-spectrum pathogen detection, increased throughput, expanded redundancy, smaller sample volumes, and rapid inclusion of additional targets. The PLx-RVP assay is a multiplexed molecular diagnostic system that was formulated for this study to detect 17 respiratory-pathogen targets simultaneously. The assay took approximately 4 h to analyze a batch of extracted samples and used only 6.0 μl of total extracted sample. This compares favorably with the standard algorithm used by the WSLH, which tests for fewer targets, does not typically detect coinfections, can take as long as 10 days before results are finalized, and requires more sample (both extracted and nonextracted). The time savings, in particular, can be important for surveillance networks during epidemics, when testing rates rise. It will be even more critical when the next influenza pandemic strikes.
Yet with molecular multiplexed testing come concerns about inadequate sensitivity, amplicon carryover or cross-contamination, and complexity. Sensitivity concerns regarding molecular tests can be attributed to poor primer designs, target sequence variability, and inadequate extraction procedures. Primer design is more critical for multiplexed testing, since mixed primer sets should not interact with one another; such interaction leads to primer dimer issues, which in turn can lead to poor sensitivity. Target sequence variability and drift are concerns when new, unknown variants arise in nature. We found this to be the case with the PLx-RVP assay. The assay was quickly updated for influenza virus B detection when almost 50% of the positive samples tested negative by the PLx-RVP assay. Isolates were sequenced from many of the samples, and we determined that unknown influenza virus B strains had entered the population. What we found most interesting was that many of these influenza virus B strains should have been detected by the PLx-RVP system, since the primers aligned with the strain sequences. Our only explanation was that the sequence structure, influenced by sequence differences downstream or upstream from the targeted region, was interfering with detection. Nucleic acid structure differences have been reported for other infectious disease targets (10). To minimize the effects that structure or sequence variations can have on assay sensitivity, multiplex assay developers have the capability of targeting multiple regions with multiple primer sets. One can also minimize such effects by regular sequence analysis and, when needed, primer updates.
Crossover contamination is always an issue with open testing systems. As determined by the no-target control reactions, no crossover contamination was observed in this study. This may be due to the fact that the PCR and all subsequent steps of the PLx-RVP assay are contained in the same well; that is, no transfers of the PCR amplicon take place. Because of this, the assay is much less complex than assays requiring multiple transfers, washings, and other manipulations. The use of the expanded genetic alphabet allows this: standard triphosphates do not need to be removed or diluted, since labels are placed on amplification products using the added base pair. In addition, washing and stringent temperature and buffering conditions are not required, since the coding to the solid support is accomplished using short DNA sequences (EraCodes) not found in nature (37).
Complete resolution of the discrepant results of this study would require confirmatory testing with additional molecular tests. The retesting of a subset of specimens using redesigned primers allowed a partial resolution of discrepancies. The results of the PLx-RVP assay, version 2, for influenza viruses A and B were highly concordant with the WSLH results. Increased diagnostic yields of the PLx RVP assay relative to culture or DFA were observed for RSV, Ad, and HRV, an outcome similar to those recently reported for similar multiplexed tests for respiratory viruses (27, 34). In contrast, detection of PIV by the PLx-RVP assay was no better than that by culture, a finding consistent with reports of other multiplexed molecular methods (28). Unfortunately, there was insufficient specimen volume remaining for additional analysis. Apparent false-positive results by the PLx-RVP assay may indeed be true positives, and the discrepancy may be attributed to increased diagnostic yield, as recently reported for similar multiplexed tests for respiratory viruses (27, 34).
Finally, the results in this study show that multiplexed molecular assays can be transferred from the research laboratory to the clinical arena (hospital and public health laboratory settings). The test we developed was initially designed for asthma research (27) but was later modified to screen for respiratory diseases in clinical settings. Additional targets could be added to this panel, including bocavirus, CoV HKU1, and perhaps bacterial pathogens. The flexibility to add more targets is a distinct advantage of MultiCode technology and suggests the possibility that additional designer panels could be formulated in the future to detect pathogens specific for patient characteristics. The type of agents detected will depend on such factors as patient age, symptoms, history, and, as in the case of cystic fibrosis, patient genetics. Even the time of year should be factored in when one is deciding what multiplex to run. The ability to add targets or make designer panels may be the future. Since a number of illnesses can be caused by multiple infectious agents, multiplexed molecular testing should become commonplace.
Supplementary Material
Acknowledgments
We thank Gregory A. Storch, Max Q. Arens, and Richard S. Buller at the Departments of Pediatrics, Medicine, and Molecular Microbiology at Washington University School of Medicine, St. Louis, MO, for developing and running RT-PCR confirmation assays on samples positive for CoV and HMPV by the PLx-RVP assay. We thank Christin Kiesner at the WSLH for performing the PLx-RVP assay and data analysis. MultiCode-PLx-RVP reagents used in the course of these studies were provided free of charge by EraGen Biosciences, Inc.
Footnotes
Published ahead of print on 10 October 2007.
Supplemental material for this article may be found at http://jsm.asm.org/.
REFERENCES
- 1.Bellau-Pujol, S., A. Vabret, L. Legrand, J. Dina, S. Gouarin, J. Petitjean-Lecherbonnier, B. Pozzetto, C. Ginevra, and F. Freymuth. 2005. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods 126:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin, G., S. Cote, P. Dery, G. De Serres, and M. G. Bergeron. 2004. Multiplex real-time PCR assay for detection of influenza and human respiratory syncytial viruses. J. Clin. Microbiol. 42:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridges, C. B., K. Fukuda, T. M. Uyeki, N. J. Cox, and J. A. Singleton. 2002. Prevention and control of influenza. Recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. Recommend. Rep. 51:1-31. [PubMed] [Google Scholar]
- 4.Brunstein, J., and E. Thomas. 2006. Direct screening of clinical specimens for multiple respiratory pathogens using the Genaco Respiratory Panels 1 and 2. Diagn. Mol. Pathol. 15:169-173. [DOI] [PubMed] [Google Scholar]
- 5.Bustin, S. A., and R. Mueller. 2005. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. (London) 109:365-379. [DOI] [PubMed] [Google Scholar]
- 6.Cattoli, G., and I. Capua. 2006. Molecular diagnosis of avian influenza during an outbreak. Dev. Biol. (Basel) 124:99-105. [PubMed] [Google Scholar]
- 7.Church, D. L., H. D. Davies, C. Mitton, H. Semeniuk, M. Logue, C. Maxwell, and C. Donaldson. 2002. Clinical and economic evaluation of rapid influenza A virus testing in nursing homes in Calgary, Canada. Clin. Infect. Dis. 34:790-795. [DOI] [PubMed] [Google Scholar]
- 7a.CLSI. 2002. User protocol for evaluation of qualitative test performance. Document EP12-A. CLSI, Wayne, PA.
- 8.Coiras, M. T., J. C. Aguilar, M. L. Garcia, I. Casas, and P. Perez-Brena. 2004. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J. Med. Virol. 72:484-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coiras, M. T., M. R. Lopez-Huertas, G. Lopez-Campos, J. C. Aguilar, and P. Perez-Brena. 2005. Oligonucleotide array for simultaneous detection of respiratory viruses using a reverse-line blot hybridization assay. J. Med. Virol. 76:256-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, F., H. T. Allawi, T. Anderson, B. P. Neri, and V. I. Lyamichev. 2001. Secondary structure prediction and structure-specific sequence analysis of single-stranded DNA. Nucleic Acids Res. 29:3248-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donoso Mantke, O., H. Schmitz, H. Zeller, P. Heyman, A. Papa, M. Niedrig, et al. 2005. Quality assurance for the diagnostics of viral diseases to enhance the emergency preparedness in Europe. Euro Surveill. 10:102-106. [PubMed] [Google Scholar]
- 12.Doyle, W. J., and C. M. Alper. 2007. Use of diagnostic algorithms and new technologies to study the incidence and prevalence of viral upper respiratory tract infections and their complications in high risk populations. Curr. Opin. Allergy Clin. Immunol. 7:11-16. [DOI] [PubMed] [Google Scholar]
- 13.Druce, J., T. Tran, H. Kelly, M. Kaye, D. Chibo, R. Kostecki, A. Amiri, M. Catton, and C. Birch. 2005. Laboratory diagnosis and surveillance of human respiratory viruses by PCR in Victoria, Australia, 2002-2003. J. Med. Virol. 75:122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fauci, A. S., N. A. Touchette, and G. K. Folkers. 2005. Emerging infectious diseases: a 10-year perspective from the National Institute of Allergy and Infectious Diseases. Emerg. Infect. Dis. 11:519-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouchier, R. A., G. F. Rimmelzwaan, T. Kuiken, and A. D. Osterhaus. 2005. Newer respiratory virus infections: human metapneumovirus, avian influenza virus, and human coronaviruses. Curr. Opin. Infect. Dis. 18:141-146. [DOI] [PubMed] [Google Scholar]
- 16.Frisbie, B., Y. W. Tang, M. Griffin, K. Poehling, P. F. Wright, K. Holland, and K. M. Edwards. 2004. Surveillance of childhood influenza virus infection: what is the best diagnostic method to use for archival samples? J. Clin. Microbiol. 42:1181-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillim-Ross, L., and K. Subbarao. 2006. Emerging respiratory viruses: challenges and vaccine strategies. Clin. Microbiol. Rev. 19:614-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greaves, F. 2004. What are the most appropriate methods of surveillance for monitoring an emerging respiratory infection such as SARS? J. Public Health (Oxford) 26:288-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gröndahl, B., W. Puppe, J. Weigl, and H. J. Schmitt. 2005. Comparison of the BD Directigen Flu A+B Kit and the Abbott TestPack RSV with a multiplex RT-PCR ELISA for rapid detection of influenza viruses and respiratory syncytial virus. Clin. Microbiol. Infect. 11:848-850. [DOI] [PubMed] [Google Scholar]
- 20.Horejsh, D., F. Martini, F. Poccia, G. Ippolito, A. Di Caro, and M. R. Capobianchi. 2005. A molecular beacon, bead-based assay for the detection of nucleic acids by flow cytometry. Nucleic Acids Res. 33:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, S. C., D. J. Marshall, G. Harms, C. M. Miller, C. B. Sherrill, E. L. Beaty, S. A. Lederer, E. B. Roesch, G. Madsen, G. L. Hoffman, R. H. Laessig, G. J. Kopish, M. W. Baker, S. A. Benner, P. M. Farrell, and J. R. Prudent. 2004. Multiplexed genetic analysis using an expanded genetic alphabet. Clin. Chem. 50:2019-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye, M., S. Skidmore, H. Osman, M. Weinbren, and R. Warren. 2006. Surveillance of respiratory virus infections in adult hospital admissions using rapid methods. Epidemiol. Infect. 134:792-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kehl, S. C., K. J. Henrickson, W. Hua, and J. Fan. 2001. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J. Clin. Microbiol. 39:1696-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessler, N., O. Ferraris, K. Palmer, W. Marsh, and A. Steel. 2004. Use of the DNA flow-thru chip, a three-dimensional biochip, for typing and subtyping of influenza viruses. J. Clin. Microbiol. 42:2173-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lashley, F. R. 2004. Emerging infectious diseases: vulnerabilities, contributing factors and approaches. Expert Rev. Anti-Infect. Ther. 2:299-316. [DOI] [PubMed] [Google Scholar]
- 26.Lednicky, J. A., and J. O. Rayner. 2006. Uncommon respiratory pathogens. Curr. Opin. Pulm. Med. 12:235-239. [DOI] [PubMed] [Google Scholar]
- 27.Lee, W. M., K. Grindle, T. Pappas, D. J. Marshall, M. J. Moser, E. L. Beaty, P. A. Shult, J. R. Prudent, and J. E. Gern. 2007. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J. Clin. Microbiol. 45:2626-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, H., M. A. McCormac, R. W. Estes, S. E. Sefers, R. K. Dare, J. D. Chappell, D. D. Erdman, P. F. Wright, and Y. W. Tang. 2007. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J. Clin. Microbiol. 45:2105-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maertzdorf, J., C. K. Wang, J. B. Brown, J. D. Quinto, M. Chu, M. de Graaf, B. G. van den Hoogen, R. Spaete, A. D. Osterhaus, and R. A. Fouchier. 2004. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J. Clin. Microbiol. 42:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahony, J., S. Chong, F. Merante, S. Yaghoubian, T. Sinha, C. Lisle, and R. Janeczko. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J. Clin. Microbiol. 45:2965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahony, J. B., A. Petrich, L. Louie, X. Song, S. Chong, M. Smieja, M. Chernesky, M. Loeb, and S. Richardson. 2004. Performance and cost evaluation of one commercial and six in-house conventional and real-time reverse transcription-PCR assays for detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 42:1471-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.NIH. 2004. Morbidity and mortality: 2004 chart book on cardiovascular, lung, and blood diseases. U.S. Department of Health and Human Services, Washington, DC.
- 33.Nokso-Koivisto, J., T. Hovi, and A. Pitkaranta. 2006. Viral upper respiratory tract infections in young children with emphasis on acute otitis media. Int. J. Pediatr. Otorhinolaryngol. 70:1333-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolte, F. S., D. J. Marshall, C. Rasberry, S. Schievelbein, G. G. Banks, G. A. Storch, M. Q. Arens, R. S. Buller, and J. R. Prudent. 2007. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J. Clin. Microbiol. 45:2779-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaller, M. A., A. F. Ehrhardt, and R. N. Jones. 2001. Frequency of pathogen occurrence and antimicrobial susceptibility among community-acquired respiratory tract infections in the respiratory surveillance program study: microbiology from the medical office practice environment. Am. J. Med. 111(Suppl. 9A):4S-12S, 36S-38S. [DOI] [PubMed] [Google Scholar]
- 36.Pietz, B. C., M. B. Warden, B. K. DuChateau, and T. M. Ellis. 2005. Multiplex genotyping of human minor histocompatibility antigens. Hum. Immunol. 66:1174-1182. [DOI] [PubMed] [Google Scholar]
- 37.Prudent, J. R. 2006. Using expanded genetic alphabets to simplify high-throughput genetic testing. Expert Rev. Mol. Diagn. 6:245-252. [DOI] [PubMed] [Google Scholar]
- 38.Puppe, W., J. A. Weigl, G. Aron, B. Grondahl, H. J. Schmitt, H. G. Niesters, and J. Groen. 2004. Evaluation of a multiplex reverse transcriptase PCR ELISA for the detection of nine respiratory tract pathogens. J. Clin. Virol. 30:165-174. [DOI] [PubMed] [Google Scholar]
- 39.Simonsen, L., L. A. Conn, R. W. Pinner, and S. Teutsch. 1998. Trends in infectious disease hospitalizations in the United States, 1980-1994. Arch. Intern. Med. 158:1923-1928. [DOI] [PubMed] [Google Scholar]
- 40.Singer, V. L., L. J. Jones, S. T. Yue, and R. P. Haugland. 1997. Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal. Biochem. 249:228-238. [DOI] [PubMed] [Google Scholar]
- 41.Smith, A. B., V. Mock, R. Melear, P. Colarusso, and D. E. Willis. 2003. Rapid detection of influenza A and B viruses in clinical specimens by LightCycler real time RT-PCR. J. Clin. Virol. 28:51-58. [DOI] [PubMed] [Google Scholar]
- 42.Smith, N. M., J. S. Bresee, D. K. Shay, T. M. Uyeki, N. J. Cox, and R. A. Strikas. 2006. Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. Recommend. Rep. 55:1-42. [PubMed] [Google Scholar]
- 43.Stockton, J., J. S. Ellis, M. Saville, J. P. Clewley, and M. C. Zambon. 1998. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J. Clin. Microbiol. 36:2990-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Syrmis, M. W., D. M. Whiley, M. Thomas, I. M. Mackay, J. Williamson, D. J. Siebert, M. D. Nissen, and T. P. Sloots. 2004. A sensitive, specific, and cost-effective multiplex reverse transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J. Mol. Diagn. 6:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tapper, M. L. 2006. Emerging viral diseases and infectious disease risks. Haemophilia 12(Suppl. 1):3-7, 26-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Templeton, K. E., S. A. Scheltinga, M. F. Beersma, A. C. Kroes, and E. C. Claas. 2004. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 42:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallace, L. A., T. C. Collins, J. D. Douglas, S. McIntyre, J. Millar, and W. F. Carman. 2004. Virological surveillance of influenza-like illness in the community using PCR and serology. J. Clin. Virol. 31:40-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yam, W. C., K. H. Chan, K. H. Chow, L. L. Poon, H. Y. Lam, K. Y. Yuen, W. H. Seto, and J. S. Peiris. 2005. Clinical evaluation of real-time PCR assays for rapid diagnosis of SARS coronavirus during outbreak and post-epidemic periods. J. Clin. Virol. 33:19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, S., and R. E. Rothman. 2004. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 4:337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.